Abstract
Cardiomyopathy is associated with altered expression of genes encoding contractile proteins. Here we show that Trbp (Tarbp2), an RNA binding protein, is required for normal heart function. Cardiac-specific inactivation of Trbp (TrbpcKO) caused progressive cardiomyopathy and lethal heart failure. Trbp loss of function resulted in upregulation of Sox6, repression of genes encoding normal cardiac slow-twitch myofiber proteins, and pathologically increased expression of skeletal fast-twitch myofiber genes. Remarkably, knockdown of Sox6 fully rescued the Trbp mutant phenotype, whereas Sox6 overexpression phenocopied the TrbpcKO phenotype. Trbp inactivation was mechanistically linked to Sox6 upregulation through altered processing of miR-208a, which is a direct inhibitor of Sox6. Transgenic overexpression of miR-208a sufficiently repressed Sox6, restored the balance of fast- and slow- twitch myofiber gene expression, and rescued cardiac function in TrbpcKO mice. Together, our studies reveal a novel Trbp-mediated microRNA processing mechanism in regulating a linear genetic cascade essential for normal heart function.
Heart failure and cardiovascular disease remain the leading cause of mortality and morbidity in the human population. Genetic mutations and altered expression of many genes encoding contractile proteins in the heart have been associated with cardiac malfunction and cardiovascular diseases1–9. MicroRNAs (miRNAs), a class of ~22 nucleotide-long RNAs, are often conceptualized as “fine-tuners” of gene expression, whose functional influences are due to the accumulative effects of coordinated modulation of multiple downstream mRNA transcripts10–12; Thus, it is believed that the functional impact of a single miRNA:mRNA interaction is generally limited. miRNAs play integral roles in regulating cardiac development and function13–25. However, relatively little is known about the modulation of miRNA biogenesis and its functional consequences in the heart. Trbp, or TARBP2, was initially identified as a RNA binding protein (RBP) involved in HIV pathogenesis26,27. Trbp was found to interact with Dicer to regulate miRNA biogenesis28–32. However, the in vivo functional significance and regulatory pathway of Trbp remains poorly understood.
In this study, we specifically inactivated Trbp in murine cardiomyocytes. We found that loss of Trbp in the heart severely impaired cardiac contraction. Interestingly, cardiac-specific Trbp deletion only altered the expression of a small fraction of miRNAs in the heart. Unexpectedly, the function of Trbp appears to be primarily mediated by a single miRNA, the heart-specific miR-208a, which is abolished in Trbp mutant hearts. miR-208a regulates Sox6, which controls the balance of slow- and fast- twitch muscle gene isoforms. Restoration of miR-208a or Sox6 to normal levels corrected the cardiac defects caused by Trbp cardiac inactivation. Together, our study uncovered an unanticipated linear Trbp-miR-208a-Sox6 pathway in the heart, in which a single miRNA:mRNA axis profoundly influences cardiac contraction.
RESULTS
Cardiac-specific deletion of Trbp impairs cardiac function
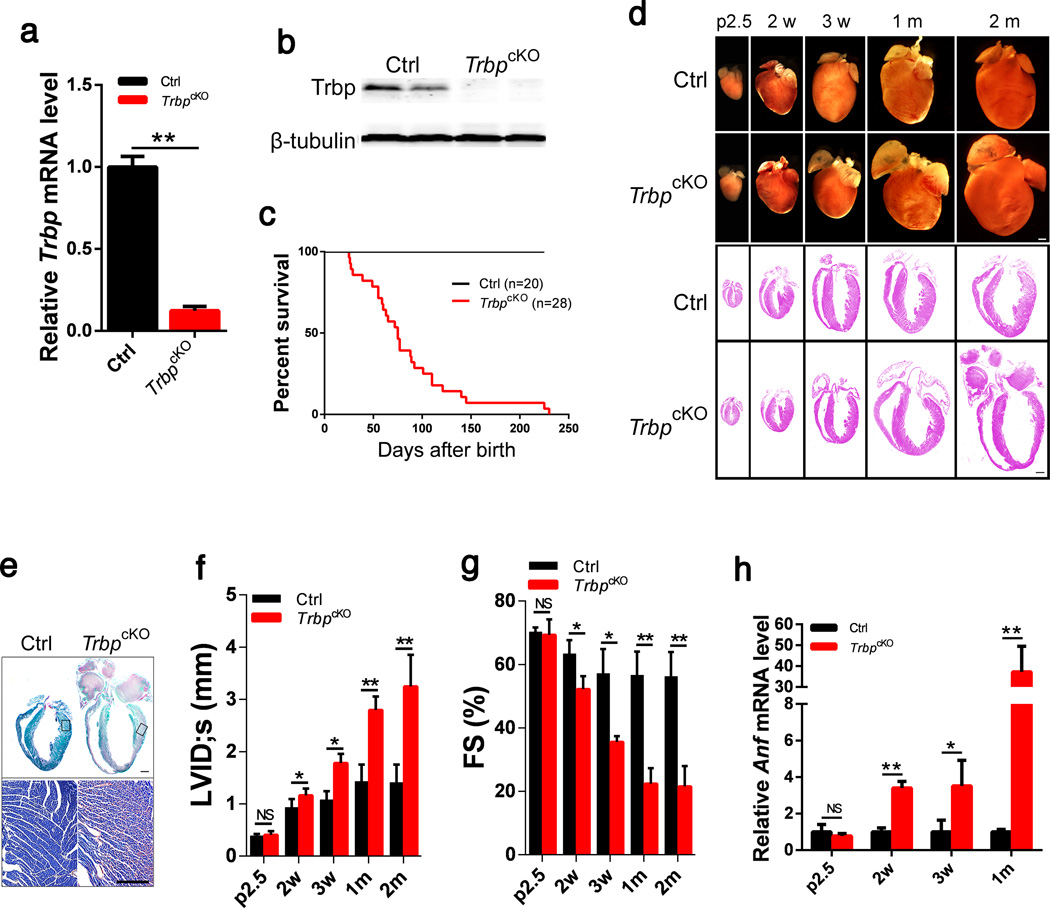
We generated a floxed allele of the Trbp gene, Trbpflox (Trbpfl), in which exons 3–9 of the Trbp gene are flanked by loxP sites (Supplementary Fig.1). The Trbpfl mice were first bred with EIIA-CreTG mice33 to globally ablate Trbp (Trbp−/−). Trbp null mice were smaller and died before weaning (Supplementary Fig. 1), consistent with a previous report34. Next, we generated mice in which the cTNT-Cre transgene35 mediated cardiac-specific Trbp inactivation (cTNT-Cre;Trbpfl/fl, abbreviated as TrbpcKO mice). We confirmed the deletion of the Trbp gene and marked downregulation of Trbp mRNA and protein in TrbpcKO hearts (Fig. 1a, b). TrbpcKO mice were born at the expected normal Mendelian ratio, suggesting lack of embryonic lethality (Supplementary Table 1). However, mutant mice exhibited significantly reduced postnatal viability. More than 80% of TrbpcKO mice died by 4 months of age, and none of the TrbpcKO mice survived beyond 8 months (Fig. 1c).
Figure 1. Cardiac-specific Trbp knockout resulted in contraction defects in the heart.
a) qRT-PCR of Trbp mRNA levels in the hearts of postnatal day 2.5 cTNT-CreTG;Trbpfl/fl (TrbpcKO) and control littermate (Ctrl) mice. n = 3.
b) Trbp protein levels in the hearts of postnatal day 2.5 TrbpcKO and control mice were assayed with Western blot. β-tubulin serves as a loading control.
c) Kaplan-Meier survival curves of TrbpcKO and control mice. P<0.01.
d) Gross morphology and histology of TrbpcKO and control hearts at different time points. p2.5: postnatal day 2.5; 2w: 2 weeks after birth; 3w: 3 weeks after birth; 1m: 1 month after birth; 2m: 2 month after birth. Bars = 1.0 mm.
e) Fast green and Sirius red staining of TrbpcKO and control hearts from 2-month old mice. The portion of the heart represented in high magnification is boxed. Bar = 1.0 mm (upper panel); Bar = 500 µm (lower panel).
f) Echocardiography of left ventricle internal dimension at systolic (LVID;s) in TrbpcKO and control mice at indicated time points. n = 3–6.
g) Fractional shortening (FS%) of TrbpcKO and control mice at indicated time points. n = 3–6.
h) Anf mRNA level in the hearts of TrbpcKO and control mice at indicated time points. n = 3. Values are expressed as the mean ± SD. NS: not significant, *: P<0.05, **: P<0.01.
TrbpcKO hearts did not display gross morphological defects at postnatal day 2.5 (p2.5) and 2 weeks (Fig. 1d). However, the atria were dilated in TrbpcKO mice at 3 weeks after birth (Fig. 1d). By one month, TrbpcKO hearts start to display significant dilation in both atrial and ventricular chambers, and the dilation was severe in mutant mice that survived to 2 months of age (Fig. 1d). Histological analyses confirmed the above observations (Fig. 1d). We found a striking increase in cardiac fibrosis in TrbpcKO hearts in 2-month old mice (Fig. 1e). TrbpcKO cardiomyocyte size was comparable with that of controls (data not shown), suggesting that deletion of Trbp does not affect myocyte growth.
We used echocardiography to measure cardiac function of TrbpcKO and control mice at various ages. At p2.5, TrbpcKO heart function was comparable to that of control littermates (Fig. 1g; Supplementary Table 2). Left ventricular fractional shortening (FS%), a measure of systolic heart function, was significantly decreased in TrbpcKO mice by 2 weeks of age, and it dropped precipitously so that mice had severe systolic dysfunction by 1 month of age (Fig. 1f, g; Supplementary Table 2). Expression of cardiomyopathy marker atrial natriuretic factor (Anf) became progressively elevated to over 30 times control levels in TrbpcKO hearts (Fig. 1h), supporting the view that TrbpcKO mice suffer from heart failure.
Deletion of Trbp alters fast- and slow-twitch myofiber gene expression
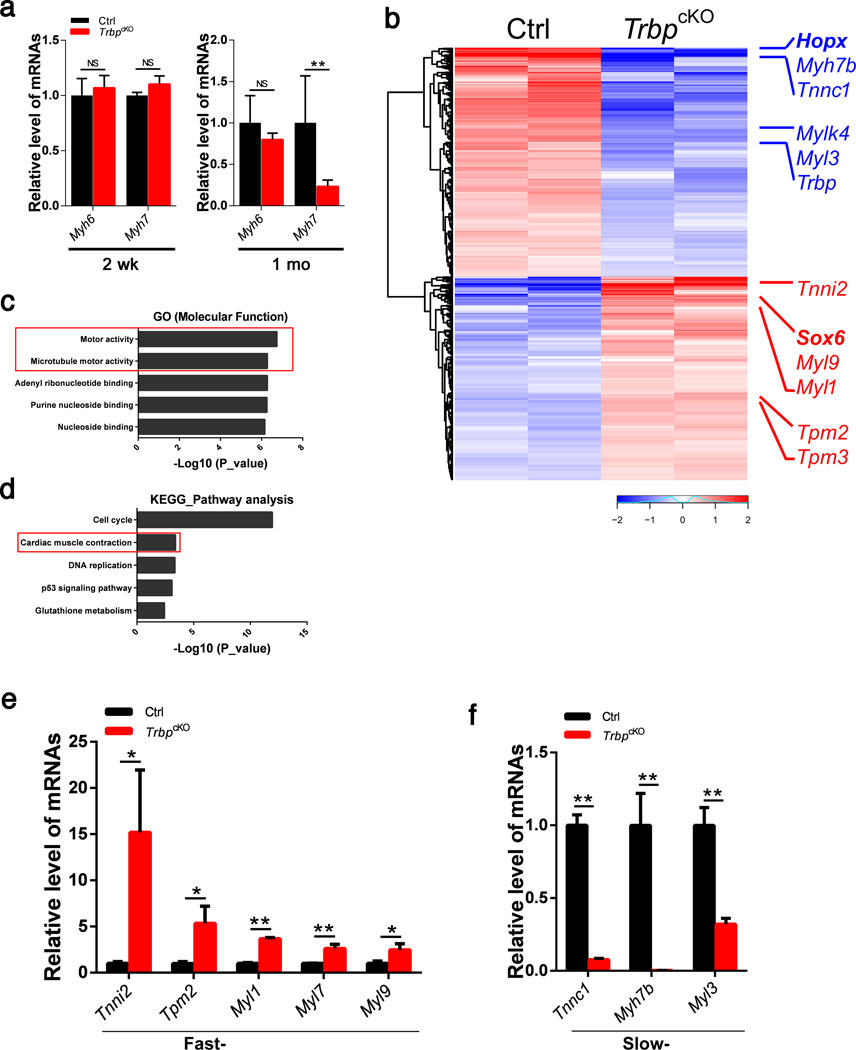
We asked whether the expression of genes encoding adult and fetal isoforms of myosin heavy chains, which are often associated with cardiomyopathy36,37, is altered in TrbpcKO hearts. Unexpectedly, the expression of Myh6 and Myh7 was not significantly different between TrbpcKO and control hearts at two weeks of age. By 1 month of age, the expression of Myh7, which is normally decreased in adult hearts and reactivated in pathological cardiac conditions, was further decreased while the expression of Myh6 remained unchanged in TrbpcKO hearts (Fig. 2a).
Figure 2. Genome-wide identification of dysregulated mRNAs in TrbpcKO hearts.
a) qRT-PCR of Myh6 and Myh7 mRNA levels in TrbpcKO and control hearts at 2 weeks (n = 3) and 1 month (n = 6).
b) Hierarchical clustering heatmap of 351 upregulated and 395 downregulated transcripts from 2-week old TrbpcKO and control hearts. Multiple fast-twitch (red) and slow-twitch (blue) myofiber genes and Hopx, Sox6 genes are marked.
c) The top 5 functional categories from Gene Ontology (GO, Molecular Function) analyses of dysregulated genes in TrbpcKO hearts.
d) The top 5 categories from KEGG pathway analysis of dysregulated genes in TrbpcKO hearts.
e) qRT-PCR of fast-twitch myofiber genes in 2-week old TrbpcKO hearts. n = 3.
f) qRT-PCR of slow-twitch myofiber genes in 2-week old TrbpcKO hearts. n = 3. Values are expressed as the mean ± SD. NS: not significant, *: P<0.05, **: P<0.01.
We performed genome-wide RNA sequencing (RNA-seq) to profile the transcriptome of TrbpcKO and control hearts. We identified a set of 559 genes that were differentially expressed in TrbpcKO hearts (Fig. 2b). 268 genes were up-regulated, and 291 were down-regulated. Trbp itself was among the most drastically down-regulated genes. Gene Ontology (GO) analyses of dysregulated genes in TrbpcKO hearts revealed that many of these genes, including Myh7b, Myl1, Myl3, and Myl9, are involved in “motor activity” and “microtubule motor activity” (Fig. 2c). Using molecular pathways annotated in the Kyoto Encyclopedia of Genes and Genomes (KEGG), we found that the molecular pathway related to “cell cycle” was the most significantly dysregulated (Fig. 2d). We were able to verify the altered expression of some of these genes, including Cdca8, Ckap2l, Cenp-f and Cenp-p, by qRT-PCR in 2-week old hearts (Supplementary Fig. 2). Prompted by this observation, we asked whether the expression of these cell cycle related genes was also altered in TrbpcKO hearts during other developmental stages and if their deregulated pattern is correlated with the observed cardiac pathology. However, their expression is not altered in either p2.5 or 1-month-old TrbpcKO hearts (Supplementary Fig. 2). Next, we tested whether the cell cycle of myocytes was altered in TrbpcKO hearts. However, the staining pattern of phosphorylated Histone 3 (PH3), a marker of mitosis, appeared normal in the mutant hearts. We also quantified the distribution of mono- and bi-nucleated cardiomyocytes in TrbpcKO hearts and found no significant difference when compared with that of control hearts (data not shown). These analyses suggest that dysregulation of these cell cycle genes is unlikely relevant to the cardiac defects in the TrbpcKO hearts.
Remarkably, we found that genes encoding fast skeletal muscle contractile proteins, including Tnni2, Myl1, and Myl9 were significantly up-regulated in the mutant hearts, while genes encoding Myh7b, Tnnc1 and Myl3, slow-twitch muscle contractile proteins, were dramatically down-regulated (Fig. 2b). A proper and balanced expression pattern of fast-, and slow- twitch troponin and myosin isoforms is required for the normal performance of skeletal muscle. However, their altered expression in cardiac muscle has not been well documented, nor has their pathophysiological significance been determined. We validated the deregulation of these candidate genes in the hearts of TrbpcKO and control littermate mice (Fig. 2e,f). These results indicate that the cardiac dysfunction in TrbpcKO mutants might be attributed to the deregulation of genes encoding fast- and slow- twitch myofibers.
Reintroduction of Trbp completely rescues cardiac function in TrbpcKO mice
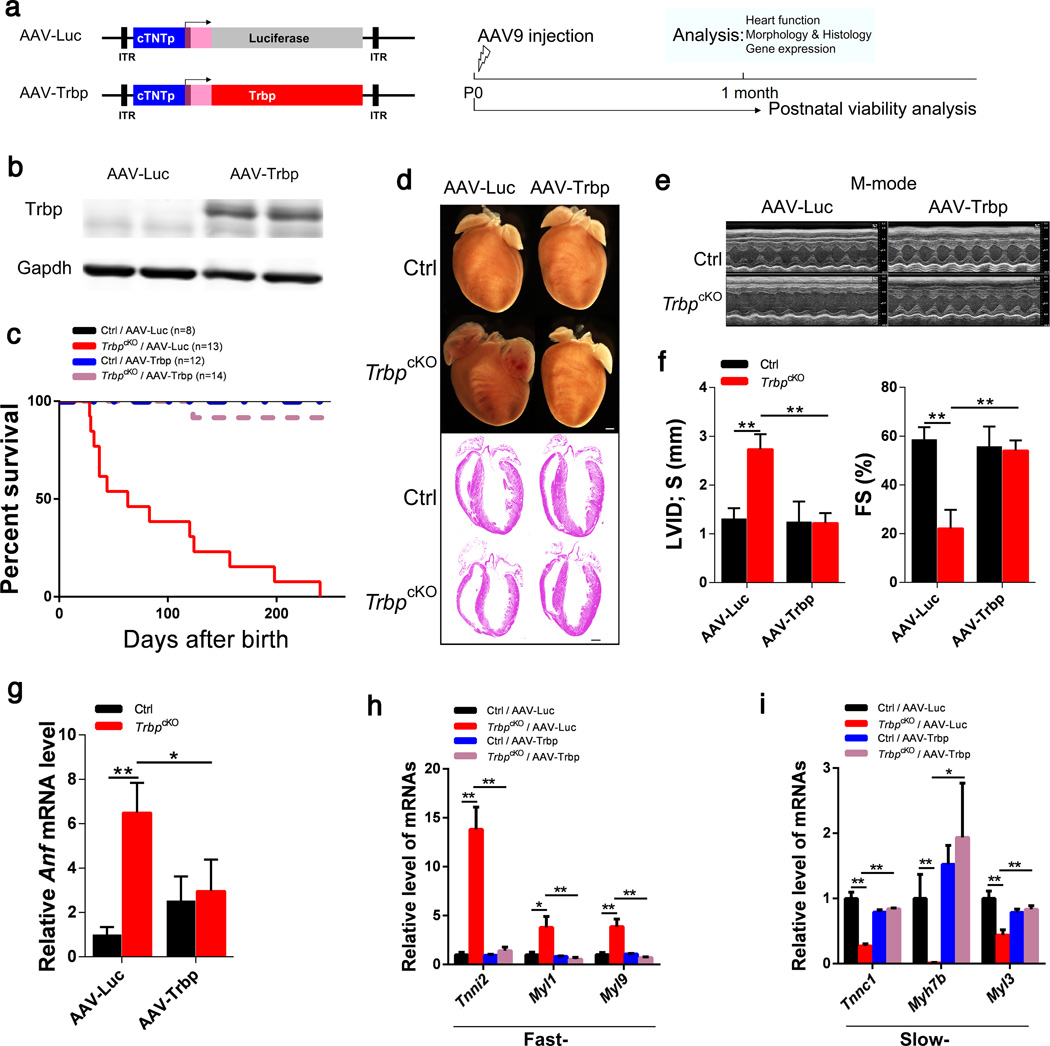
To verify that the loss of Trbp expression in the heart is the cause of cardiac defects, we performed a rescue experiment in which we re-introduced exogenous Trbp into TrbpcKO mice using an adeno-associated virus (AAV)-mediated delivery system38,39 (Fig. 3a). Efficient expression of exogenous Trbp protein in the heart was confirmed by Western blot (Fig. 3b). Re-expression of Trbp, but not the control AAV9.cTNT::Luciferase (AAV-Luc), in the heart completely restored the viability of mutant mice (Fig. 3c). AAV9.cTNT::Trbp (AAV-Trbp) suppressed chamber dilatation and restored normal cardiac morphology in TrbpcKO mice (Fig. 3d). Morphological correction was confirmed on histological sections (Fig. 3d). Echocardiographic analysis showed that Trbp overexpression in control hearts was well tolerated and fully rescued TrbpcKO heart function (Fig. 3e, f; Supplementary Table 3). AAV-Trbp reduced Anf expression to control levels, consistent with rescue of heart failure (Fig. 3g). Remarkably, the expression of fast- and slow- twitch myofiber genes was also fully restored to control levels (Fig. 3h, i). Together, these experiments confirm that Trbp loss of function causes the cardiac abnormalities and mortality observed in TrbpcKO mice.
Figure 3. Re-introduction of Trbp in TrbpcKO hearts rescued cardiac defects.
a) Schematic of AAV9 constructs and the experimental procedure.
b) Western blot to detect Trbp protein expression in 1-month old hearts. Gapdh serves as a loading control.
c) Kaplan-Meier survival curves of AAV-Trbp and AAV-Luc control injected TrbpcKO and control mice (TrbpcKO /AAV-Luc VS. TrbpcKO /AAV-Trbp, P<0.01).
d) Gross morphology and histology of heart samples from 1-month old TrbpcKO and control mice injected with AAV-Trbp or AAV-Luc control. Bars = 1.0 mm.
e) M-mode echocardiography images of 1-month old TrbpcKO and control mice injected with AAV-Trbp or AAV-Luc control.
f) Left ventricle internal dimension at systolic (LVID;s) and fractional shortening (FS%) of 1-month old TrbpcKO and control mice injected with AAV-Trbp or AAV-Luc control (n = 5–14).
g) Anf mRNA level in the hearts of 1-month old TrbpcKO and control mice injected with AAV-Trbp or AAV-Luc control. n = 3.
h) qRT-PCR of fast-twitch myofiber genes in 1-month old TrbpcKO and control mice injected with AAV-Trbp or AAV-Luc control. n = 3.
i) qRT-PCR of slow-twitch myofiber genes in 1-month old TrbpcKO and control mice injected with AAV-Trbp or AAV-Luc control. n = 3. Values are expressed as the mean ± SD. NS: not significant, *: P<0.05, **: P<0.01.
Sox6 functions downstream of Trbp in the heart
We next asked how Trbp regulates the expression of fast- and slow-twitch myofiber genes in the heart. We reasoned that key transcriptional regulators are likely responsible for the observed dysregulation of cardiac gene expression in TrbpcKO hearts. Among the most dramatically dysregulated transcription factors was Hopx, a homeodomain only transcription factor previously linked to cardiac development and cardiomyopathy40–43. RNA-seq analysis showed that the expression of Hopx is significantly reduced in TrbpcKO hearts (Fig. 2b), and we confirmed this observation using qRT-PCR (Supplementary Fig. 3). Next, we asked if rescue of Hopx expression would ameliorate the TrbpcKO phenotype. We generated the cardiac-specific AAV9.cTNT::Hopx (AAV-Hopx) virus and injected it into neonatal TrbpcKO mice (Supplementary Fig. 3). However, overexpression of Hopx failed to rescue animal survival or cardiac function of TrbpcKO mice (Supplementary Fig. 3; Supplementary Table 3). These results suggest that Hopx misexpression alone is unlikely to account for the TrbpcKO phenotype.
RNA-seq indicated that the expression of another transcriptional regulator, Sox6, was significantly up-regulated in TrbpcKO hearts (Fig. 2b). Sox6 has previously been shown to be a key regulator of many of the fast- and slow-twitch myofiber genes in skeletal muscle17, 44,45. However, such a regulatory network and its functional consequences have not been established in the heart. We verified Sox6 upregulation in the hearts of TrbpcKO mice at p2.5, 2 weeks and 1 month (Supplementary Fig. 4a). Interestingly, Sox6 was reduced to control levels in AAV-Trbp transduced TrbpcKO hearts (Supplementary Fig. 4b), concurrent with the correction of fast- and slow-twitch myofiber gene expression in the rescued hearts.
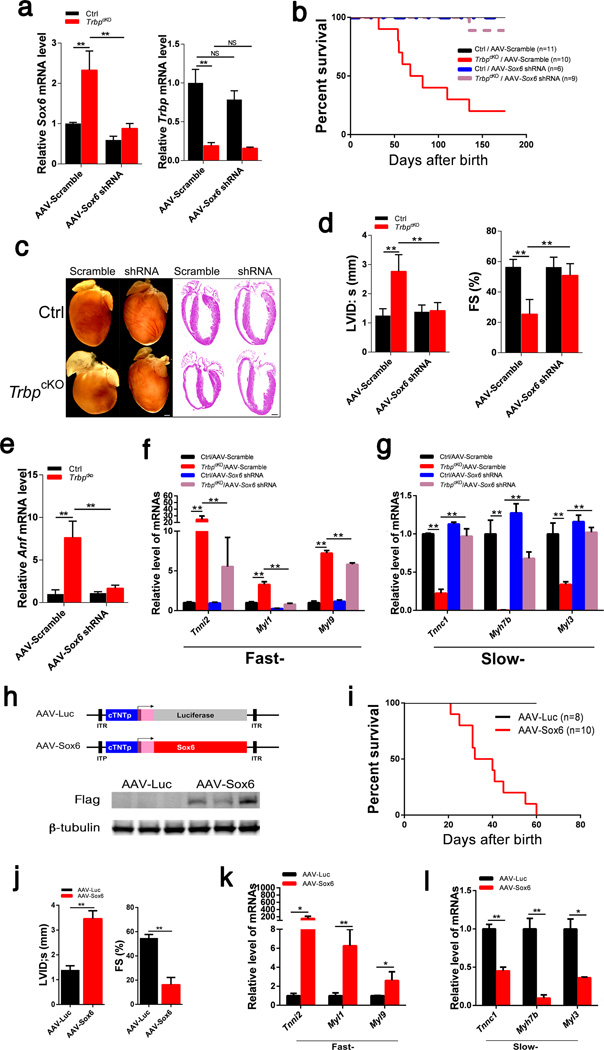
We hypothesized that Sox6 is the key transcriptional regulator down-stream of Trbp that mediates its function in regulating the expression of fast- and slow-twitch myofiber genes. We generated AAV9-shRNA to knockdown Sox6 in vivo (Fig. 4a), predicting that it would suppress the TrbpcKO phenotypic defects in the heart. Remarkably, knockdown of Sox6 completely rescued the viability of TrbpcKO mice (Fig. 4b). Morphological and histological analyses indicated that the TrbpcKO hearts treated with AAV-Sox6 shRNA were indistinguishable from those of control mice, whereas AAV-Scramble had no effects (Fig. 4c). Functional measurements showed that AAV-Sox6 shRNA normalized the systolic LV internal dimension (LVID;s) and LV fractional shortening (FS%), further confirming the full rescue of TrbpcKO mice by knockdown Sox6 (Fig. 4d; Supplementary Table 4). Furthermore, the expression of Anf was reduced to control levels (Fig. 4e). Most remarkably, the expression of fast- and slow-twitch myofiber genes, which was dramatically distorted in TrbpcKO hearts, was restored by Sox6 inhibition (Fig. 4f, g). These findings demonstrate that Sox6 upregulation provokes misexpression of myofiber genes, leading to the heart failure phenotype, and that Sox6 is a key gene downstream of Trbp that mediates its function in the heart.
Figure 4. The function of Trbp is mediated by Sox6 in the heart.
a) Expression of Sox6 (left panel) and Trbp (right panel) in the hearts of 1-month old TrbpcKO and control mice injected with AAV-Scramble or AAV-Sox6 shRNA. n = 3.
b) Kaplan-Meier survival curves (TrbpcKO /AAV-Scramble VS. TrbpcKO /AAV-Sox6 shRNA, p<0.01).
c) Gross morphology and histology of the heart samples from 1-month old mice. Bars = 1.0 mm.
d) Left ventricle internal dimension at systolic (LVID;s) and fractional shortening (FS%) of 1-month old mice. n = 6–12.
e) Expression of Anf in the hearts of 1-month old mice. n = 3.
f) Expression of fast-twitch myofiber genes in the hearts of 1-month old mice. n = 3.
g) Expression of slow-twitch myofiber genes in the hearts of 1-month old mice. n = 3.
h) Schematic of AAV9 constructs (upper panel). Western blot to detect Flag-tagged Trbp protein expression in 1-week old hearts after AAV injection (lower panel). β-tubulin serves as a loading control.
i) Kaplan-Meier survival curves of AAV-Sox6 and AAV-Luc control injected mice (p<0.01).
j) Left ventricle internal dimension at systolic (LVID;s) and fractional shortening (FS%) of 1-month old mice injected with AAV-Sox6 or AAV-Luc control. n = 6–7.
k) Expression of fast-twitch myofiber genes in the hearts of 1-month old mice injected with AAV-Sox6 or AAV-Luc control. n = 3.
l) Expression of slow-twitch myofiber genes in the hearts of 1-month old mice injected with AAV-Sox6 or AAV-Luc control. n = 3. Values are expressed as the mean ± SD. NS: not significant, *: P<0.05, **: P<0.01.
The successful rescue of TrbpcKO phenotype by Sox6 knockdown prompted us to determine whether Sox6 overexpression in wild type mice is sufficient to recapitulate the phenotypes associated with cardiac-specific Trbp loss-of function. We transduced neonatal wild type hearts with AAV9.cTNT::Sox6 (AAV-Sox6) or control AAV9.cTNT::Luciferase (AAV-Luc) (Fig. 4h). Sox6 overexpression resulted in decreased cardiac function and heart failure that resembled the TrbpcKO phenotype (Supplementary Fig. 4c). Animals with cardiac-specific overexpression of Sox6 exhibited significantly reduced viability, also resembling TrbpcKO mice (Fig. 4i). Echocardiography showed that mice overexpressing Sox6 exhibited dilated ventricles and a dramatic decrease in systolic function (Fig. 4j; Supplementary Table 5). Furthermore, the expression of fast-twitch myofiber genes was increased in AAV-Sox6 treated hearts, whereas the slow-twitch myofiber genes were downregulated, again mimicking the abnormalities seen in TrbpcKO hearts (Fig. 4k, l). These studies suggest that Sox6 modulates the expression of fast- and slow-twitch myofiber genes in the heart to induce heart failure.
Trbp regulates processing of a sub-set of miRNAs in the heart
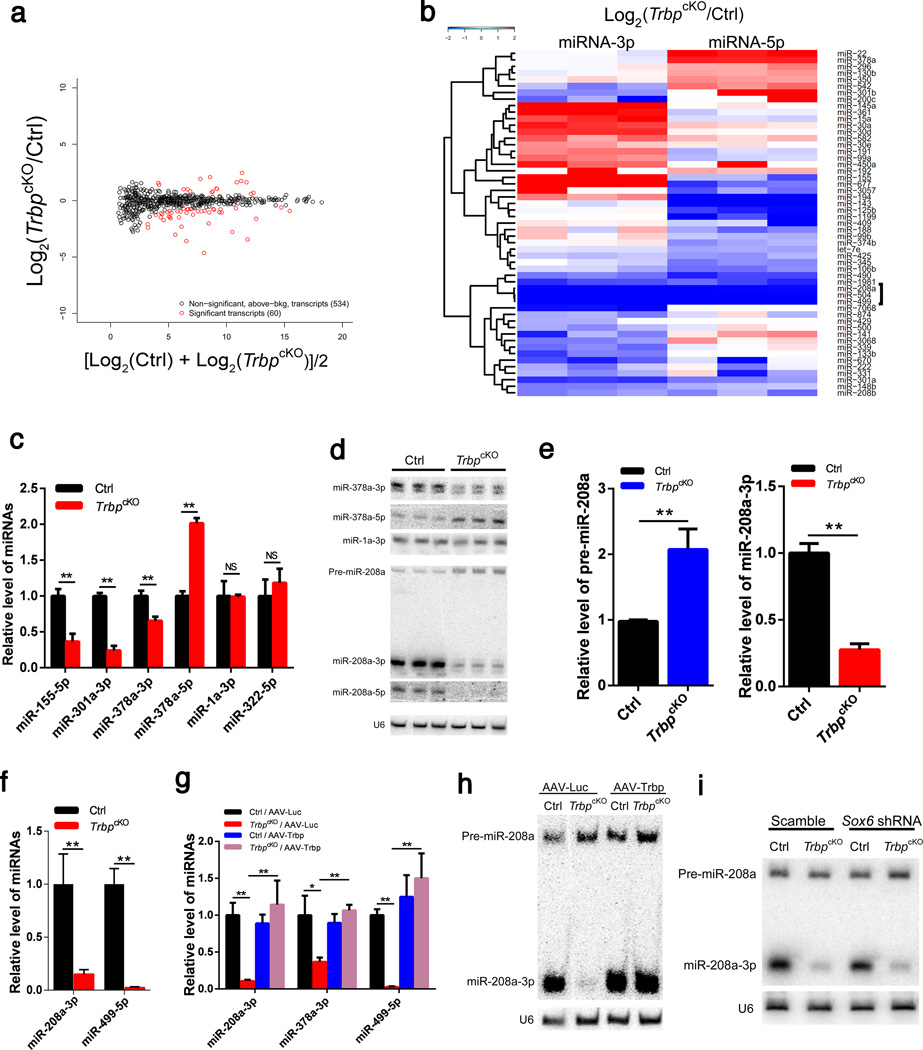
To determine the role of Trbp on miRNA processing in the heart in vivo, and to identify putative miRNAs that act downstream Trbp and participate in regulating heart function, we profiled miRNA expression in TrbpcKO hearts by small RNA-sequencing (small RNA-Seq). Profiling of small RNAs revealed 594 miRNA species expressed in the heart, which is comparable with previously reported miRNA-seq studies46,47. Overall, the expression of only 60 miRNAs species, derived from 53 miRNA precursors, was altered in TrbpcKO hearts compared to littermate controls (Fig. 5a, b), suggesting that Trbp only regulates a subset of miRNAs in the heart. We confirmed the dysregulation of miR-155-5p, miR-301a-3p, and miR-378a-5p in TrbpcKO hearts (Fig. 5c). We noticed that the levels of 5p and 3p strands of several miRNAs, including miR-378a, were distinctively regulated (Fig. 5b), implying that Trbp influences strand-specific selection during miRNA processing in the heart. As shown in Taqman-based qRT-PCR assays, the level of miR-378a-3p was decreased, whereas miR-378a-5p was increased in TrbpcKO hearts (Fig. 5c). Of note, the expression of miR-1a-3p, a muscle-specific and abundantly expressed miRNA, was not affected in TrbpcKO hearts (Fig. 5b, c). We verified the above observations using Northern blot assays (Fig. 5d).
Figure 5. Genome-wide identification of dysregulated miRNA species in TrbpcKO hearts.
a) Scatter plot of the expression of 594 miRNAs between p2.5 TrbpcKO and control hearts. 60 dysregulated miRNA species (derived from 53 miRNA genes) are marked red.
b) Hierarchical clustering heatmap of the 53 dysregulated miRNA genes between TrbpcKO (n = 3) and control (n = 3) hearts. Significantly down-regulated miR-208a cluster is marked.
c) qRT-PCR of indicated miRNAs in p2.5 TrbpcKO and control hearts. n = 3.
d) Northern blot of indicated miRNAs in p2.5 TrbpcKO and control hearts. U6 serves as a loading control.
e) Quantification of pre-miR-208a (left panel) and mature miR-208a (right panel) from Northern blots of p2.5 TrbpcKO and control hearts. n = 3.
f) qRT-PCR of miR-208a-3p and miR-499-5p in p2.5 TrbpcKO and control hearts. n = 3.
g) qRT-PCR of miR-208a-3p, miR-378a-5p and miR-499-5p in the hearts of 1-month old TrbpcKO and control mice injected with AAV-Trbp or AAV-Luc control. n = 3.
h) Northern blot of miR-208a-3p in the hearts of 1-month old TrbpcKO and control mice injected with AAV-Trbp or AAV-Luc control. Both the precursor (pre-miR-208a) and mature product (miR-208a-3p) are shown. U6 serves as a loading control.
i) Northern blot of miR-208-3p in the hearts of 1-month old TrbpcKO and control mice injected with AAV-Scramble or AAV-Sox6 shRNA. Both the precursor (pre-miR-208a) and mature product (miR-208a-3p) are shown. U6 serves as a loading control. Values are expressed as the mean ± SD. NS: not significant, *: P<0.05, **: P<0.01.
In addition to modulating strand selection of miRNAs, we noticed that the expression of several miRNAs, including miR-208a, miR-504 and miR-499, was abolished for both 5p- and 3p- strands in TrbpcKO hearts (Fig. 5b). We verified the down-regulation of miR-208a and miR-499 by qRT-PCR and Northern blot assays in p2.5 TrbpcKO hearts (Fig. 5d, e, f). Consistent with impaired miR-208a processing, Northern blots showed that levels of the precursor of miR-208a (pre-miR-208a), an immature form generated during miR-208a biogenesis, accumulated in TrbpcKO hearts, while both mature strands miR-208a-5p and miR-208a-3p were depleted (Fig. 5d). Quantification confirmed an increased level of pre-miR-208a and decreased expression of miR-208a-3p (Fig. 5e). Interestingly, we did not observe much accumulation of pre-miR-208a in the hearts of older TrbpcKO mice (Supplementary Figure 5a), suggesting a putative surveillance mechanism that modulates the steady-state level and homeostasis of miRNA precursors. Together these data demonstrate a critical and selective role for Trbp in the biogenesis of myomiRs.
Next, we examined whether re-introduction of Trbp could reestablish the expression of these miRNAs in TrbpcKO hearts. Indeed, AAV-Trbp, which fully rescued cardiac function and myofiber gene expression signatures in Trbp mutant mice, completely restored the expression level of miR-208a-3p, miR-378a-3p and miR-499 in the heart (Fig. 5g). Northern blot showed that overexpression of Trbp completely restored the expression level of mature miR-208a (Fig. 5h). However, reintroduction of Trbp into the hearts of TrbpcKO mice did not significantly increase the level of miR-208a precursors (Fig. 5h; Supplementary Fig. 5b), indicating that Trbp is likely involved in the regulation of pre-miR-208a processing. Although Sox6 knockdown also rescued cardiac function and restored the expression of fast- and slow- myofiber genes, it did not restore miR-208a expression (Fig. 5i; Supplementary Fig. 5c). Thus, Trbp regulates miR-208a upstream of Sox6.
miR-208a functions down-stream of Trbp to regulate cardiac function
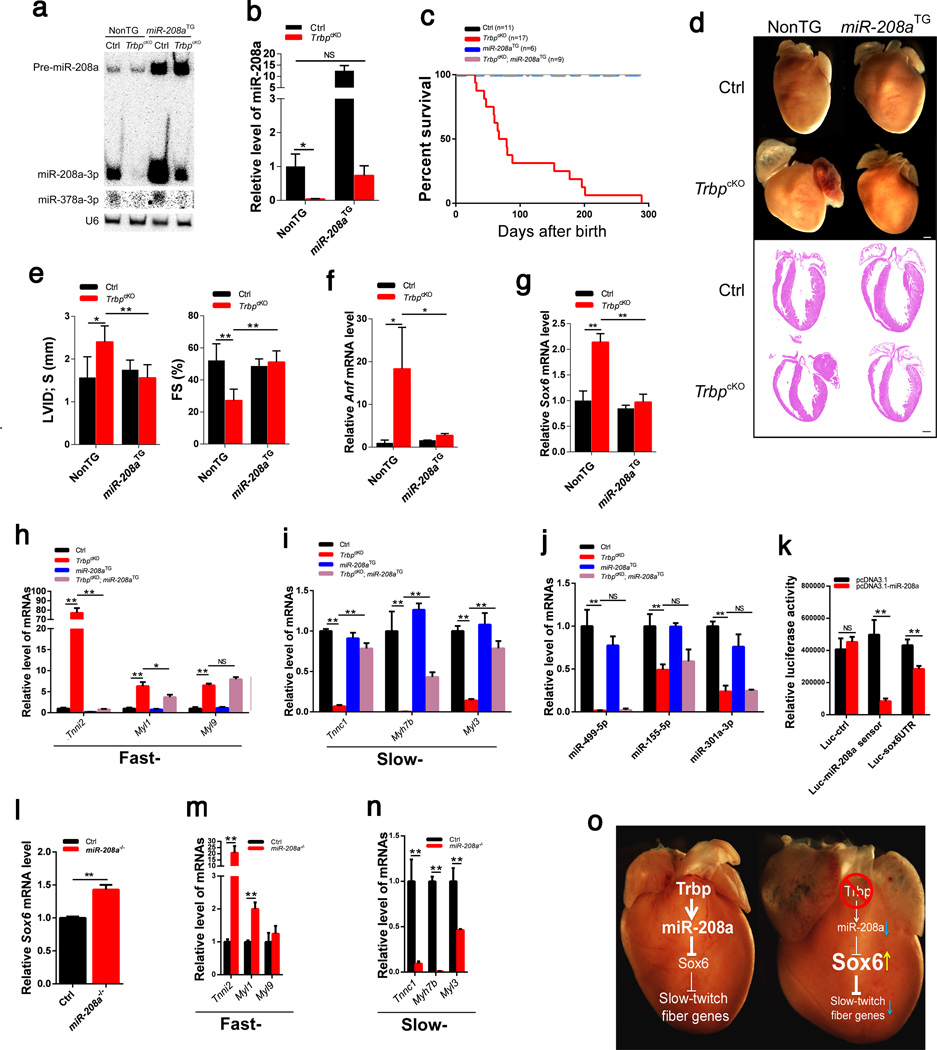
To test the hypothesis that down-regulation of miR-208a is central to heart failure pathogenesis in TrbpcKO hearts, we decided to restore miR-208a cardiac expression by crossing a miR-208a transgene (miR-208aTG) into the TrbpcKO genetic background. In control hearts with normal Trbp levels, miR-208aTG upregulated the expression of both pre-miR-208a and mature miR-208a by over 8-fold (Fig. 6a, b; Supplementary Fig. 5d), resulting in cardiac hypertrophy16. In miR-208aTG; TrbpcKO compound mice, miR-208aTG restored miR-208a to a level similar to that of control littermates, whereas pre-miR-208a remained elevated (Fig. 6a, b; Supplementary Fig. 5d). Reduced transgenic expression of miR-208a in the Trbp mutant compared to Trbp wild-type background is consistent with a key role for Trbp in miR-208a biogenesis. Remarkably, normalizing miR-208a expression fully rescued the viability of TrbpcKO mice (Fig. 6c). Heart morphology and histology of miR-208aTG;TrbpcKO compound mice were indistinguishable from those of control littermates (Fig. 6d). Echocardiographic analyses confirmed that cardiac function was completely rescued in miR-208aTG;TrbpcKO compound mice (Fig. 6e; Supplementary Table 6). Elevated expression of cardiomyopathy marker Anf found in TrbpcKO hearts was also normalized by miR-208a overexpression (Fig. 6f).
Figure 6. miR-208a is a Trbp target to mediate its function in TrbpcKO hearts.
a) Northern blot of miR-208a and miR-378a in the hearts of indicated mice. U6 serves as a loading control.
b) qRT-PCR of miR-208a in the hearts of indicated mice. n = 3.
c) Kaplan-Meier survival curves (TrbpcKOVS. TrbpcKO; miR-208aTG, P<0.01).
d) Gross morphology and histology of the heart samples from 1-month old mice. Bars = 1.0 mm.
e) Left ventricle internal dimension at systolic (LVID;s) and fractional shortening (FS%) of 1-month old mice. n = 7–9.
f) Anf mRNA level in the hearts of 1-month old mice with indicated genotypes. n = 3.
g) Expression of Sox6 in the hearts of 1-month old mice. n = 3.
h) Expression of fast-twitch myofiber genes in the hearts of 1-month old mice. n = 3.
i) Expression of slow-twitch myofiber genes in the hearts of 1-month old mice. n = 3.
j) Expression of miR-499-5p, miR-155-5p and miR-301a-3p in the hearts of 1-month old mice. n = 3.
k) Luciferase reporters were co-transfected with pcDNA-miR-208a or control and relative luciferase activity determined. n = 3.
l) Expression of Sox6 in the hearts of 2-week old mice. n = 3.
m) Expression of fast-twitch myofiber genes in the hearts of 2-week old mice. n = 3.
n) Expression of slow-twitch myofiber genes in the hearts of 2-week old mice. n = 3.
o) A working model signifying the Trbp:miR-208a:Sox6 linear pathway in the regulation of slow-twitch myofiber gene expression and cardiomyopathy. Values are expressed as the mean ± SD. NS: not significant, *: P<0.05, **: P<0.01.
Sox6 has been identified as a target of the myomiRs miR-208a/miR-208b and miR-49917. Therefore, we hypothesized that Sox6 upregulation in TrbpcKO hearts is due to miR-208a downregulation. To test this hypothesis, we asked if restoration of miR-208a in TrbpcKO hearts normalized Sox6 expression. Indeed, Sox6 level in miR- miR-208aTG; TrbpcKO hearts was comparable to that of controls (Fig. 6g). Most importantly, the expression signatures of fast- and slow-twitch myofiber genes were substantially restored in the hearts of the miR-208aTG; TrbpcKO compound mice (Fig. 6h, i). Successful restoration of TrbpcKO cardiac pathology by miR-208a transgene indicated that down-regulation of miR-208a is essential for the pathogenesis of cardiomyopathy downstream of Trbp loss-of-function.
Interestingly, expression of Myh7b, the host gene of miR-499, was substantially restored in miR-208aTG; TrbpcKO hearts (Fig. 6i), but miR-499 expression was not (Fig. 6j). Nor did miR-208a rescue the expression of miR-378a-3p (Fig. 6a), miR-155-5p, or miR-301a-3p (Fig. 6j). This is distinct from AAV-Trbp-mediated rescue, where the expression level of miR-499 was restored (Extended Data Fig. 5e), suggesting that the function of Trbp is likely mediated by miR-208a, not miR-499, in the heart. Next, we asked whether miR-208a inhibits the expression of Sox6 directly. Using a luciferase reporter assay in which the 3’ untranslated region (3’ UTR) of Sox6 gene is cloned into the reporter, we found that miR-208a represses the Sox6-3’UTR-luciferase reporter (Fig. 6k), confirming that Sox6 is a direct target of miR-208a in the heart.
To further test the hypothesis that down-regulation of miR-208a in TrbpcKO hearts is responsible for the observed cardiac defects, we examined the expression of Sox6 in the miR-208a knockout (miR-208a−/−) hearts16. Indeed, Sox6 expression level was elevated in miR-208a−/− hearts (Fig. 6l). Most importantly, we found that the expression levels of fast- and slow- twitch myofiber genes were also significantly increased and decreased, respectively, in miR-208a−/− hearts (Fig. 6m, n), further supporting the view that miR-208a directly modulates the expression and function of Sox6 and its downstream myofiber genes in the heart.
DISCUSSION
In this study, we show that the RNA binding protein Trbp is indispensible for cardiac function and postnatal survival. We demonstrate that Trbp selectively mediates the biogenesis of a small set of miRNAs in the heart. This is in sharp contrast with the cardiac-specific Dicer knockout, which alters the biogenesis of most miRNAs in the heart14. Such specificity for Trbp function in vivo in the heart is surprising, given that it has been proposed that Trbp broadly regulates miRNA processing48. We identify miR-208a as a critical target of Trbp and show that miR-208a is required to suppress Sox6 and thereby maintain the balance of fast- and slow- myofiber gene expression essential for normal heart function. Interestingly, genetic mutations of these isoforms in skeletal muscle have been associated with muscle related diseases2,3,7. Our results suggest that proper and balanced expression of fast-, and slow- twitch myofiber genes is also essential for cardiomyocyte contraction and cardiac function. We predict that mutations of these genes and/or dysregulation of their expression will be associated with cardiomyopathy in human patients.
Probably one of the most astonishing findings of this study is that a single miRNA (miR-208a) and single mRNA target (Sox6) are able to convey the function of Trbp, a RBP involved in the miRNA pathway that was thought to regulate the biogenesis and function of multiple RNA species (miRNAs and mRNAs) in the heart (Fig. 6o). Despite the establishment of the role of miR-208a in mediating Trbp function in the heart, we cannot formally rule out the contribution of other miRNAs, which are dysregulated in TrbpcKO hearts, in this process. To further support the view that the Trbp→miR-208a—|Sox6 pathway controls cardiomyopathy in the heart, we have noticed that the miR-208a null mice that we previously generated16 also exhibit dilated cardiomyopathy and heart failure, similar to what we observe in Trbp mutant mice (Ding and Wang, unpublished observation). Collectively, our study may reshape how we think about the function and functional mechanisms of miRNAs. Most importantly, investigating and understanding such regulation will enable us to identify new therapeutic targets to treat cardiovascular diseases.
METHODS
Mouse models
Trbpflox/flox (Trbpfl/fl) mice were generated by inserting LoxP sites for Cre-mediated excision flanking exons 3 to 9 as shown in the Extended Data information (Supplementary Fig.1). Trbpfl/fl mice were bred with an EIIa-CreTG line to achieve global deletion of the targeted region. To obtain cardiac-specific knockout of Trbp (TrbpcKO), Trbpfl/fl mice were bred with cTNT-CreTG mice. The miR-208a knockout line (miR-208a−/−) and miR-208a transgenic strain (α-MHC-tTA; tet-miR-208a, or miR-208aTG), were previously described16. Overexpression of miR-208a was controlled by a cardiac-specific tet-off binary system. The miR-208a transgenic mice were intercrossed with TrbpcKO mice to obtain the compound genetic background. We performed most of our experiments with 1 month-old mice. Some animals died before this age were naturally excluded from the analysis. The experiments were performed and the samples were collected without bias. All experiments with mice were performed according to protocols approved by the Institutional Animal Care and Use Committees of Boston Children's Hospital.
AAV preparation and injection
The cDNA fragments encoding Luciferase, Flag-Trbp, Flag-Hopx, or Flag-Sox6 were separately cloned into ITR-containing AAV9 plasmid (Penn Vector Core P1967) harboring the chicken cardiac TNT promoter, to yield pAAV.cTnT::Luciferase (AAV-Luc), pAAV.cTnT::Flag-Trbp (AAV-Trbp), pAAV.cTnT::Flag-Hopx (AAV-Hopx) and pAAV.cTnT::Flag-Sox6 (AAV-Sox6), respectively. To generate p.AAV.U6::shRNA constructs, DNA fragments harboring U6 promoter-driven Scramble or Sox6 shRNA cassettes were separately cloned into the ITR-containing AAV9 plasmid. AAV9 was packaged in 293T cells with AAV9:Rep-Cap and pHelper (pAd deltaF6, Penn Vector Core) and purified and concentrated by gradient centrifugation. AAV9 titer was determined by quantitative PCR. Neonatal mice were treated with AAV (1–1.5 × 1011 particles/pup for overexpression, 2.5–5 × 1011 particles/pup for knockdown) at postnatal day 0.5–1.5 by subcutaneous injection.
Histology
Mouse hearts were dissected out, rinsed with PBS and arrested in diastole with KCl and BDM buffer, then fixed in 4% paraformaldehyde (pH 7.4) overnight. After dehydration through a series of ethanol baths, samples were embedded in paraffin wax according to standard laboratory procedures. Sections of 5 µm were stained with haematoxylin and eosin (H&E), or further fixed with pre-warmed Bouins’ solution, 55°C for 1 hour, and stained with Fast Green and Sirius Red as previously described23–25. The stained sections were used for routine histological examination with light microscope.
Echocardiography
Two-dimensional and M-mode imaging was performed using a VisualSonics Vevo® 2100 Imaging System with a 40 MHz MicroScan transducer (model MS-550D) as previously described23,25. Heart rate and left ventricular (LV) dimensions were measured from 2-D short-axis under M-mode tracings at the level of the papillary muscle. Functional parameters such as percentage of fractional shortening (FS%) and left ventricular volume were calculated using the above primary measurements and accompanying software.
RNA-sequencing and data analysis
RNA from the hearts of ~2-week old mice was prepared for gene expression profiling (two biological replicates for each genotype). Total cardiac RNAs were isolated from fresh ventricular tissue using TRIzol (Invitrogen) and were twice oligo(dT)-selected using the Dynabead mRNA purification system (Invitrogen). The obtained RNA samples were fragmented and reverse transcribed, followed by second-strand cDNA synthesis. The cDNAs were end-repaired, modified, and ligated into Illumina Truseq adapter. The ligated DNAs were amplified using primers with specific barcode indexes by PCR. The obtained barcoded libraries were combined in equimolar amounts for cluster formation on a single Illumina flow cell lane, followed by single-end sequencing (SE50, HiSeq2000, TUFC genomics facility). Single-read FASTQ files were extracted from Illumina HiSeq2000 with Illumina's bcl2fastq.pl using default options. TruSeq sequencing adapters were removed from reads with cutadapt. The FASTQ files were aligned to mouse genome (mm10) using bowtie. RNA from the hearts of neonatal p2.5 mice was prepared for small RNA-seq (Three biological replicates for each genotype). RNA isolated from ventricular tissue was ligated to 3’ and 5’ Illumina Truseq adaptors for small RNAs, reverse transcribed and amplified by PCR using the primers with barcode indexes. The barcoded libraries were combined in equimolar amounts for cluster formation on a single Illumina flow cell lane, followed by single-end sequencing (SE50, HiSeq 2000, TUFC genomics facility). Read length after adapter removal shows a tight distribution mostly within 16–27nt with mode at 21–22nt. Bioinformatically, reads with length no more than 30nt are considered from mature miRNAs, and were counted and mapped to the genome using Bowtie alignment tool. Expression analysis was run in R. Differential expression analysis was performed with the edgeR library, hierarchical clustering heatmap was made with the ggplot library. GO and KEGG pathway analysis was performed with David bioinformatics sever49,50.
Quantitative RT-PCR and Northern blot analyses
For quantitative RT-PCR detecting the expression of protein-coding genes, 1 µg RNA samples were reverse-transcribed to cDNA using random primers and MMLV reverse transcriptase (Invitrogen) in 20 µl reaction system. The obtained cDNA samples were 10× diluted in nuclease-free water. For each reaction, 1 µl diluted cDNA was used with Sybergreen probes and normalized to Gapdh. For quantitative RT-PCR detecting the expression of miRNAs, 10 ng RNA samples were reverse-transcribed to cDNA using the TaqMan® MicroRNA Reverse Transcription Kit. The level of miRNAs was assayed using Taqman miRNA assay kit. U6 snoRNA was used as internal control. For reverse transcription of miRNA precursor, ~ 1 µg RNA was reverse transcribed with PrimeScript Reverse Transcriptase (CloneTech) using 10 µM of the pre-miR-208a specific anti-sense primer (pre-miR-208a-qR) and the primer for the internal control (18S rRNA, 18sr-qR). The RNA and primer mixture was heated to 80 °C for 5 min to denature the RNA, followed by 5 min incubation at 60 °C to anneal the primers. The reactions were cooled to room temperature and the remaining reagents (5× buffer, dNTPs, DTT, RNase inhibitor, PrimeScript RT) were added. The reaction proceeded for 30 min at 50 °C followed by a 5 min incubation at 70 °C to inactivate PrimeScript RT. For miRNA Northern blot analyses, an aliquot of 15–20 µg total RNA was analyzed by electrophoresis on 12% acrylamide/8 M urea gels. Electrotransfer onto nylon membrane (Hybond-N+; Amersham) was followed by UV irradiation for 5 min. Hybridization with 5’-P32-labeled probes was performed as previously described. Sequences of PCR primers and RNA probes used in this study are listed in Extend Data Table 7.
Western blot analyses
Protein lysate samples were prepared from heart tissues in RIPA buffer with proteinase inhibitors. Lysate samples (15–20 µg total protein for each) were separated by 12% SDS-PAGE, and electrophoretically transferred to PVDF membranes. Trbp protein was probed with Rabbit or Mouse anti-Trbp antibodies (AbCam, #42018, or Thermo Scientific, #LF-MA0209, 1:1,000). Flag-tagged proteins were probed with Rabbit anti-Flag M2 (Sigma, #F7425, 1:2,000). Gapdh, or β-tubulin used as loading controls, were probed with mouse anti-Gapdh (EMD Millipore, MAB374, 1:10,000) or mouse anti- β-Tubulin(Sigma, #T8328, 1:5,000) antibodies, respectively. Protein bands were visualized with Oddessay image system.
Luciferase reporter assay
The 3’UTR fragments containing putative miR-208a binding sites were cloned into pGl3cm vector to generate the Luc-Sox6 3’UTR reporter construct. Empty pcDNA3.1 plasmid (negative control) or pcDNA3.1-miR-208a was transiently transfected into HEK293 cells together with Luc-Sox6 3’UTR reporter, or empty pGL3cm (Luc-Ctrl), or Luc-miR208 sensor. Luciferase reporter assay was performed as previously described ~72 hours after transfection23,24.
Statistics
Survival curve and echocardiographic analyses are from cumulative data. Morphological and histological analyses were repeated at least three times. Two and three biological replicates were used for mRNA-seq and small RNA-seq experiments, respectively. All other experiments have been repeated at least three times. Values are expressed as the mean ± SD. Student’s t-test was performed for paired analysis (two-sided, adjusted for multiple comparisons). Log-rankMantel-cox test was performed for survival analysis with Prism 6. The null hypothesis was rejected if p < 0.05.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. David Clapham and members of the Wang laboratory for advice and support. We thank Ramon Espinoza-Lewis, Gang Wang and Fei Gu for technical support. Work in the Wang lab is supported by the March of Dimes Foundation, Muscular Dystrophy Association and the NIH (HL085635, HL116919). M Kataoka was supported by Banyu Life Science Foundation International.
Footnotes
Accession codes
AUTHOR CONTRIBUTIONS
J.D. and D.-Z.W. conceived the project, designed and analyzed the experiments, and wrote the manuscript. J.D. generated and characterized the Trbp mutant mice and performed molecular biology experiments. J.D., L.M. and P.Z. contributed to targeting vector construction and southern blotting. J.D. and J.C. contributed to the echocardiographic data acquisition and analyzing. Y.W. analyzed the RNA-sequencing data. J.D., X.H. M.N. and Z.-L.D. contributed to the morphologic and histologic data acquisition and analyzing. J.D., M.K. and Z.L. contributed to adeno-associated virus preparation and administration. W.P. supervised adeno-associated virus preparation and administration, and reviewed the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Feng H-Z, Jin J-P. Coexistence of cardiac troponin T variants reduces heart efficiency. Am. J. Physiol-Heart C. 2010;299:H97–H105. doi: 10.1152/ajpheart.01105.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimber E, Tajsharghi H, Kroksmark A-K, Oldfors A, Tulinius M. A mutation in the fast skeletal muscle troponin I gene causes myopathy and distal arthrogryposis. Neurol. 2006;67:597–601. doi: 10.1212/01.wnl.0000230168.05328.f4. [DOI] [PubMed] [Google Scholar]
- 3.Laing NG, et al. A mutation in the α tropomyosin gene TPM3 associated with autosomal dominant nemaline myopathy. Nat. Genet. 1995;9:75–79. doi: 10.1038/ng0195-75. [DOI] [PubMed] [Google Scholar]
- 4.Landstrom AP, et al. Molecular and functional characterization of novel hypertrophic cardiomyopathy susceptibility mutations inTNNC1-encoded troponin C. J. Mol. Cell. Cardiol. 2008;45:281–288. doi: 10.1016/j.yjmcc.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mogensen J, et al. Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J. Am. Coll. Cardiol. 2004;44:2033–2040. doi: 10.1016/j.jacc.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 6.Olson TM, Karst ML, Whitby FG, Driscoll DJ. Myosin light chain mutation causes autosomal recessive cardiomyopathy with mid-cavitary hypertrophy and restrictive physiology. Circulation. 2002;105:2337–2340. doi: 10.1161/01.cir.0000018444.47798.94. [DOI] [PubMed] [Google Scholar]
- 7.Tajsharghi H, Ohlsson M, Lindberg C, Oldfors A. Congenital myopathy with nemaline rods and cap structures caused by a mutation in the β-tropomyosin gene (TPM2) Arch. neurol. 2007;64:1334–1338. doi: 10.1001/archneur.64.9.1334. [DOI] [PubMed] [Google Scholar]
- 8.Yu Z-B, Wei H, Jin J-P. Chronic coexistence of two troponin T isoforms in adult transgenic mouse cardiomyocytes decreased contractile kinetics and caused dilatative remodeling. Am. J. Physiol-Cell Physiol. 2012;303:C24–C32. doi: 10.1152/ajpcell.00026.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petchey LK, et al. Loss of Prox1 in striated muscle causes slow to fast skeletal muscle fiber conversion and dilated cardiomyopathy. Proc. Natl. Acad. Sci. U S A. 2014;111:9515–9520. doi: 10.1073/pnas.1406191111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP, Chen C-Z. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 13.van Rooij E, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 14.Chen J-F, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc. Natl. Acad. Sci. U S A. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu N, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callis TE, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Rooij E, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heidersbach A, et al. microRNA-1 regulates sarcomere formation and suppresses smooth muscle gene expression in the mammalian heart. Elife. 2013;2:e01323. doi: 10.7554/eLife.01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 21.Morton SU, et al. microRNA-138 modulates cardiac patterning during embryonic development. Proc. Natl. Acad. Sci. U S A. 2008;105:17830–17835. doi: 10.1073/pnas.0804673105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shieh JT, Huang Y, Gilmore J, Srivastava D. Elevated miR-499 levels blunt the cardiac stress response. PLoS ONE. 2011;6:e19481. doi: 10.1371/journal.pone.0019481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang ZP, et al. MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ. Res. 2013;112:1234–1243. doi: 10.1161/CIRCRESAHA.112.300682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seok HY, et al. Loss of MicroRNA-155 protects the heart from pathological cardiac hypertrophy. Circ. Res. 2014;114:1585–1595. doi: 10.1161/CIRCRESAHA.114.303784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, et al. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ. Res. 2013;112:1557–1566. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatignol A BC, Jeang KT. Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsI kinase and Drosophila staufen. Mol. Cell Biol. 1993;13:2193–2202. doi: 10.1128/mcb.13.4.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gatignol A, Buckler-White A, Berkhout B, Jeang K-T. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 28.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haase AD, et al. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukunaga R, et al. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012;151:533–546. doi: 10.1016/j.cell.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee HY, Doudna JA. TRBP alters human precursor microRNA processing in vitro. RNA. 2012;18:2012–2019. doi: 10.1261/rna.035501.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y, et al. Deletion of Human tarbp2 Reveals Cellular MicroRNA Targets and Cell-Cycle Function of TRBP. Cell Rep. 2014;9:1061–1074. doi: 10.1016/j.celrep.2014.09.039. [DOI] [PubMed] [Google Scholar]
- 33.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong J, Peters AH, Lee K, Braun RE. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat. Genet. 1999;22:171–174. doi: 10.1038/9684. [DOI] [PubMed] [Google Scholar]
- 35.Jiao K, et al. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Creemers EE, Wilde AA, Pinto YM. Heart failure: advances through genomics. Nat. Rev. Genet. 2011;12:357–362. doi: 10.1038/nrg2983. [DOI] [PubMed] [Google Scholar]
- 37.van Bilsen M, Chien KR. Growth and hypertrophy of the heart: towards an understanding of cardiac specific and inducible gene expression. Cardiovasc. Res. 1993;27:1140–1149. doi: 10.1093/cvr/27.7.1140. [DOI] [PubMed] [Google Scholar]
- 38.Jiang J, Wakimoto H, Seidman JG, Seidman CE. Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science. 2013;342:111–114. doi: 10.1126/science.1236921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Z, et al. Cardiac-specific YAP activation improves cardiac function and survival in an experimental murine MI model. Circ. Res. 2014;115:354–363. doi: 10.1161/CIRCRESAHA.115.303632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin CH, et al. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell. 2002;110:725–735. doi: 10.1016/s0092-8674(02)00933-9. [DOI] [PubMed] [Google Scholar]
- 41.Trivedi CM, Cappola TP, Margulies KB, Epstein JA. Homeodomain Only Protein X. is down-regulated in human heart failure. J. Mol.. Cell. Cardiol. 2011;50:1056–1058. doi: 10.1016/j.yjmcc.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trivedi CM, et al. Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation. Dev. Cell. 2010;19:450–459. doi: 10.1016/j.devcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen F, et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110:713–723. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- 44.An CI, Dong Y, Hagiwara N. Genome-wide mapping of Sox6 binding sites in skeletal muscle reveals both direct and indirect regulation of muscle terminal differentiation by Sox6. BMC Dev. Biol. 2011;11:59. doi: 10.1186/1471-213X-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quiat D, et al. Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor Sox6. Proc Natl Acad Sci U S A. 2011;108:10196–10201. doi: 10.1073/pnas.1107413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee LW, et al. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA. 2010;16:2170–2180. doi: 10.1261/rna.2225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matkovich SJ, Hu Y, Dorn GW. Regulation of cardiac microRNAs by cardiac microRNAs. Circ. Res. 2013;113:62–71. doi: 10.1161/CIRCRESAHA.113.300975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Vito C, et al. A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer Cell. 2012;21:807–821. doi: 10.1016/j.ccr.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 49.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 50.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.