Epigenetics can contribute to pathogenic mechanisms in autoimmunity. We recently identified an imprinted DNA methylation pattern in rheumatoid arthritis (RA) fibroblast-like synoviocytes (FLS) involving multiple genes in pathways implicated in cell migration, matrix regulation and immune responses.1,2 To understand when alterations in DNA methylation occur in RA and the specificity of the methylation changes in RA, we compared differentially methylated loci (DMLs) of early RA (ERA), juvenile idiopathic arthritis (JIA) to longstanding RA (LRA) and osteoarthritis (OA).
Genomic DNA from fibroblast-like synoviocytes (FLS) was isolated from 4 ERA (1–13 months of symptoms; all female; average age 52) as previously described. 1, 2, 3 All ERA patients were anti-citrullinated peptide antibody positive. Two were treated with methotrexate alone and one was treated with methotrexate and abatacept. One patient was not taking anti-rheumatic drugs. Patients met the 2010 classification criteria for RA and samples were obtained at the time of arthroscopic biopsy.4, 5 FLS from patients with JIA (n=3; 2 female, one male; average age 31) was isolated from synovium obtained at the time of arthroplasty. Previously reported 11 LRA and 11 OA were used as controls.5 DNA methylation level was measured using the Illumina Infinium HumanMethylation450 chip across 485,512 loci and the methylation level at individual locus was reported as β values (see Supplementary Methods).
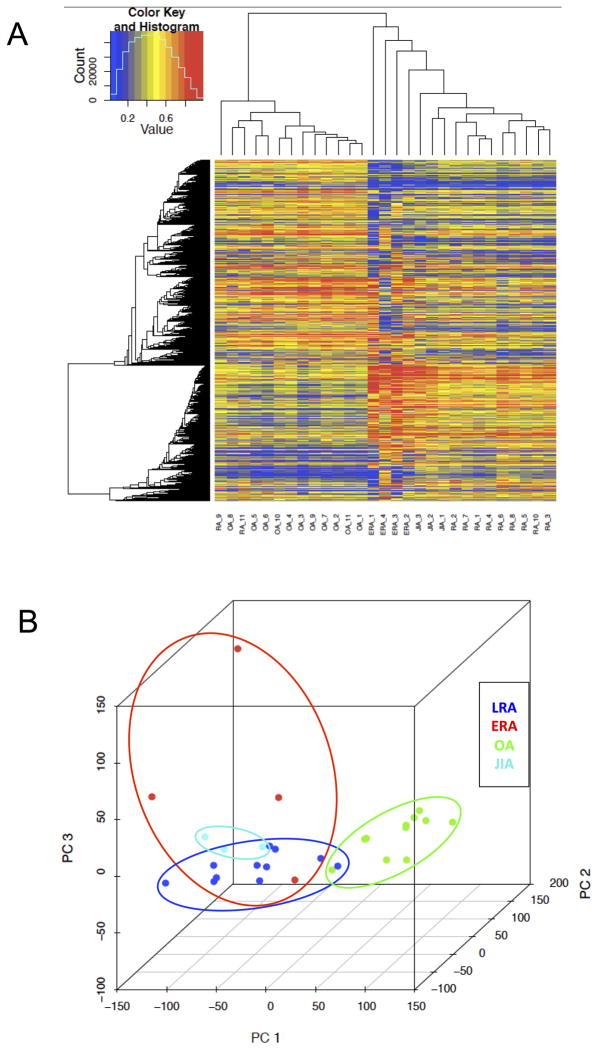
Small but statistically significant global hypomethylation in LRA compared with ERA (average β value per CpG loci = 0.163 in LRA and 0.168 in ERA; p value=0.0046) was detected by summing β values in gene promoter region. Unbiased hierarchical clustering and principal component analysis (PCA) revealed relationships of ERA and JIA to LRA and OA FLS lines (Figure 1A and B, respectively) based on previously identified 15,220 DMLs between 11 LRA and 11 OA2. ERA and JIA clustered with LRA and segregated from OA. ERA and LRA patterns partially overlapped, which could be consistent with a transition of ERA to LRA methylation. FLS from JIA also segregated with the LRA group but formed a separate subgroup. With methylome comparisons between LRA and ERA using Welch’s t-test, we identified 20,776 DMLs (8,059 hypomethylated and 12,717 hypermethylated in LRA compared with ERA) in 5,469 DMGs (average differences of β values > 0.1 and q values < 0.05). Pathway enrichment analysis evaluated by Ingenuity pathway analysis (IPA) identified five potentially enriched pathways (Wnt/β-catenin signaling, enrichment ratio (ER) = 0.249; Integrin signaling, ER = 0.233; RAR activation, ER = 0.233; PDGF signaling, ER = 0.286; Superpathway of D-myo-inositol (1,4,5)-trisphosphate metabolism, ER = 0.417) with hypomethylated DMGs in LRA (BH-adjusted p values < 0.1). In addition, 340 CpG loci that most distinguished ERA from LRA were identified by comparing the entire methylation datasets by random forest method and confirmed that ERA and LRA could be distinguished from each other (Supplementary Figure 1).
Figure 1.
Analysis of DNA methylation patterns in early and longstanding rheumatoid arthritis. (A) Hierarchical clustering of longstanding RA, (LRA), early RA (ERA), juvenile idiopathic arthritis (JIA) and osteoarthritis (OA) based on differentially methylated loci. (B) Analysis of the first three principal components of longstanding RA, (LRA), early RA (ERA), juvenile idiopathic arthritis (JIA) and osteoarthritis (OA) based on differentially methylated loci. Note that the inflammatory arthritis samples mainly cluster together, and that ERA segregates from JIA.
Inflammatory arthritis genome-wide DNA methylation patterns differed from OA and clustered together, suggesting that there are common epigenetic elements of these diseases. However, the FLS formed subgroups implying that the variations are disease specific. Of interest, the methylome of ERA demonstrated a distinctive pattern compared with long-standing disease. These data indicate that differential methylation of RA FLS might occur early and evolve over time. Differentially methylated pathways between ERA and LRA involved cell migration, differentiation, and adhesion, which raises the intriguing possibility that the transition to chronic RA involves epigenetic changes that alter synoviocyte hyperplasia and aggressive behavior. One obvious limitation of the study is that the number of samples is small, in part because obtaining cell lines from ERA biopsies is challenging. Despite that, these unique marks can potentially provide information on mechanisms of disease and how RA evolves over time as well as identifying therapeutic targets based on the duration and type of synovitis.
Supplementary Material
Acknowledgments
Funded in part by grants from the Rheumatology Research Foundation, the Arthritis Foundation, the National Institutes of Arthritis and Musculoskeletal and Skin Diseases R01 AR065466, and IMI EU-funded project BeTheCure no. 115142 and FP7 Euro-TEAM consortium n° 305549
Footnotes
AUTHOR CONTRIBUTIONS
Substantial contributions to study conception and design: Rizi Ai, John W. Whitaker, David L. Boyle, Paul Peter Tak, Danielle M. Gerlag, Wei Wang and Gary S. Firestein
Substantial contributions to acquisition of data: Rizi Ai, David L. Boyle, Danielle Gerlag, Paul-Peter Tak
Substantial contributions to analysis and interpretation of data: Rizi Ai, John W. Whitaker, David L. Boyle, Paul Peter Tak, Danielle M. Gerlag, Wei Wang and Gary S. Firestein
Drafting the article or revising it critically for important intellectual content: Rizi Ai, John W. Whitaker, David L. Boyle, Paul Peter Tak, Danielle M. Gerlag, Wei Wang and Gary S. Firestein
References
- 1.Nakano K, Whitaker JW, Boyle DL, Wang W, Firestein GS. DNA methylome signature in rheumatoid arthritis. Ann Rheum Dis. 2013;72:110–7. doi: 10.1136/annrheumdis-2012-201526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitaker JW, Shoemaker R, Boyle DL, Hillman J, Anderson D, Wang W, et al. An imprinted rheumatoid arthritis methylome signature reflects pathogenic phenotype. Genome Med. 2013;5:40. doi: 10.1186/gm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartok B, Firestein GS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev. 2010;233:233–55. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van de Sande MG, de Hair MJ, Schuller Y, van de Sande GP, Wijbrandts CA, Dinant HJ, et al. The features of the synovium in early rheumatoid arthritis according to the 2010 ACR/EULAR classification criteria. PLoS One. 2012;7:e36668. doi: 10.1371/journal.pone.0036668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62:2569–81. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.