Abstract
Superpositively charged mutants of green fluorescent protein (GFP) demonstrated a dramatically improved chemical exchange saturation transfer (CEST) MRI contrast compared to their wild type counterparts. The mutants +36 GFP and +48 GFP were successfully expressed in mammalian cells and retained part of their fluorescence, making them a new potential bimodal reporter gene.
The discovery, isolation, cloning, and expression of green fluorescent protein (GFP)1 has revolutionized the field of reporter genes, allowing scientists to visualize cellular processes in real-time. Since then, directed mutagenesis of GFP has provided us with multicolor fluorescent reporter genes,2 further expanding the use of GFP for the detection of multiple cell types. However, the limited depth of light penetration for such imaging reporters calls for alternative strategies for imaging reporter gene expression. Recent advances in the field of molecular magnetic resonance imaging (MRI) have increased our ability to monitor gene expression in deep tissue, using various MRI contrast mechanisms.3 One such example is the Lysine–Rich–Protein (LRP),3b a prototype artificial reporter gene that produces MRI contrast based on the chemical exchange saturation transfer (CEST) mechanism.4 The positively charged amino acids (mostly lysine and arginine) in peptides and proteins enable their use as CEST-based contrast agents5 or reporter genes.6 In an effort to develop alternative CEST reporter genes, with a defined structure and controlled expression level, we investigated whether superpositively charged mutants7 of GFP could be used as CEST reporter genes, based on their high content of lysine and arginine residues.
E. coli optimized genes encoding to wild type (wt) GFP (total charge of −7) and its superpositively charged variants (+36 and +48) were transformed into BL21 chemically competent E. coli cells. Recombinant wt and mutated GFP proteins, fused to histidine tags, were expressed and purified using immobilized metal affinity chromatography, followed by dialysis with 10 mM PBS, pH = 7.2 containing 2 M NaCl as previously described.7a Protein solutions were concentrated and aliquots were stored at −80 °C for further experiments. After evaluation of the protein purification and fluorescence (Fig. 1a), the pH of the protein solutions was adjusted to 7.2. CEST experiments were performed using variable levels of saturation power (B1) (Fig. 1). Both +36 GFP and +48 GFP showed a significantly higher CEST contrast compared to wt GFP under all conditions. Fig. 1b demonstrates the MTR asymmetry (MTRasym) maps obtained from 1.25 mg mL−1 pure protein solution at pH = 7.2, with the saturation pulse applied at Δω = 1.8 ppm offset from the frequency of the water protons.
Figure 1.
CEST MRI of GFP proteins. (a) Fluorescence of the examined GFP proteins and their purity, as determined by SDS-polyacrylamide gel electrophoresis. (b) MTRasym maps obtained for a saturation pulse at Δω = 1.8 ppm frequency offset. Shown is the dependency of CEST contrast on B1 power. CEST data from 1.25 mg mL−1 pure protein solutions were acquired at 11.7 T, 37 °C, pH = 7.2, and B1 = 4000 ms.
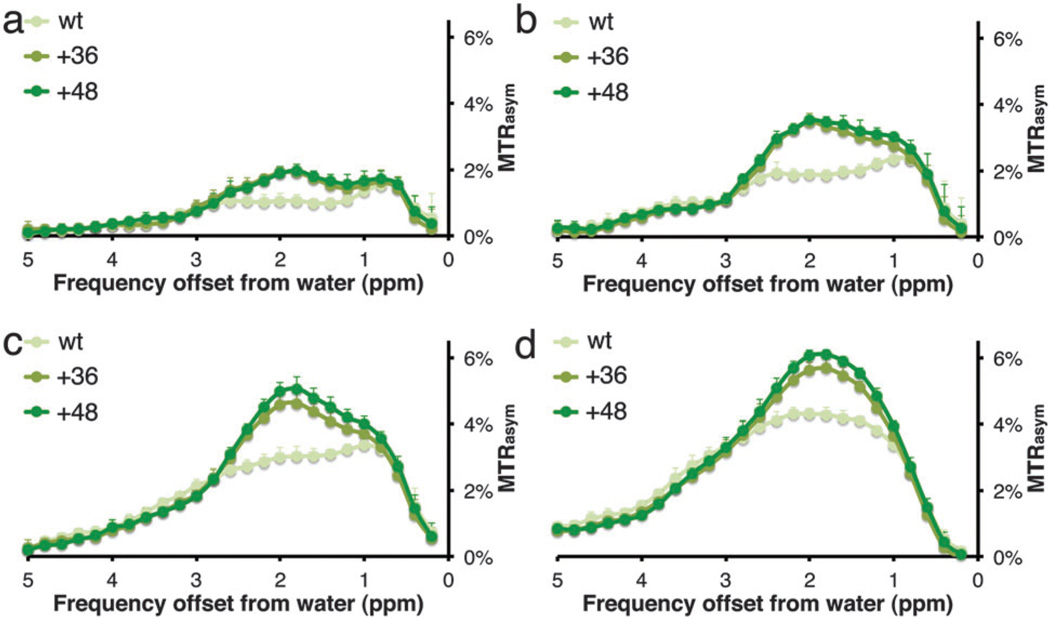
The MTRasym plots of the three examined proteins at different B1 values are shown in Fig. 2. It can be seen that the highest CEST contrast was obtained when the saturation pulse was applied at the frequency offset of the guanidine exchangeable protons of the arginine amino acids, i.e., Δω = 1.8 ppm. This characteristic MTRasym signature was previously demonstrated for arginine-rich synthetic peptides8 and proteins (either naturally occurring salmon5a or human protamine6). From Fig. 1 and 2, it is clear that when a stronger B1 saturation pulse is used, a higher CEST contrast can be obtained for all three GFP proteins. When 7.2 µT was used as B1 saturation pulse, a higher CEST contrast is obtained from +48 GFP compared to +36 GFP. At a lower power (i.e., 3.6 µT) the relative CEST effect is much higher between the mutated GFPs (either +48 or +36) and the wt GFP (see Fig. S1 and S2 and Table S1, ESI†). It should be noted that, although strong saturation pulses may increase the CEST contrast, this might also trigger a higher magnetization transfer (MT) effect from biological tissues, which should be processed and filtered properly. In addition, the back-exchange process that reduces the CEST effect is also a major factor at stronger-than-optimal B1.4
Figure 2.
MTRasym plots of GFP proteins as a function of applied saturation pulse (B1) power: (a) 2.4 µT; (b) 3.6 µT; (c) 4.7 µT; and (d) 7.2 µT. Data from 1.25 mg mL−1 pure protein solutions were acquired at 11.7 T, 37 °C, pH = 7.2, and B1 = 4000 ms. N = 7 for each sample, error bars represent standard deviation.
Although GFP has been suggested previously as a reporter platform for MRI, based on the magnetization transfer contrast mechanism,9 our data demonstrate the much higher specificity when using supercharged GFP reporters and CEST instead of MT as the contrast mechanism. The strong CEST peak at the 1.8 ppm frequency offset, which is characteristic for arginine-rich proteins, makes the supercharged GFP mutants unique for MRI applications, with well-characterized imaging features compared to native proteins with a more normal distribution of amino acids.
Interestingly, no difference in CEST contrast (i.e., MTRasym values) could be obtained when the saturation pulse was performed at the frequency offset of the amide protons (i.e., Δω = 3.6 ppm) for either of the mutants, compared to wt GFP (see Fig. 2). It is known that high CEST contrast from the amide protons of lysine-rich proteins is obtained only where the contributing NH-proton is that of the amide bond between neighbouring positively charged amino acids10 or in other well-defined amino acids sequences.5a Therefore, no CEST contribution was observed from the added lysine amino acids (see Table 1). In addition, the amine exchangeable protons of the lysine side chain are protonated at physiological pH (−NH3+) and exchange too fast with water protons in order to be observed in CEST experiments performed at 11.7 T. Therefore, the only amino acids from the mutants contributing to the CEST contrast are the water-exposed additional arginines (see Table 1). Although lysine is two times more abundant in +48 GFP (42 lysine/protein) than in wt GFP (20 lysine/protein), no difference was observed at Δω = 3.6 ppm offset. However, as shown in Fig. 1 and 2 and in Table 1, an increasing number of arginine residues do, indeed, contribute to higher CEST contrast at 1.8 ppm from both +36 GFP (20 arginine/protein) and +48 GFP (21 arginine/protein). As demonstrated for several other CEST probes, the addition of exchangeable protons does not always increase the obtained CEST contrast.5a,10 This may be explained by the occurrence of a back exchange from saturated water protons to the CEST probes at higher concentrations. In addition, the exchange rate (and hence CEST contrast) between exchangeable protons from the same functional groups located at different protein positions may alter the CEST contrast. As a result, it is difficult to predict the increase in contrast that could be obtained from additional exchangeable protons, such as from the extra arginines in this study.
Table 1.
Number of positively charged amino acids and their measured MTRasym value (1.25 mg mL−1; B1 = 4.7 µT). N = 7 for each sampled protein
| No. of lysines |
MTRasym 3.6 ppm |
No. of arginines |
MTRasym 1.8 ppm |
|
|---|---|---|---|---|
| wt GFP | 20 | 1.3 ± 0.1% | 7 | 3.0 ± 0.3% |
| +36 GFP | 36 | 1.2 ± 0.3% | 20 | 4.6 ± 0.3% |
| +48 GFP | 42 | 1.2 ± 0.3% | 21 | 5.1 ± 0.3% |
Gene optimization is essential for the expression of CEST arginine-rich proteins in bacteria.6,8 To enable successful reporter gene expression in eukaryotes, the genes encoding for wt, +36, and +48 GFP were optimized to enable the expression in mammalian cells (for optimized gene sequences, see ESI†). Human embryonic kidney (HEK) 293T cells were transfected with wt, +36, or +48 GFP mammalian-optimized genes to assess their potential as bimodal reporter genes. All three variants of GFP were expressed in HEK 293 cells 24 hours post transfection (Fig. 3). In addition to wt GFP, both CEST-generating GFP mutants still exhibited cellular fluorescence, albeit lower for +48 compared to +36, a phenomenon that should be further explored. One possible explanation for the observed reduction in cellular fluorescence is that superpositively charged proteins can bind to negatively charged entities such as nucleic acids, which may lead to their aggregation.7a
Figure 3.
Fluorescent microscopy images of human embryonic kidney cells (HEK293T) without transfection (a) and 24 h after transfection with optimized genes encoding for (b) wt, (c) +36, and (d) +48. Bar: 100 µm.
Other strategies have been suggested for imaging gene expression with CEST MRI,11 in which improved contrast specificity can be obtained by administering probes that increase the chemical shift offsets of exchangeable protons.12 However, the necessity of probe administration and sufficient accumulation may be difficult or not possible in certain applications. This includes studying the central nervous system, where the blood brain barrier may prevent the uptake of injected imaging probes. Therefore, imaging reporters that can be directly detected by endogenous expression are, in some cases, most desirable,6,9,13 including supercharged GFPs.
Superpositively-charged GFP mutants have potential for use as CEST MRI reporter genes while retaining their optical properties. The replacement of specific amino acids with arginine can increase the CEST signal, while maintaining the protein structure and function. These findings suggest that molecular engineering of existing proteins may be used to create a new generation of molecular probes. Such a bi-modal probe, if furthered optimized as a robust reporter gene system, may allow fluorescent pre-sorting of the cells of interest prior to in vivo administration and MRI detection, and may provide intrinsic validation of CEST MRI by post-mortem fluorescence.
Supplementary Material
Acknowledgments
The study was supported in part by 2 RO1 NS045062, MSCRFF-0151-00, and MSCRFII-0042. We are grateful to Dr David R. Liu for providing the supercharged GFP plasmids.
Footnotes
Electronic supplementary information (ESI) available: Experimental methodologies, mammalian-optimised genes sequences and figures. See DOI: 10.1039/c4cc10195b
Notes and references
- 1.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Science. 1994;263:802. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 2.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Nat. Biotechnol. 2004;22:1567. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 3.(a) Cohen B, Ziv K, Plaks V, Israely T, Kalchenko V, Harmelin A, Benjamin LE, Neeman M. Nat. Med. 2007;13:498. doi: 10.1038/nm1497. [DOI] [PubMed] [Google Scholar]; (b) Gilad AA, McMahon MT, Walczak P, Winnard PT, Jr, Raman V, van Laarhoven HW, Skoglund CM, Bulte JW, van Zijl PC. Nat. Biotechnol. 2007;25:217. doi: 10.1038/nbt1277. [DOI] [PubMed] [Google Scholar]; (c) Genove G, DeMarco U, Xu H, Goins WF, Ahrens ET. Nat. Med. 2005;11:450. doi: 10.1038/nm1208. [DOI] [PubMed] [Google Scholar]; (d) Louie AY, Huber MM, Ahrens ET, Rothbacher U, Moats R, Jacobs RE, Fraser SE, Meade TJ. Nat. Biotechnol. 2000;18:321. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- 4.van Zijl PC, Yadav NN. Magn. Reson. Med. 2011;65:927. doi: 10.1002/mrm.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) McMahon MT, Gilad AA, DeLiso MA, Berman SM, Bulte JW, van Zijl PC. Magn. Reson. Med. 2008;60:803. doi: 10.1002/mrm.21683. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Oskolkov N, Bar-Shir A, Chan KW, Song X, van Zijl PC, Bulte JW, Gilad AA, McMahon MT. ACS Macro Lett. 2015;4:34. doi: 10.1021/mz500681y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bar-Shir A, Liu G, Chan KW, Oskolkov N, Song X, Yadav NN, Walczak P, McMahon MT, van Zijl PC, Bulte JW, Gilad AA. ACS Chem. Biol. 2014;9:134. doi: 10.1021/cb400617q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Lawrence MS, Phillips KJ, Liu DR. J. Am. Chem. Soc. 2007;129:10110. doi: 10.1021/ja071641y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cronican JJ, Thompson DB, Beier KT, McNaughton BR, Cepko CL, Liu DR. ACS Chem. Biol. 2010;5:747. doi: 10.1021/cb1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) McNaughton BR, Cronican JJ, Thompson DB, Liu DR. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6111. doi: 10.1073/pnas.0807883106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Airan RD, Bar-Shir A, Liu G, Pelled G, McMahon MT, van Zijl PC, Bulte JW, Gilad AA. Magn. Reson. Med. 2012;68:1919. doi: 10.1002/mrm.24483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Torres CJ, Massaad CA, Hilsenbeck SG, Serrano F, Pautler RG. NeuroImage. 2010;50:375. doi: 10.1016/j.neuroimage.2009.12.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goffeney N, Bulte JW, Duyn J, Bryant LH, Jr, van Zijl PC. J. Am. Chem. Soc. 2001;123:8628. doi: 10.1021/ja0158455. [DOI] [PubMed] [Google Scholar]
- 11.(a) Bar-Shir A, Liu G, Liang Y, Yadav NN, McMahon MT, Walczak P, Nimmagadda S, Pomper MG, Tallman KA, Greenberg MM, van Zijl PC, Bulte JW, Gilad AA. J. Am. Chem. Soc. 2013;135:1617. doi: 10.1021/ja312353e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu G, Liang Y, Bar-Shir A, Chan KW, Galpoththawela CS, Bernard SM, Tse T, Yadav NN, Walczak P, McMahon MT, Bulte JW, van Zijl PC, Gilad AA. J. Am. Chem. Soc. 2011;133:16326. doi: 10.1021/ja204701x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Jamin Y, Eykyn TR, Poon E, Springer CJ, Robinson SP. Mol. Imaging Biol. 2014;16:152. doi: 10.1007/s11307-013-0680-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Shir A, Liu G, Greenberg MM, Bulte JW, Gilad AA. Nat. Protoc. 2013;8:2380. doi: 10.1038/nprot.2013.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Vandsburger MH, Radoul M, Addadi Y, Mpofu S, Cohen B, Eilam R, Neeman M. Radiology. 2013;268:790. doi: 10.1148/radiol.13122053. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Iordanova B, Ahrens ET. NeuroImage. 2012;59:1004. doi: 10.1016/j.neuroimage.2011.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.