Abstract
Proteoliposome reconstitution is a standard method to stabilize purified transmembrane proteins in membranes for structural and functional assays. Here we quantified intrareconstitution heterogeneities in single proteoliposomes using fluorescence microscopy. Our results suggest that compositional heterogeneities can severely skew ensemble-average proteoliposome measurements but also enable ultraminiaturized high-content screens. We took advantage of this screening capability to map the oligomerization energy of the β2-adrenergic receptor using ~109-fold less protein than conventional assays.
Proteoliposomes comprise one of the most common model-membrane systems for reconstituting and studying transmembrane proteins outside the complex cellular milieu1-3 and have thus been instrumental in elucidating the structure and function of this important class of proteins1-3. However, the reproducible production of proteoliposomes with homogeneous protein and lipid composition remains a great challenge and is often a limiting step1,4-8. Such intrasample compositional heterogeneity translates to a broad distribution of physicochemical properties; hence, ensemble-based functional readouts that by definition ignore the underlying distribution are prone to artifacts from hidden bias4-7,9.
Pronounced heterogeneity in protein-to-lipid ratio (P/L) within proteoliposome preparations has been observed using density-gradient centrifugation and electron microscopy. Gradient centrifugation is typically employed to qualitatively characterize sample heterogeneity4,5,7,8 and in some instances to separate fractions that, however, still comprise highly heterogeneous proteoliposome populations10. Here we employed single-particle quantitative fluorescence microscopy to directly map the distribution of P/L within proteoliposome samples with superior resolution. We then exploited this proteoliposome heterogeneity to perform quantitative high-content analysis (HCA) on the nanoscale.
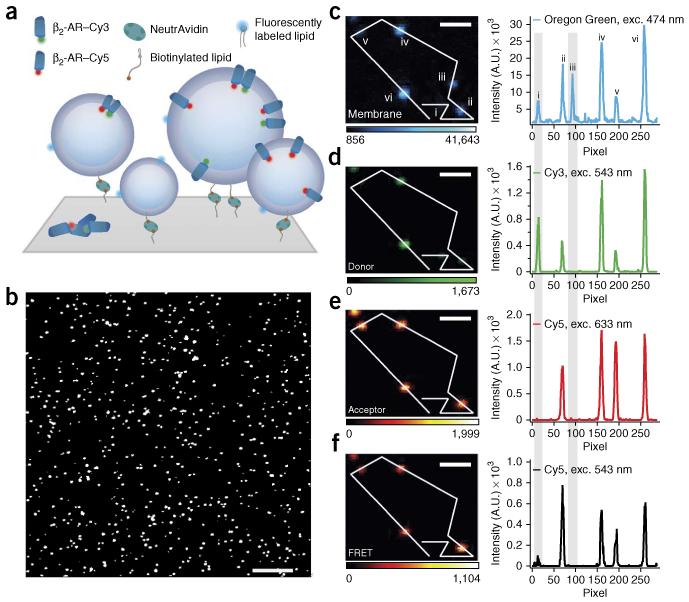
We and others11,12 have imaged liposomes at the single-particle level by tethering them on a surface at dilute densities (Fig. 1a,b; see ref. 13 for a recent review) and have demonstrated that under controlled tethering conditions, liposomes remain spherical14 and maintain native membrane permeability to ions15. Here, we fluorescently labeled the lipid phase (with Oregon Green DHPE) and the transmembrane proteins (with Cy3 or Cy5), which allowed the accurate quantitative assessment of P/L stoichiometry per proteoliposome. As a proof of concept, we focused on G protein–coupled receptors (GPCRs), which comprise the most common clinical drug targets16,17.
Figure 1.
Surface immobilization and fluorescence microscopy imaging allow for single-proteoliposome characterization. (a) Proteoliposomes tethered through a biotin-NeutrAvidin linker to a polymer-passivated (PLL-g-PEG/PLL-g-PEG-biotin) glass surface. Proteoliposomes are labeled with a lipid-coupled dye (Oregon Green DHPE) and harbor GPCRs labeled with either Cy3 or Cy5 for quantification of receptor oligomerization by FRET. (b–f) Micrographs of typical β2-AR proteoliposome samples with nominal 1:1,000 protein-to-lipid ratio. (b) Magnified version of a typical confocal image. The assay allows high-throughput sampling of ~1,000 proteoliposomes per frame. (c–f) Micrographs and line scans showing high signal to noise for Oregon Green, Cy3, Cy5 and FRET. Gray shading highlights, respectively, an example of a proteoliposomes that carry only donor-labeled receptors (liposome i) and an example of an empty liposome (liposome iii). Exc., excitation wavelength. Color scales represent intensity in arbitrary units. Scale bars, 10 μm (b) and 1.2 μm (c–f).
We studied three prototypical GPCRs: the β2-adrenergic receptor (β2-AR), the cannabinoid receptor type 1 (CB1) and opsin. We used maleimide chemistry to fluorescently label single reactive cysteines (Supplementary Fig. 1 and Online Methods) with Cy3 or Cy5 fluorophores, which allowed us to measure the protein amount per proteoliposome. Reconstitution in proteoliposomes was performed using standard, well-documented protocols (gel chromatography, removal of detergent using Bio-Beads and rapid dilution; Online Methods). Mixing receptors labeled with Cy3 (donor, D) and Cy5 (acceptor, A) in a nominal 1:1 ratio allowed us to measure fluorescence resonance energy transfer (FRET) and quantify receptor oligomerization within single nanoscale proteoliposomes.
Fluorescence confocal microscopy allowed us to measure thousands of proteoliposomes in parallel (Fig. 1b). We typically imaged four intensity signals per proteoliposome, namely those of the lipid dye, D, A and FRET, with very high signal-to-noise ratio (~30–800; Online Methods and Fig. 1c–f). Using automated particle tracking, we localized each proteoliposome that appeared as a diffraction-limited spot (Fig. 1c–f) and fit the four fluorescent signals with a two-dimensional (2D) Gaussian function to extract integrated intensities.
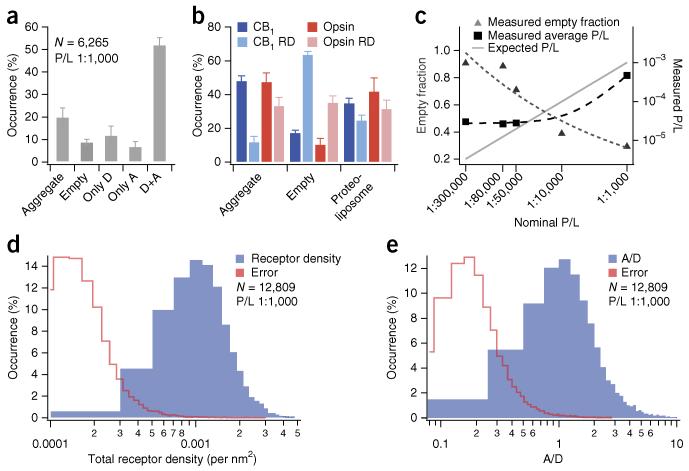
When we colocalized the intensity spots of the three fluorescent labels, we found that 9% ± 1% (error represents s.d. of technical uncertainties unless otherwise stated) of the identified lipid intensities did not colocalize with any β2-AR intensity (Figs. 1c–f and 2a). This verifies the existence of a population of empty liposomes, as has been reported4,5. Furthermore, we observed that 20% ± 4% of receptor intensities did not colocalize with any lipid signal, indicating a population of nonreconstituted receptor aggregates. Using statistical analysis (Online Methods), we verified that the observation of empty liposomes and nonreconstituted receptor aggregates could not simply be due to partial labeling of receptors or liposomes.
Figure 2.
Direct imaging of single nanoscale proteoliposomes allows for high-content analysis of intrasample compositional heterogeneities. (a) Protein aggregates, empty liposomes and proteoliposomes harboring only donor (D), only acceptor (A) and both donor and acceptor (D+A) subpopulations within a β2-AR reconstitution sample with nominal 1:1,000 protein-to-lipid ratio (P/L). Only proteoliposomes having both β2-AR–Cy3 and β2-AR–Cy5 (52% ± 3%) are selected for FRET analysis. Data in the panel include N = 6,265 particles. Error bars indicate s.d. of technical replicates from 3 independent experiments. (b) Protein aggregates, empty liposomes and proteoliposome subpopulations within reconstitution samples of CB1 and opsin receptors (both nominal P/L = 1:1,000) reconstituted by removal of detergent by either Bio-Beads (darker shades) or rapid dilution (RD; lighter shades). Data in the panel include N = 11,076 particles (CB1), N = 11,585 (CB1 RD), N = 10,701 (Opsin), N = 15,943 (Opsin RD). Error bars for darker bars: s.d. of technical replicates for ≥8 microscope chamber positions. Error bars for lighter bars: s.d. of technical replicates from 3 independent microscopy experiments. (c) Effect of titrating the nominal P/L (data were collected for both β2-AR R333C and 265C; Online Methods). Data were fit to power functions to guide the eye (dashed lines). (d) Histogram of observed receptor densities for individual proteoliposomes (β2-AR, nominal P/L = 1:1,000). (e) Histogram displaying A/D ratios on individual proteoliposomes (β2-AR, nominal P/L = 1:1,000). All histograms include 12,809 proteoliposomes from 7 independent experiments. In d and e the red traces indicate histograms of absolute errors from single proteoliposomes.
Within the population of identified proteoliposomes (those with lipid and receptor intensity colocalized) we found three subpopulations. One fraction of proteoliposomes had only A-labeled β2-AR (7% ± 2%), whereas another fraction had only D-labeled receptor (12% ± 4%; Figs. 1c–f and 2a). Only 52% ± 3% of the liposomes contained both A- and D-labeled receptor as intended. Further independent measurements of CB1 and opsin (Fig. 2b), each reconstituted via two alternative preparation routes (Online Methods), corroborated the observations with β2-AR reconstituted via gel chromatography. Together these results provide a quantitative insight into the commonly held notion that proteoliposome samples are likely to comprise substantial subpopulations with heterogeneous transmembrane protein composition2,7,9-12.
Next we investigated the effect of titrating the nominal P/L on liposomal compositional heterogeneities. We assessed the average measured P/L using single-molecule photobleaching to determine the mean number of receptors and assumed a mean proteoliposome diameter of 100 nm (Supplementary Fig. 2 and Online Methods). We quantified the fraction of empty liposomes and the average measured P/L in five different reconstitutions of β2-AR (Fig. 2c). Interestingly, the measured receptor density did not scale linearly with the titrated density as one would expect for homogeneous samples. In the lower range of protein concentrations, the measured P/L remained essentially constant, and the effect of diluting receptor concentration manifested as an increased fraction of empty liposomes (Fig. 2c). If the incorporation of receptors in liposomes was completely random, it should have been governed by Poissonian statistics. However, our results show that receptor incorporation cannot be rationalized as a single stochastic or Poissonian process (Supplementary Discussion) and that the existence of empty liposomes is not simply due to P/L dilution (Supplementary Fig. 3).
To more accurately determine heterogeneities in the absolute β2-AR surface density, we calculated the diameters and surface areas of individual proteoliposomes using a previously published calibration procedure14 (Online Methods). We generally observed non-Gaussian distributions of β2-AR densities that spanned an order of magnitude (~0.3 × 10−3 –3 × 10−3 receptors/nm2; Fig. 2d). The samples also spanned a range of different proteoliposome diameters (Supplementary Fig. 4a). Surprisingly, we found a strong correlation between receptor density and proteoliposome size (Supplementary Fig. 4b), a result suggesting that receptor spontaneous curvature induces a preference for curved membrane geometry. A large variation was also observed in the range of A/D ratios (~0.2–4), revealing a spread in compositional heterogeneities substantially larger than the errors of receptor density and A/D ratios on individual proteoliposomes (Fig. 2d,e and Online Methods).
Next we investigated whether these compositional heterogeneities could be exploited to perform HCA screens as a function of proteoliposome composition. As proof of concept, we focused on the self-association of GPCRs (see refs. 16,18 and references therein), which is proposed to be important for the development of improved therapeutic strategies18.
GPCR self-association was first commonly studied using biochemical separation of receptor complexes. However, because such approaches (for example, coimmunoprecipitation and cross-linking) are prone to artifacts arising from membrane lysis19 that can perturb native protein-protein interactions, a number of powerful real-time optical methods were developed19. The latter methods have provided valuable information on the effective self-association in the biologically relevant context of the cell membrane. In comparison, our in vitro methodology, being unimpaired by complex cellular effectors including confining cytoskeleton elements, rafts and receptor-interacting proteins, gives more precise insight on the inherent propensity of receptors to self-associate. Furthermore, because absolute calibration of receptor densities is facilitated in vitro, we were able to use proteoliposome heterogeneities to effortlessly construct 2D binding curves and extract an equilibrium association constant. Additionally, by screening on the single-proteoliposome level, substantially less (~109-fold) protein sample was required than that needed for ensemble-averaged experiments.
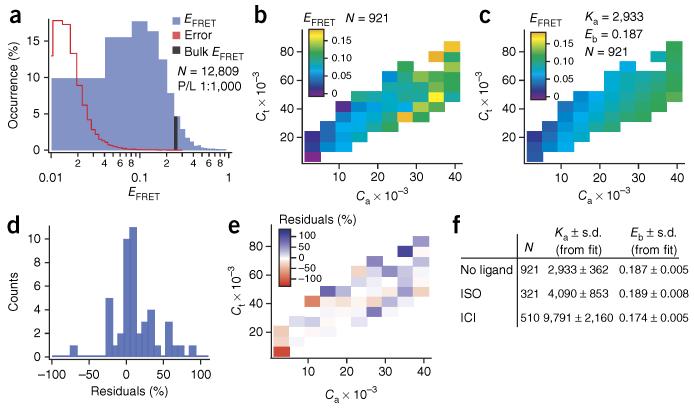
Measurements on ~13,000 individual proteoliposomes containing β2-AR revealed a histogram of FRET efficiencies (EFRET) that peaked at ~0.1 but spanned nearly all possible values between 0 and 1 (Fig. 3a). Interestingly, only ~4% of the single proteoliposomes (Fig. 3a and Supplementary Fig. 5) actually had an EFRET equal to the calculated ensemble-average EFRET of this sample (Online Methods), a result demonstrating the severe bias that compositional heterogeneities would introduce on an ensemble-average readout of FRET with a spectrophotometer, for example.
Figure 3.
Quantification of β2-AR association energy using heterogeneities in a single reconstitution sample and ~6 pg of receptor. (a) Histogram of single proteoliposome FRET efficiencies (EFRET) (blue) and corresponding histogram of EFRET errors (red; histogram is shown in linear scale in Supplementary Fig. 5). The black bar marks the calculated ensemble-average EFRET. Nominal protein-to-lipid ratio (P/L) = 1:1,000. (b) EFRET (color scale) as a function of the total reduced receptor density (Ct) and reduced acceptor density (Ca) for a population of 921 proteoliposomes with a narrow size distribution of 120–130 nm (nominal P/L = 1:1,000). The unitless reduced densities correspond to receptor surface density multiplied by the square of the Förster radius (R02) (Supplementary Note). For better visualization, EFRET values of single proteoliposomes in b,c are binned, and a weighted average of each bin is displayed. (c) Weighted fit of the data going into b (without binning) with the theoretical model (equations (11), (14) and (16) in the Supplementary Note). From the fit we extract two fitting parameters: the dimer association constant Ka and the FRET efficiency within a dimer Ebound (Eb). Bins are constructed to match those in panel b. (d,e) Residuals representing the difference between the fit in c and experimental data in b, here displayed as the percentage deviation, suggest no systematic deviations from the theory. (f) Table displaying Ka and Eb obtained by fitting the theoretical scheme (equations (11), (14) and (16)) to proteoliposomes incubated with no ligand, a saturating amount of agonist (isoproterenol (ISO), 10 μM) or an inverse agonist (ICI 118,551, 500 nM).
The dependence of EFRET on surface density due to both random collisions and reversible specific association has been treated theoretically before in two dimensions. Here we combined three previously developed and tested theoretical schemes20-22 to extract the association energy for β2-AR (Supplementary Note). Because the theory accounts specifically for random collisions, we could verify that our experimentally measured EFRET always exceeded the EFRET assigned to random encounters, thus demonstrating specific oligomerization (Supplementary Fig. 6).
EFRET could be presented in a 3D plot covering a broad range of reduced acceptor density Ca and reduced total receptor density Ct (Fig. 3b). Next we fit the combined theory expressed in equations (11), (14) and (16) (Supplementary Note) in a weighted fit and extracted the only two free parameters: the dimer association constant, Ka = 2,933 ± 362 (s.d. from fit), and the EFRET within a dimer, Ebound = 0.187 ± 0.005 (s.d. from fit) (Fig. 3c). Ka is given here in mole fraction units, allowing direct conversion to Gibbs’ free energy. The corresponding dissociation constant Kd is 517 ± 64 copies/μm2 (Supplementary Discussion). A comparison of data (Fig. 3b) and fit (Fig. 3c) demonstrates that the experimental data follow the theoretical trends with no substantial systematic deviations (residuals are randomly distributed around 0; Fig. 3d,e). We thus concluded that this transient dimerization model is a reasonable approximation to the concentration-dependent tendency of β2-AR to self-associate and that supported proteoliposomes are a suitable model system for investigating diffusion of transmembrane proteins.
Our findings demonstrated an inherent ability of β2-AR monomers to interact that is governed by the law of mass action. Consequently, the proportion of dimers in a given cell, at a given site on the membrane, is intimately tied to the local protein density (Supplementary Discussion). Knowledge of Ka allowed us to calculate the corresponding association energy of −19.5 ± 1 kJ/mol (or −8kBT (where temperature T = 20 °C and kB is the Boltzmann constant); equation (13) in the Supplementary Note). The β2-AR complexes are thus stabilized with a substantial energy contribution above thermal fluctuations (Supplementary Discussion).
We also tested the influence of ligands using an agonist (isoproterenol) and an inverse agonist (ICI 118,551) and observed an approximately threefold increase of Ka in the latter case (Fig. 3f and Supplementary Fig. 7). These data suggest that ligands have the potential to modulate the inherent propensity of β2-AR to self-associate and warrant further study both in vitro and in vivo. In sum, experimental methods to determine GPCR association energies are essential for quantitatively comparing the interaction strength of different homomeric and heteromeric GPCR complexes or oligomerization mutants, and they serve as reference and validation for both theoretical calculations and simulations.
Here we demonstrated how the powerful concept of HCA23 that was developed for single-cell measurements can be applied to single nanoscale proteoliposomes. The plethora of commercially available fluorescent dyes suggests that numerous proteoliposome properties could be analyzed quantitatively (including stoichiometry of lipids, sterols, transmembrane proteins, cofactor proteins and drug-candidate ligands) and correlated with readouts of transmembrane protein structure or function. More broadly, intrasample compositional and structural heterogeneity is a recurring problem in the production of most complex supramolecular architectures, including all types of colloids, emulsions, micelles, nanodiscs, etc.9,24. Historically, tremendous effort has been invested in developing laborious homogenization or separation procedures for such samples, with varying success24. Our method brings forward the general concept of exploiting heterogeneities in micro- and nanoparticle particulates for quantitative HCA and thus transforms a potentially prohibitive problem into an enabling technology.
ONLINE METHODS
Proteoliposome preparation
A functional single reactive cysteine mutant (β2-AR–Δ5-R333C25 or β2-AR–365-265C26) of β2-AR was expressed, purified and labeled with either Cy3- or Cy5-maleimide (Amersham Biosciences) as described previously25 (see Supplementary Fig. 8 for elution profiles of the purified protein). Liposomes of the lipid composition DOPC/CHS/DOPG/Oregon Green DHPE/DSPE-PEG2000-biotin (79.4:10:10:0.5:0.1) (Avanti polar lipids, Steraloids Inc., Invitrogen) were prepared by evaporating chloroform under argon and were dried 1 h under vacuum to prepare a thin lipid film. The film was resuspended in buffer (20 mM HEPES, 100 mM NaCl, 1% octylglucoside, pH 7.5), and the lipid-detergent mixture was formed by sonication for 1 h in an ice-water bath. Lipid-detergent mixture and labeled β2-AR were mixed in the desired protein-to-lipid ratio. The lipid-receptor mixture and sample buffer (20 mM HEPES, 100 mM NaCl, pH 7.5) (until 300 μl) were mixed and kept on ice for 2 h. Proteoliposomes were formed by the removal of detergent on a Sephadex G-50 (fine) column (25 × 0.8 cm). Four different samples were prepared: empty liposomes, only Cy3–β2-AR reconstituted, only Cy5–β2-AR reconstituted and both Cy3–β2-AR and Cy5–β2-AR reconstituted in a 1:1 ratio.
Single cysteine-reactive mutants of thermostable 2C-282C-opsin (θ′-316C)27,28 and CB1 (θ-A342C)29 were expressed, purified and labeled with either Cy3- or Cy5-maleimide (Amersham Biosciences) (see Supplementary Fig. 8 for elution profiles of the purified protein). Liposome samples were prepared as previously described30. In brief, receptor (solubilized in 0.05% n-dodecyl-β-d-maltoside) and lipids (solubilized in 0.5 M sodium cholate) were mixed with approximately two-thirds vol of Bio-Beads SM-2 (Bio-Rad) overnight at 4 °C. Both reconstitutions were prepared with 1:1 ratio of Cy3-opsin:Cy5-opsin or Cy3-CB1:Cy5-CB1, and the total protein-to-lipid ratio for each reconstitution was set at 1:1,000. Bio-Beads were removed by centrifugation (1,000g, 1 min) yielding proteoliposome preparations of POPC/POPG/ DHPE Oregon Green/DSPE-PEG2000-biotin (59.85:39.85:0.2:0.1) (Avanti Polar Lipids, Invitrogen) in 20 mM HEPES, pH 7.3, 150 mM NaCl, 2 mM MgCl2 and 1 mM EDTA.
The rapid dilution reconstitution of opsin and CB1 followed previously published protocols4. In brief, 200 μl total volume (~130 pmol receptor, ~130 nmol lipid) in ~0.6% (w/v) sodium cholate were diluted tenfold dropwise to instantaneously lower the sodium cholate concentration well below the critical micelle concentration of 0.388–0.603% (w/v). The remaining monomeric sodium cholate was removed by dialysis (10,000 MWCO) against 500 ml excess buffer (20 mM HEPES, pH 7.3, 150 mM NaCl, 2 mM MgCl2 and 1 mM EDTA) containing 250 mg Bio-Beads for 24 h, with three buffer-exchange steps at times ~1–2 h, ~4–6 h, ~16–18 h.
Receptor constructs
For all receptors used in this study, surface-exposed reactive cysteines had to be either mutated or truncated in order to create specific cysteine labeling cites. In all cases we confirmed that labeling did not alter receptor functionality. For specific purification, characterization and labeling conditions, we refer to the respective citations.
β2-AR labeled at H8 position R333C25
This construct was termed Δ5. Five cysteines were mutated and substituted respectively with C77V, C265A, C327S, C378A and C406A. Subsequently, a single cysteine was introduced at position R333C for specific labeling.
β2-AR labeled at TM6 position 265C26,31
This construct was termed β2-AR-365. 48 amino acids in the C terminus were truncated. Specific labeling was performed at the endogenous cysteine 265C.
Opsin labeled at helix 8 position 316C27,28
The construct termed θ′ was created using a well-characterized nonreactive labeling construct (140S, 167S, 222S, 264S, 322S and 323S)28. Into this construct two cysteines were introduced (N2C, D282C) to thermally stabilize the apo-protein opsin27. The endogenous 316C was used for specific labeling.
CB1 labeled at TM6 position A342C29
In the minimal cysteine truncated purification construct (Δ88/Δ417) termed θ (ref. 29), only 2 of the 13 endogenous cysteines (Cys257 and Cys264) were retained to ensure a functional receptor32. A specific cysteine for labeling was introduced at A342C.
Single proteoliposome immobilization and characterization by confocal fluorescence microscopy
Proteoliposomes were immobilized on a passivated glass surface in home-built microscope chambers and imaged by confocal fluorescence microscopy. Chamber parts were cleaned extensively using ethanol and Milli-Q water (MQ; Millipore). Glass slides (thickness 170 ± 10 μm) were cleaned by consecutive rounds of sonication by 2% (v/v) Helmanex following three washes (×3) with MQ and ×2 with methanol. Glass slides were dried in nitrogen flow, plasma etched for 2 min, mounted in a microscope chamber and incubated with a mixture of 1,000:6 PLL-g-PEG and PLL-g-PEG-biotin (SuSoS) (1 g/l) in surface buffer (15 mM HEPES, pH 5.6) for 30 min. After carefully washing with sample buffer (20 mM HEPES, 100 mM NaCl, pH 7.5), we incubated the surfaces with 0.1 g/l NeutrAvidin (Life Technologies) in surface buffer for 10 min after additional washing (×10) with sample buffer. Proteoliposome surface density was controlled by addition of 4 μl (0.05 g/l) proteoliposomes to 80 μl chamber volume. The chamber was washed ×10 in sample buffer when the surface density reached approximately 500 proteoliposomes per frame.
Images were acquired on a Leica TCS-SP5 inverted confocal microscope setup using a HCX PL APO oil-immersion objective (100× magnification, 1.4 numerical aperture (NA)) (Leica). Oregon Green DHPE, Cy3–β2-AR and Cy5–β2-AR were excited by a 476-nm laser line, a 543-nm laser line and a 633-nm laser line, respectively. Oregon Green DHPE intensity was collected by a photomultiplier tube (PMT), and emission was filtered out in the range of 486–530 nm. Donor and acceptor emission were detected by avalanche photodiodes (APDs). The emission signals were cut off at 625 nm between APD1 and APD2 with a dichroic mirror BS625 (Chroma Technology). Donor and acceptor emissions were filtered by the use of band-pass filters: for Cy3, BrightLine HC 585/40 (Semrock) and for Cy5, HQ675/55M (Chroma). Images were acquired in the format 1,024 × 1,024 pixels, each pixel corresponding to 50.5-nm sample length, bit depth of 16, with a scan speed of 400 Hz.
The signal-to-noise (S/N) ratio was calculated by integrating the intensity in a region of interest (ROI) around a proteoliposome (Iproteoliposome) and in an ROI of identical size placed in the background (Ibackground) next to the proteoliposome. The S/N ratio was assessed as Iproteoliposome/Ibackground. In three randomly chosen images from three independent experiments, the S/N ratio of the lowest and the highest intensities were determined. An average of the three lowest and an average of the three highest S/N ratios are stated as the range of S/N ratios found in the experiments.
Data analysis and quantification of FRET efficiencies
Software written in Igor Pro v. 6.01 (WaveMetrics and Supplementary Software) was used to treat all collected images. The software identified intensity signals above a user-defined threshold (>60) and fitted a 2D Gaussian bell in order to determine the integrated intensity of each label and assign an xy center position of all diffraction-limited intensity spots (Fig. 1b). A circularity cutoff of 0.5 (minor axis divided by major axis) was applied to reject spurious intensity signals. Colocalization was defined as particles in separate color channels having centers within a distance of 3 pixels. For two consecutive images the software accepted particles having centers within this defined pixel distance. For calculation of FRET efficiencies, four signals (Fig. 1c–f) are required to colocalize.
The FRET efficiencies were calculated according to equation (1)33
| (1) |
where
| (2) |
ID and IAFRET are the donor and acceptor intensity excited by donor laser line (exc. 543 nm). IA is the acceptor intensity excited by acceptor laser line (exc. 633 nm), and the superscript 0 denotes raw uncorrected intensities. For correct assessment of FRET efficiencies, IA0,FRET must be corrected for two signal contaminations α and β. β denotes the donor emission that leaks into the acceptor detector channel, and α denotes the small amount of acceptor that is directly excited by donor laser line33 (Supplementary Fig. 5a). β was determined using a control sample carrying only Cy3–β2-AR as the ratio of donor intensity arising in the acceptor channel (exc. 543 nm) to the donor intensity in the donor channel (exc. 543 nm). β (11.1% ± 0.06%, ±s.e.m.) was quantified for 1,130 single proteoliposomes (Supplementary Fig. 5b). α was determined using a control sample carrying only Cy5–β2-AR as the ratio of acceptor intensity (exc. 543 nm) in the acceptor channel to the intensity of the acceptor excited at 633 nm. α (9.8% ± 0.1%, ±s.e.m.) was quantified for 729 single proteoliposomes (Supplementary Fig. 5c). Critically, both β and α were determined under the exact same image conditions (intensity levels, etc.) as those of the FRET samples (both Cy3–β2-AR and Cy5–β2-AR reconstituted in a 1:1 ratio).
The control sample of empty proteoliposomes was furthermore imaged in both donor and acceptor channels (Fig. 1d–f). Only ID had to be corrected for a small contribution of Oregon Green DHPE emission in the donor channel upon donor excitation (denoted ω). ω was calculated as the ratio of Oregon Green DHPE intensity in donor channel (exc. 543 nm) to Oregon Green DHPE intensity excited by 476 nm. ω (0.9% ± 0.02%, ±s.e.m.) was quantified for 729 single proteoliposomes (Supplementary Fig. 5d).
In order to decouple the FRET efficiency from instrumental and photophysical effects, we corrected for differences in fluorophore quantum yields (ΦA/ΦD) and detection efficiencies (ηA/ηD) through a γfactor (equation (2))34. Fluorophore quantum yields of Cy3 (ΦCy3 = 0.35) and Cy5 (ΦCy5 = 0.65) were accessed experimentally using the comparative method35 (equation (3)) and two well-described references: cresyl violet (Φref = 0.54 in absolute methanol)36 and rhodamine 800 (Φref = 0.19 in absolute ethanol)37. The fluorophore quantum yield was calculated as
| (3) |
Fsample and Fref being the integrated fluorescence emission of either Cy3–β2-AR and cresyl violet or Cy5–β2-AR and rhodamine 800, respectively. Asample and Aref are the absorbance at excitation wavelengths, and nsample and nref are the refractive indices of the fluorophore solvents.
Fluorescence emission spectra were recorded with a Horiba Jobin Yvon Fluoromax-4 spectrometer. For all spectra, the same Hellma 45-μl microcuvette was used, slit width was kept constant at 2 nm for excitations and integration time was 0.12 s. Cy3 and cresyl violet were excited at 543 nm, and emission was detected in the wavelength interval 553–750 nm. Cy5 and rhodamine 800 were excited at 633 nm, and emission was detected in the range of 643–800 nm. Absorbance spectra were collected with a PerkinElmer Lambda 800 UV/VIS spectrometer. For all spectra, the same Hellma 50-μl absorbance microcuvette was used, absorbance was measured in the wavelength range 200–750 nm, the slit width of detection were kept constant at 2 nm and integration time was 0.12 s.
Differences in detection efficiencies (ηA/ηD) between acceptor and donor detection units were quantified by a theoretical approach as described previously34. The difference between detecting efficiencies (ηA/ηD = 1.18) was calculated as the ratio of the integrated reduced emission after passing the normalized Cy3 and Cy5 emission spectra through each element in the optical pathway: (i) beam splitter, (ii) filter and (iii) APD (Supplementary Fig. 9). On the final γfactor (2.2) we estimated an error of 10%.
We composed a pseudo bulk FRET efficiency according to equation (4)
| (4) |
In the sums of IAFRET, ID and IA, we counted all protein signals, including signals from protein aggregates and proteoliposomes carrying only donor or only acceptor.
To estimate the total amount of protein needed for a miniaturized screen, we used the fact that a microscope experiment required 1.5 × 106 liposomes given a microscope chamber of 5 mm in diameter and a surface density of 7.5 × 1010 liposomes/m2. Assuming each liposome carries 50 receptors of 47,058.1 g/mol each, this corresponds to 5.9 pg of protein.
Quantification of proteoliposome size and receptor densities
Proteoliposome diameters were calculated as described previously14,38. Because the number of fluorophores incorporated in the membrane (Oregon Green DHPE) is proportional to the proteoliposome surface area Aliposome and thereby related to diameter Dliposome through equation (5), a conversion from diffraction-limited intensity spots to physical proteoliposome size was possible.
| (5) |
The calibration factor Ccal was determined by the use of a calibration sample, in which empty liposomes were extruded ×20 through two 50-nm filters (Millipore) to produce a narrow size distribution. The calibration sample was examined first by dynamic light scattering (DLS) to obtain a mean diameter and then by confocal microscopy using identical imaging conditions to those for the normal FRET samples (both Cy3–β2-AR and Cy5–β2-AR were reconstituted in a 1:1 ratio). The mean of the integrated Oregon Green DHPE intensity spots was correlated to the mean radius found by DLS to obtain Ccal. When Ccal was determined, all intensities were converted to diameters by equation (5). DLS measurements were performed on an ALV-5000 correlator equipped with a 633-nm laser. The concentration of liposomes were 0.1 g/l, and all data were collected at T = 20 °C.
For calibration of receptor densities, we used the fluorescence intensities from either Cy3–β2-AR or Cy5–β2-AR and the calculated single proteoliposome surface area according to equation (5). The integrated intensity of the labeled receptor IA/D is proportional to the number of fluorophores, and thus the number of proteins, as each β2-AR carries one label. This holds true for acceptor fluorophores excited by acceptor laser line (633 nm); however, donor intensities are quenched by FRET. To recover the unquenched donor intensity, we added the corrected acceptor FRET intensity to ID (ref. 33), using an IAFRET that was decoupled from instrumental and photophysical effects through the γfactor.
To calibrate absolute receptor densities, we collected single-molecule bleaching traces for samples carrying only β2-AR–Cy3 and only β2-AR–Cy5. Bleaching movies were acquired on a Leica DMI6000 TIRF setup using an oil-immersion objective HCX PL APO CS (100× magnification, 1.46 NA) (Leica). Oregon Green DHPE, β2-AR–Cy3 and β2-AR–Cy5 were excited by a 488-nm laser line, a 561-nm laser line and a 635-nm laser line, respectively. Oregon Green emission was filtered through a filter cube with a dichroic mirror Q495LP and a band-pass filter HQ525/50m. Cy3 emission was filtered through a filter cube with a dichroic mirror T565LP and a band-pass filter ET605/70m. Cy5 emission was filtered through a filter cube with a dichroic mirror Q660LP and a band-pass filter HQ700/75m. All filters and dichroic mirrors were from Chroma Technology. The fluorescence intensity was collected on an electron-multiplying Andor iXon 897 camera. Images were acquired in the format of 512 pixels × 512 pixels, each pixel corresponding to a 160-nm sample length, bit depth of 14 and 250-ms exposure time. Each frame was transferred in 0.304 s, and bleaching series were acquired for 900 frames.
Single-molecule bleaching trace intensities were extracted by software written in Igor Pro v. 6.01 (WaveMetrics). Single-molecule bleaching step intensities were quantified by subtracting the average step intensity by the average background intensity (Supplementary Fig. 2). For a narrow size distribution ranging from 0 to 60 nm, the mean number of receptors was assessed by dividing the unbleached starting intensity by the mean bleaching step intensity39. The mean number of receptors was correlated to the mean intensity obtained by confocal microscopy for the same narrow size range (0–60 nm), and this factor was used to access the number of receptors on all individual proteoliposomes.
Error propagation and the significance of the observed heterogeneities
Mapping the heterogeneities of the proteoliposome samples involved quantifying the fluorescence intensity of the receptor labels and liposome diameter. The uncertainty associated with quantifying the fluorescence intensity signals in the micrographs by fitting 2D Gaussians was accounted for by propagating, for each single proteoliposome, the errors on the fit coefficients from the 2D Gaussian for both lipid and protein signals to the final errors on receptor density, A/D ratio and EFRET.
In addition to uncertainty originating from noise in the micrographs, non-ideal (i.e., heterogeneous) staining of each liposome with the lipid marker can potentially add to the total error on estimating liposome sizes. However, in a previous publication38, we showed experimentally that the employed fluorescence-based size calibration is in excellent agreement with cryo-TEM and dynamic light scattering measurements on the same liposome samples (within a margin of 7–9% on the average diameter), thus ruling out significant bias due to imperfect staining of the lipid phase. This total 7–9% error margin included contributions from both intensity assessment and imperfect lipid staining.
In the present work we were particularly conservative in the error assessment and added to the errors from Gaussian fitting an additional 5% error to account for non-ideal staining of single liposomes with the lipid marker. As reported in Figure 2d,e, the total (cumulated) error on the receptor densities and A/D ratios amounted to 10–14% on average, i.e., ~50% greater than the errors measured in Kunding et al.38.
Partial labeling of protein and liposomes
To estimate the highest probability of finding a proteoliposome having only unlabeled receptors, we used the fact that only 0.02% of all proteoliposomes (β2-AR nominal P/L = 1:1,000) contained fewer than four receptors. This number was used as an upper limit. A labeling efficiency of 0.8 ± 0.2 (±s.e.m.) means that the chance of randomly finding an unlabeled receptor is 0.2 ± 0.2 (±s.e.m.). We therefore calculated the chance of finding four unlabeled receptors in a proteoliposome to be Punlabeled = (0.2)4 = 0.0016. Taking into account the error in the labeling efficiency (0.2 s.e.m.), we conclude that the chance of randomly finding a liposome with only unlabeled receptors is ≤0.0032.
To estimate the probability of finding labeled receptors in an unlabeled proteoliposome, we used a lower proteoliposome size limit of 30 nm in diameter and a lipid head-group area of 0.67 nm2 (ref. 40). In this case 0.5% mol of Oregon Green DHPE (for the β2-AR samples) and 0.2% (for opsin and CB1) would result in >42 and >16 dye molecules, respectively. From Poisson statistics, the probability of finding a completely unlabeled proteoliposome is 5.7 × 10−19 for β2-AR and 4.1 × 10−8 for opsin and CB1 proteoliposomes, respectively. Because 0.5% of all particles included in the histograms of Figure 2d have a diameter below 40 nm, these probabilities would be even smaller for the vast majority of the proteoliposomes. We thus conclude that our finding of protein aggregates persists beyond insufficient labeling of the lipid phase of the proteoliposomes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Lundbeck Foundation, the Danish Councils for Independent and Strategic Research and the University of Copenhagen programs of excellence “Single molecule Nanoscience,” “BioScaRT” and “UNIK-Synthetic Biology.” We thank E.M. Danielsen and H. Flyvbjerg for helpful discussion.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
D.S. conceived of the strategy and was responsible for the overall project management and supervision. D.S., S.M.C. and S.M. designed all experiments. Fluorescence measurements, data analysis and theoretical calculations were performed by S.M. S.V. assisted with ligand binding experiments. J.J.F. and S.G.F.R. expressed, purified, labeled and reconstituted β2-AR; J.F.F. expressed, purified, labeled and reconstituted CB1 and opsin. S.K.J. and S.M.C. developed analysis algorithms and wrote Igor scripts. M.K. supported theoretical fitting and calculations. D.S., S.M. and S.M.C. wrote the paper. All authors discussed the results and commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Rigaud JL, Lévy D. In: Methods in Enzymology: Liposomes, Part B. Duzgunes N, editor. Vol. 372. Elsevier; 2003. pp. 65–86. [DOI] [PubMed] [Google Scholar]
- 2.Seddon AM, Curnow P, Booth PJ. Biochim. Biophys. Acta. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Serebryany E, Zhu GA, Yan ECY. Biochim. Biophys. Acta. 2012;1818:225–233. doi: 10.1016/j.bbamem.2011.07.047. [DOI] [PubMed] [Google Scholar]
- 4.Niu SL, Doctrow B, Mitchell DC. Biochemistry. 2009;48:156–163. doi: 10.1021/bi801835s. [DOI] [PubMed] [Google Scholar]
- 5.Mansoor SE, Palczewski K, Farrens DL. Proc. Natl. Acad. Sci. USA. 2006;103:3060–3065. doi: 10.1073/pnas.0511010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsen J, Hatzakis NS, Stamou D. J. Am. Chem. Soc. 2011;133:10685–10687. doi: 10.1021/ja203984j. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, et al. Biophys. J. 2006;90:2062–2074. doi: 10.1529/biophysj.105.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura T, et al. J. Biol. Chem. 2012;287:4076–4087. doi: 10.1074/jbc.M111.268425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bomholt J, et al. Langmuir. 2011;27:866–869. doi: 10.1021/la1035163. [DOI] [PubMed] [Google Scholar]
- 10.Cowell GM, Tranum-Jensen J, Sjöström H, Norén O. Biochem. J. 1986;237:455–461. doi: 10.1042/bj2370455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diao J, et al. Nat. Protoc. 2012;7:921–934. doi: 10.1038/nprot.2012.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grasso L, et al. PLoS ONE. 2013;8:e70929. doi: 10.1371/journal.pone.0070929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen AL, Lohr C, Christensen SM, Stamou D. Lab Chip. 2013;13:3613–3625. doi: 10.1039/c3lc50492a. [DOI] [PubMed] [Google Scholar]
- 14.Bendix PM, Pedersen MS, Stamou D. Proc. Natl. Acad. Sci. USA. 2009;106:12341–12346. doi: 10.1073/pnas.0903052106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stamou D, Duschl C, Delamarche E, Vogel H. Angew. Chem. Int. Ed. Engl. 2003;42:5580–5583. doi: 10.1002/anie.200351866. [DOI] [PubMed] [Google Scholar]
- 16.Audet M, Bouvier M. Cell. 2012;151:14–23. doi: 10.1016/j.cell.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Overington JP, Al-Lazikani B, Hopkins AL. Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 18.Milligan G. Mol. Pharmacol. 2013;84:158–169. doi: 10.1124/mol.113.084780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciruela F, Vilardaga JP, Fernández-Dueñas V. Trends Biotechnol. 2010;28:407–415. doi: 10.1016/j.tibtech.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolber PK, Hudson BS. Biophys. J. 1979;28:197–210. doi: 10.1016/S0006-3495(79)85171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adair BD, Engelman DM. Biochemistry. 1994;33:5539–5544. doi: 10.1021/bi00184a024. [DOI] [PubMed] [Google Scholar]
- 22.Yano Y, Matsuzaki K. Biochemistry. 2006;45:3370–3378. doi: 10.1021/bi0522854. [DOI] [PubMed] [Google Scholar]
- 23.Leslie M. Science. 2011;331:24–26. doi: 10.1126/science.331.6013.24-a. [DOI] [PubMed] [Google Scholar]
- 24.Caruso F. Colloids and Colloid Assemblies: Synthesis, Modification, Organization and Utilization of Colloid Particles. Wiley; 2006. [Google Scholar]
- 25.Fung JJ, et al. EMBO J. 2009;28:3315–3328. doi: 10.1038/emboj.2009.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmussen SGF, et al. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 27.Xie G, Gross AK, Oprian DD. Biochemistry. 2003;42:1995–2001. doi: 10.1021/bi020611z. [DOI] [PubMed] [Google Scholar]
- 28.Ridge KD, Lu Z, Liu X, Khorana HG. Biochemistry. 1995;34:3261–3267. doi: 10.1021/bi00010a016. [DOI] [PubMed] [Google Scholar]
- 29.Fay JF, Farrens DL. J. Biol. Chem. 2012;287:33873–33882. doi: 10.1074/jbc.M112.352328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsukamoto H, Sinha A, DeWitt M, Farrens DL. J. Mol. Biol. 2010;399:501–511. doi: 10.1016/j.jmb.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghanouni P, et al. J. Biol. Chem. 2001;276:24433–24436. doi: 10.1074/jbc.C100162200. [DOI] [PubMed] [Google Scholar]
- 32.Fay JF, Dunham TD, Farrens DL. Biochemistry. 2005;44:8757–8769. doi: 10.1021/bi0472651. [DOI] [PubMed] [Google Scholar]
- 33.Periasamy A, Wallrabe H, Chen Y, Barroso M. Methods Cell Biol. 2008;89:569–598. doi: 10.1016/S0091-679X(08)00622-5. [DOI] [PubMed] [Google Scholar]
- 34.McCann JJ, Choi UB, Zheng LQ, Weninger K, Bowen ME. Biophys. J. 2010;99:961–970. doi: 10.1016/j.bpj.2010.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd edn. Springer; 2006. [Google Scholar]
- 36.Magde D, Brannon JH, Cremers TL, Olmsted J. J. Phys. Chem. 1979;83:696–699. [Google Scholar]
- 37.Sauer M, et al. J. Fluoresc. 1995;5:247–261. doi: 10.1007/BF00723896. [DOI] [PubMed] [Google Scholar]
- 38.Kunding AH, Mortensen MW, Christensen SM, Stamou D. Biophys. J. 2008;95:1176–1188. doi: 10.1529/biophysj.108.128819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulbrich MH, Isacoff EY. Nat. Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marrink SJ, de Vries AH, Mark AE. J. Phys. Chem. B. 2004;108:750–760. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.