Abstract
Trimethyllysine 72 (tmK72) has been suggested to play a role in sterically constraining the heme crevice dynamics of yeast iso-1-cytochrome c mediated by the Ω-loop D cooperative substructure (residues 70 to 85). A tmK72A mutation causes a gain in peroxidase activity, a function of cytochrome c that is important early in apoptosis. More than one higher energy state is accessible for the Ω-loop D substructure via tier 0 dynamics. Two of these are alkaline conformers mediated by Lys73 and Lys79. In the current work, the effect of the tmK72A mutation on the thermodynamic and kinetic properties of wild type iso-1-cytochrome c (yWT versus WT*) and on variants carrying a K73H mutation (yWT/K73H versus WT*/K73H) is studied. Whereas the tmK72A mutation confers increased peroxidase activity in wild type yeast iso-1-cytochrome c and increased dynamics for formation of a previously studied His79-heme alkaline conformer, the tmK72A mutation speeds return of the His73-heme alkaline conformer to the native state through destabilization of the His73-heme alkaline conformer relative to the native conformer. These opposing behaviors demonstrate that the response of the dynamics of a protein substructure to mutation depends on the nature of the perturbation to the substructure. For a protein substructure which mediates more than one function of a protein through multiple non-native structures, a mutation could change the partitioning between these functions. The current results suggest that the tier 0 dynamics of Ω-loop D that mediates peroxidase activity has similarities to the tier 0 dynamics required to form the His79-heme alkaline conformer.
Keywords: Cooperative substructure dynamics, Cytochrome c, Alkaline conformational transition, Conformationally-gated electron transfer
Introduction
Tier 0 dynamics, which involves interconverison between conformers separated by a thermal barrier, is particularly important in protein function, often controlling access to functionally active conformers [1]. A detailed understanding of the structural factors that control tier 0 dynamics and thus the function of proteins is essential. Cytochrome c (Cytc) has long been a protein of interest due to both its function in electron transport [2] and its role in apoptosis [3]. Of particular interest with regard to these functions are the heme crevice dynamics, which can be studied with the alkaline conformational transition of Cytc [4]. The alkaline conformational transition is an example of tier 0 dynamics because the barrier between the native and alkaline states is significantly larger than ambient thermal energy [1, 5].
Here, we use yeast iso-1-cytochrome c (iso-1-Cytc) as a vehicle to probe the structural factors controlling protein dynamics. Residues 70-85 of iso-1-Cytc comprise the heme crevice loop, Ω-loop D (Fig. 1), which lies across one face of the heme. These residues are highly conserved across Cytc from different species [2, 6]. Met80 is bound to the sixth coordination site of the heme in the native iso-1-Cytc conformer [7]. During the tier 0 alkaline conformational transition of the ferric state of Cytc, the Met80-heme ligand is replaced by lysine residues within Ω-loop D. These conformational changes are of particular biological interest because replacement of Met80-heme ligation with Lys-heme ligation can modulate electron transfer properties, possibly optimizing electron flow in the Electron Transport chain [4, 8-12]. The Met80 ligand must also leave the 6th coordination site of the heme in order for peroxidase activity to occur in the beginning stages of apoptosis [3]. Apoptosis can be induced in yeast, causing release of Cytc from the mitochondria. However, the proteins needed to form an apoptosome in the cytoplasm and induce the caspase cascade, which leads to cell death in higher eukaryotes, are missing [16]. Given the high degree of conservation of Ω-loop D in Cytc (13 of 16 residues are conserved from yeast to mammals, [2, 6]), studies on the dynamics of this loop in iso-1-Cytc provide a relevant model for heme crevice dynamics as it relates to peroxidase activity early in apoptosis, prior to release of cytochrome c from the mitochondria.
Fig. 1.
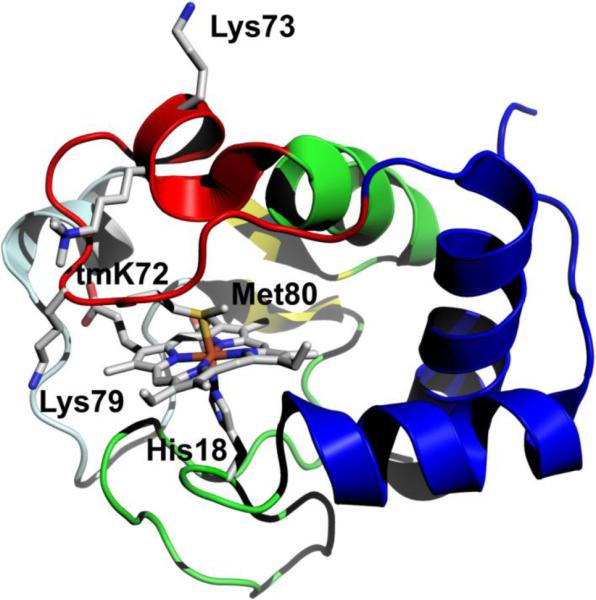
Structure of iso-1-Cytc (pdb code: 2YCC, [7]). The loop shown in red is Ω-loop D and provides Met80 as a heme ligand in the native state. Lysines 73 and 79 provide the alkaline state ligands for wild type iso-1-Cytc. Lysine 72 is trimethylated, tmK72, when iso-1-Cytc is expressed in its native host yeast. tmK72 can be seen to lie across the surface of the heme crevice loop, Ω-loop D. The cooperative substructures of Cytc, as defined by Englander and coworkers [13-15], from least to most stable are color-coded: pale cyan, red, yellow, green and blue
An increase in pH triggers ionization events leading to replacement of the native state Met80-heme ligand by either Lys73 or Lys79 in the alkaline conformational transition [17, 18]. The alkaline conformational transition of wild-type iso-1-Cytc expressed from yeast (yWT) is a monophasic conformational transition with either Lys73 or Lys79 replacing Met80 with similar, but not identical, kinetics and thermodynamics [18, 19]. Variants with a Lys73 to histidine mutation exhibit a biphasic alkaline transition where His73-heme ligation occurs near neutral pH, followed by Lys79-heme ligation above pH 8 [9, 20-22]. Not only does incorporating His into Ω-loop D enable the distinction between residues replacing Met80, but additionally the shift in the apparent pKa, pKapp, of the alkaline transition allows the conformational change to be monitored more effectively because it can be done near neutral pH [20, 23]. Thus, characterization of the dynamics of His-heme conformers allows for thorough investigation of the effect of point mutations on Cytc dynamics as it relates to function.
In yWT iso-1-Cytc, the lysine at residue 72 undergoes posttranslational modification to trimethyllysine (tmK). During expression from Escherichia coli Lys72 is not trimethylated. Lack of trimethylation allows Lys72 to become a heme ligand in the alkaline conformational transition [24]. In order to prevent Lys72-heme ligation during the alkaline conformational transition, residue 72 can be mutated to alanine [24]. Analysis of the native conformer of yeast iso-1-Cytc (pdb code: 2YCC), shows that loss of a lysine residue at position 72 also results in a loss of contacts between residue 72 and residues Thr78, Met80 and Ala81 on the opposite side of Ω-loop D (Fig. 1) [7]. We have recently reported the crystal structure of tmK72A iso-1-Cytc, which indicates that reducing the steric size of the residue at position 72 favors formation of an alternate conformer with hydroxide replacing Met80 as the heme ligand near pH 9 [25]. Also, we have previously reported increased heme crevice dynamics for the His79-heme alkaline conformational transition for a K79H variant with a tmK72A mutation compared to yWT/K79H iso-1-Cytc which contains tmK72 [26, 27]. For horse Cytc, spin-labeling of Lys72 demonstrates that this residue is relatively immobile suggesting that the residue at this position may be important for modulating the dynamics of Ω-loop D in Cytc across a broad range of species [28]. Furthermore, the tmK72A mutation leads to enhanced peroxidase activity for iso-1-Cytc [25]. Thus, the residue at position 72 may modulate the dynamics of Ω-loop D necessary for peroxidase activity.
Both residues 73 and 79 belong to Ω-loop D, which is the second least stable of five cooperative substructures (see Fig. 1), which have been identified in Cytc using native state H/D exchange methods [13-15]. Although both residues 73 and 79 belong to the same cooperative substructure of Cytc, it is unclear whether dynamics mediated by deformation of a cooperative substructure from two different points in the substructure will be affected in the same way by a point mutation. Greater understanding of the dynamics of cooperative substructures is important for understanding how tier 0 dynamics affect protein function, in general. In the case of Cytc, the effect of the tmK72A mutation on Ω-loop D mediated from position 73 versus 79 could provide greater insight into the nature of the dynamics required for peroxidase function at the onset of apoptosis. Previous work has shown that the mechanisms of the His79-mediated alkaline transition and the His73-mediated alkaline transition differ [9, 21-23, 26, 27, 29, 30]. Additionally, denaturant m-values, which correlate to the surface area exposed by partial or full unfolding of a protein [15, 31, 32], are smaller for formation of the His79-heme alkaline conformer than for the His73-heme alkaline conformer indicating less structural rearrangement during formation of the His79-heme conformer than for the His73-heme conformer [20, 22, 23, 33]. Similarly, the enthalpies of the alkaline transition mediated by Lys73 versus Lys79, also differ significantly [18]. These differences between both the kinetic and thermodynamic properties of alkaline transitions of Cytc mediated by two different sequence positions along the Ω-loop D substructure, suggest that the effects of the tmK72A mutation on the dynamics of a K73H variant of iso-1-Cytc could differ from those of a K79H variant.
In order to investigate the effect of the tmK72A mutation on the dynamics of the His73-mediated versus His79-mediated alkaline transition, we have characterized the thermodynamic and kinetic properties of the His73-mediated alkaline conformational of tmK72A/K73H iso-1-Cytc allowing comparison with previous work on K73H variants of iso-1-Cytc. The data show that the His73-mediated alkaline transition is less favorable in the presence of the tmK72A mutation and that the dynamics of return to the native state from the His73-heme alkaline conformer are increased. This result contrasts markedly with the increase in stability of the His79-heme alkaline conformer relative to the native state of iso-1-Cytc and the increase in the dynamics of formation of the His79-heme alkaline conformer upon introduction of the tmK72A mutation [26, 27].
Materials and methods
Preparation of iso-1-Cytc variants
Two variants of iso-1-Cytc were used in this work: WT*, which contains C102S and K72A mutations and WT*/K73H, which adds the K73H mutation to the WT* background. The C102S mutation prevents disulfide dimerization during physical studies. The K72A mutation is commonly used in Escherichia coli-expressed iso-1-Cytc to prevent Lys72, which is trimethylated in its native host, Saccharomyces cerevisiae, from acting as an alkaline state ligand [24]. Both proteins were expressed from the pRbs_BTR1 vector [34], which is a derivative of the pBTR1 vector [24, 35] with an optimized ribosomal binding site [36]. This vector co-expresses the iso-1-Cytc (CYC1) and the heme lyase (CYC3) genes from S. cerevisiae, so that heme insertion occurs efficiently in the cytoplasm of E. coli. The pRbs_BTR1 vector was originally prepared carrying the TM variant (H26N, H33N, H39Q, C102S) of iso-1-Cytc [34]. Conversion to the WT* variant was accomplished in four steps. A K72A mutation was first introduced into this vector using the unique restriction site elimination method [37] with single-stranded pRbs_BTR1 DNA as template. The K72A oligonucleotide and the selection primer EcoRV−AatII+ (see Table S1, eliminates a unique EcoRV site and creates a unique AatII site in pRbs_BTR1) were used to introduce the K72A mutation. The remaining mutations were introduced using double-stranded pRbs_ BTR1 DNA as template and the PCR-based QuikChange protocol (Agilent Technologies). The wild type histidines of iso-1-Cytc were reintroduced into the CYC1 gene in the order His26, His39 and His33 using the primer pairs, N26H and N26H-r, Q39H and Q39H-r, and N33H and N33H-r (Table S1), respectively. The pRbs_BTR1 vector containing the WT* variant was used as the template to prepare the WT*/K73H variant with the QuikChange protocol and the primer pair K73H and K73H-r (Table S1). Each mutation to the CYC1 gene was confirmed using dideoxy sequencing of the entire coding region of the gene. All sequencing was carried out at the Murdock DNA Sequencing Facility at the University of Montana.
Protein expression and purification
Protein was expressed in BL21(DE3) E. coli cells from the pRbs_BTR1 vector carrying either WT* or WT*/K73H iso-1-Cytc, as described previously [26, 34]. Protein was purified as previously reported [26, 38, 39]. Briefly, after breaking the cells with a French Pressure Cell (SLM Aminco), the lysate was cleared by centrifugation. Contaminants were precipitated by adjusting the lysate to 50% ammonium sulfate. After centrifugation, the supernatant was dialyzed against 12.5 mM sodium phosphate buffer, pH 7.2, 1 mM EDTA, 2 mM β-mercaptoethanol (β-ME). Iso-1-Cytc was batch absorbed to CM-Sepharose Fast Flow resin equilibrated with 50 mM sodium phosphate buffer, pH 7.2, 1 mM EDTA and 2 mM β-ME, then protein was eluted with a linear gradient of 0-0.8 M NaCl in 50 mM sodium phosphate buffer, pH 7.2, 1 mM EDTA and 2 mM β-ME. Aliquots of about 2 mg/mL protein were flash frozen in liquid nitrogen and stored at -80 °C. Thawed aliquots were purified by cation-exchange HPLC using an Agilent Technologies 1200 series HPLC with a BioRad UNO S6 Column (catalog no. 720-0023). After collection of the iso-1-Cytc peak, samples were concentrated by ultrafiltration before oxidation with K3[Fe(CN)6]. A G25 Sephadex column was used to separate the oxidized protein from the oxidizing agent.
The molecular weight of the WT* and WT*/K73H variants were measured to be 12,592.9 ± 0.1 g/mol (expected, 12,595.1 g/mol) and 12,603.3 ± 0.7 g/mol (expected, 12,604.1 g/mol) using a Bruker microflex MALDI-ToF mass spectrometer. Protein Calibration Standard I (Bruker Part No. 206355) was used to calibrate the mass spectrometer with an enhanced cubic calibration routine.
Global stability measurements by guanidine hydrochloride denaturation
Stability measurements by guanidine hydrochloride (GdnHCl) denaturation at 25 °C and pH 7.5 in 20 mM Tris and 40 mM NaCl buffer were monitored, as previously described [22], using an Applied Photophysics Chirascan circular dichroism (CD) spectrometer coupled to a Hamilton MICROLAB 500 Titrator. The free energy of unfolding in the absence of denaturant, ΔGu°′(H2O), and the m-value were extracted from nonlinear least squares fits (SigmaPlot v. 7.0) of the data employing a two-state model assuming a linear free energy relationship [40], as previously described [41]. In these fits, the native state baseline was assumed to be independent of GdnHCl concentration [41]. Parameters are the average and standard deviation of a minimum of three independent trials.
pH titrations to measure the alkaline conformational transition of iso-1-Cytc variants
pH titrations were carried out at 22 ± 1 °C to monitor the alkaline conformational transition of each variant. Titrations were followed at 695 nm, as previously reported [42], in 100 mM NaCl using a Beckman Coulter DU 800 UV/Vis spectrophotometer. 600 μL of a 2x stock solution containing 400 μM oxidized protein in 200 mM NaCl was prepared. 400 μL of the 2x stock solution was combined with 400 μL of MilliQ water and the solution was mixed with a 1000 μL pipetman. pH was adjusted by adding equal volumes of the 2x stock solution and either NaOH or HCl solutions of appropriate concentration and measured with a Denver Instrument UB-10 pH/mV meter using an Accumet semimicro calomel pH probe (Fischer Scientific Cat. No. 13-620-293). Baseline corrections were made by subtracting the absorbance at 750 nm, A750, from the absorbance at 695 nm, A695, yielding A695corr. Corrected molar extinction coefficients, ε695corr, were then calculated from A695corr using concentration evaluated from absorbance at 570 nm and 580 nm near pH 5 [23] and the known molar extinction coefficients for these wavelengths [43].
The monophasic ε695corr versus pH data from the WT* iso-1-Cytc variant were fit to Eq. 1,
| (1) |
using nonlinear least squares methods (SigmaPlot v. 7.0). Eq. 1 is a modified form of the Henderson-Hasselbalch equation, which allows the numbers of protons, n, linked to the conformational change to be evaluated from the fit [20]. In Eq. 1, εN is the corrected extinction coefficient of the native state Met80-heme conformer at 695 nm, εalk is the corrected extinction coefficient of either the Lys73- or Lys79-heme bound alkaline state conformer at 695 nm, and pKapp is the apparent acid dissociation constant for the alkaline conformational transition.
The biphasic ε695corr versus pH data from the WT*/K73H variant were fit to a model (Eq. 2) which includes the ionization triggers for formation of both the His73-heme and Lys79-heme
| (2) |
conformers [20]. In Eq. 2, pKa(His) and pKa(Lys) are the acid dissociation constants for the ionizing trigger groups corresponding to population of the His73-heme and Lys79-heme conformers, respectively. pKC1(His) and pKC2(Lys) are the equilibrium constants corresponding to replacement of the Met80-heme ligand by the fully deprotonated His73 and Lys79 ligands, respectively. pKa(Lys) was set to 10.8 as previously determined for the conformational change to the Lys79-heme alkaline state conformer [18]. Reported parameters from fits to Eqs. 1 and 2 are the average and standard deviation of a minimum of three independent trials.
pH jump stopped-flow kinetic experiments
All stopped-flow mixing experiments were carried out at 25 °C using an Applied Photophysics SX20 stopped-flow spectrometer, as previously reported [23]. Both long (50 – 350 s) and short (1 s) time scale data acquisitions were monitored at 406 nm, collecting 5000 points logarithmically for each. Upward pH jump mixing experiments began with 20 μM protein in 0.1 M NaCl at pH 5 followed by 1:1 mixing with 20 mM buffer in 0.1 M NaCl to achieve the desired pH. Upward pH jump experiments were carried out in steps of 0.25 pH units over the pH range of 6 – 10 for WT*/K73H. Downward pH jump experiments were executed in the same way but began at pH 8.05 and ended at pH values ranging from 5.0 – 6.5 in steps of 0.25 pH units. Buffers used were as follows: acetic acid (pH 5-5.25), MES (pH 5.5-6.5), NaH2PO4 (pH 6.75-7.5), Tris (pH 7.75-8.75), and H3BO3 (pH 9-10). In all cases the effluent was collected and the actual pH after mixing was determined with a Denver Instrument UB-10 pH/mV meter using an Accumet semimicro calomel pH probe. A minimum of 5 and 8 trials were collected for long and short time scale data acquisitions, respectively, for each pH jump experiment.
Heme reduction by hexaammineruthenium(II) chloride followed by stopped-flow
Conformationally-gated electron transfer (gated ET) experiments were performed by following reduction of oxidized WT*/K73H Cytc at 550 nm, a wavelength that is very sensitive to the redox state of Cytc, with an Applied Photophysics SX20 stopped-flow spectrometer using previously established methods [9, 23, 29, 44]. The reducing reagent, hexaammineruthenium(II) chloride (a6Ru2+), was prepared by reduction of hexaammineruthenium(III) chloride (Strem Chemicals) with zinc [45]. IR spectra confirmed formation of a6Ru2+ [46] and the product was stored under argon at -20 °C. Immediately before use, a6Ru2+ was dissolved under an argon atmosphere in argon-purged 100 mM NaCl and 10 mM buffer: acetic acid (pH 5), MES (pH 6), NaH2PO4 (pH 7), Tris (pH 8), and H3BO3 (pH 9). The concentration of a6Ru2+ was determined spectrophotometrically using a Beckman Coulter DU 800 UV/Vis Spectrophotometer with extinction coefficients at 390 nm of 35 M−1cm−1 [47] and at 400 nm of 30 M−1cm−1 [48]. Absorbance at 550 nm was used to correct for variation in background absorbance. The concentration of a6Ru2+ was evaluated immediately prior to and following stopped-flow mixing. The average and standard deviation of these two measurements is reported as the a6Ru2+ concentration. At the lowest a6Ru2+ concentration, absorbance at 275 nm was also used to evaluate concentration using ε275 of 632 M−1cm−1, which is the average of the reported extinction coefficients of 624 M−1cm−1 [48] and 640 M−1cm−1 [47]. Iso-1-Cytc solutions were also prepared in argon-purged buffers. Gas tight syringes were used to transfer iso-1-Cytc and a6Ru2+ solutions to the stopped-flow and UV-Vis instruments. Argon-purged 10 mM buffer in 100 mM NaCl was used to flush the stopped-flow unit prior to use. Heme reduction was followed at 25 °C via 1:1 stopped-flow mixing. Final protein concentration after mixing was 5 μM and a6Ru2+ concentrations were approximately 0.6, 1.25, 2.5, 5, and 10 mM.
Kinetic traces were collected logarithmically with 5000 data points for time periods of 1 to 300 seconds. Data were fit to the appropriate exponential equation using SigmaPlot (version 7.0). For pH 7 and above, a quadruple exponential function was used to fit the data as two slow phases are present under these conditions (Fig. S5). Below pH 7, these slow phases are absent and the data are best fit to a two exponential function (Fig. S5). To accurately capture the slow phases, data were collected on a 150 to 300 second timescale for pH 7, 8 and 9. As the slower phases were absent at pH 5 and 6, long timescale runs were not necessary and data were collected on a 1 to 5 second time scale.
At pH 6-9, one second data sets were also collected using drive pressure holds to reduce recoil noise from the drive syringe, which otherwise can cause artifacts on a timescale similar to the two faster phases. To help constrain the upper baseline of the pH 7 to 9 short timescale data sets, the data were fit to a triple exponential equation and the slow phase rate constant was set equal to kobs,4 from the longer timescale fits. kobs,4 was used over kobs,3 because the associated amplitude was much larger for kobs,4 than for kobs,3. Typically, 5 and 8 trials were collected for long and short time scale data acquisitions, respectively, for each gated ET experiment.
Results
Global stability of E. coli-expressed iso-1-Cytc variants
The global stabilities of the WT* and WT*/K73H iso-1-Cytc variants were determined by GdnHCl denaturation at 25 °C monitored by CD spectroscopy. Denaturation curves of WT* and WT*/K73H variants in Fig. 2 show that the midpoint GdnHCl concentration for unfolding, Cm, is similar for both variants. The unfolding transition of WT*/K73H is also broader than that of WT* (Fig. 2). The K73H mutation causes a decrease of about 1 kcal mol−1 M−1 in the m-value (rate of change of ΔGu with respect to GdnHCl concentration), as seen in Table 1. Previously we have demonstrated that GdnHCl unfolding of K73H variants progresses from a partially unfolded His73-heme conformer at pH 7.5, which is responsible for the decreased m-value [20, 22, 33]. In the case of WT*/K73H, the free energy of unfolding in the absence of denaturant, ΔGu°′(H2O), is about 25% lower than that of WT* iso-1-Cytc (Table 1). Comparison with the yeast expressed variants, yWT and yWT/K73H, show that the effect of the tmK72A mutation on global stability is modest [33, 49] (Table 1).
Fig. 2.
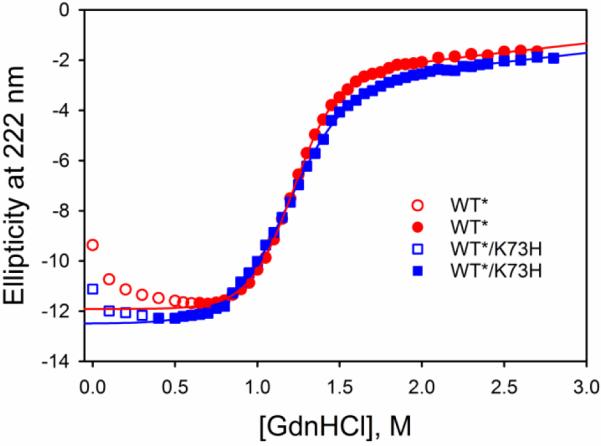
GdnHCl denaturation curves for the WT* (red circles) and WT*/K73H (blue squares) variants of iso-1-Cytc. Unfolding was monitored at 222 nm as GdnHCl was titrated into iso-1-Cytc buffered with 40 mM NaCl, 20 mM Tris buffer, pH 7.5 at 25 °C. Data in open symbols were not included in the fit to a two-state model (solid curves), described in the Materials and methods.
Table 1.
Thermodynamic parameters for iso-1-Cytc variants obtained by GdnHCl denaturation monitored by CD at 25 °C and pH 7.5.a
Alkaline conformational transition of WT* and WT*/K73H variants of iso-1-Cytc
Local unfolding of iso-1-Cytc variants at alkaline pH was monitored at 695 nm, an absorbance band which reports on the presence of the native Met80-heme ligand [2, 50, 51]. WT* iso-1-Cytc has a monophasic alkaline conformational transition (Fig. 3). The pKapp for WT* was found to be 8.66 ± 0.01, comparable to the value previously reported for yWT of 8.6 ± 0.1 with either a C102T or a C102S mutation [18, 20]. The number of protons, n, involved in the conformational change, as determined from a fit of the data to Eq. 1 (Materials and methods), is near 1 (1.15 ± 0.02) for WT* iso-1-Cytc, as expected.
Fig. 3.

ε695corr vs pH data for the WT* (red circles) and WT*/K73H (blue squares) variants of iso-1-Cytc, which monitor the alkaline conformational transition of iso-1-Cytc. WT* iso-1-Cytc displays a monophasic transition (solid red curve is a fit to Eq. 1 in Materials and methods) and WT*/K73H shows a biphasic transition (solid blue curve is a fit to Eq. 2 in Materials and methods). Data were acquired in 100 mM NaCl at 22 ± 1 °C.
WT*/K73H iso-1-Cytc has a biphasic alkaline conformational transition (Fig. 3). Between pH 6 and 8 a His73-heme ligated conformer populates followed by a Lys79-heme ligated state between pH 8 and 10. Thus, the K73H mutation allows the conformational changes mediated by the residues at positions 73 and 79 to be distinguished readily. WT*/K73H pH titration data were fit using Eq. 2 (Materials and methods). Within error, the pKa(His) values, which are consistent with the ionization of His73, are similar for both WT*/K73H and yWT/K73H (see Table 2). The conformational equilibrium constant for the alkaline conformational transition mediated by His73, pKC1(His), is more positive for WT*/K73H than for yWT/K73H [20], indicating that the alkaline conformer is less favored when tmK72 is replaced by Ala (Table 2).
Table 2.
Thermodynamic parameters for the alkaline transition of iso-1-Cytc variants obtained by pH titrations at 695 nm.a
| Variant | pKC1(His) | pKC2(Lys) | pKa(His) |
|---|---|---|---|
| WT*/K73H | 0.67 ± 0.05 | −2.04 ± 0.13 | 6.72 ± 0.10 |
| yWT/K73Hb | 0.28 ± 0.01 | −2.18 ± 0.11 | 6.60 ± 0.06 |
| WT*/K79Hc | −1.04 ± 0.04 (−1.06 ± 0.05)d | −2.8 ± 0.2 | 6.35 ± 0.04 |
| yWT/K79He | −0.99 ± 0.07 (−0.91 ± 0.03)d | −3.25 ± 0.24 | 6.62 ± 0.08 |
Parameters are the average and standard deviation of a minimum of three trials
Parameters are from ref. [20]
Parameters are from ref. [27]. To fit equilibrium pH titration data more reliably, amplitude data from gated ET experiments were used to estimate the extinction coefficient of the native conformer
Amplitude data from gated ET experiments at pH 7.5 were used to estimate pKC1(His) [27]
The extinction coefficient at 695 nm for fully native yWT/K79H was estimated using amplitudes from gated ET data reported in ref. [27] for yWT/K79H and used to refit equilibrium pH titration data for the yWT/K79H variant from ref. [23], as described in the Electronic supplementary material
Dynamics of the His73-mediated alkaline transition by pH jump methods
In order to elucidate the kinetics of the alkaline transition of the WT*/K73H iso-1-Cytc variant, stopped-flow pH jump methods were employed. Both upward and downward pH jumps were monitored at 406 nm to follow heme-ligand changes produced from Ω-loop D rearrangement. The kinetic data are consistent with three kinetic phases below pH 8 (Tables S2-S6 and Fig. S1). Above approximately pH 8, a fourth kinetic phase becomes evident (Table S3 and Fig. S1). The fast kinetic phase (Fig. S1, observed rate constant, kobs,1) occurs on a 100 millisecond timescale. The amplitude of the fast phase increases from pH 6 to pH 8, as expected for the His73-heme alkaline conformer based on thermodynamic data, which shows (see Fig. 3) that equilibrium population of the His73-heme alkaline conformer occurs over this pH range. We also see an increase in amplitude from pH 9 to pH 10 (Table S2).
Near pH 8 and above, an intermediate 1 s time scale phase is observed (Fig. S2, observed rate constant, kobs,2). Within error, kobs,2 is invariant with pH. Although the amplitude is low, an increase in amplitude between pH 8 and 10 is evident. A similar low amplitude intermediate phase from stopped-flow pH jump experiments that only populates above pH 7.3 has previously been reported for a K79H variant carrying the tmK72A mutation [26]. Stopped-flow conformationally-gated electron transfer (gated ET) experiments on two iso-1-Cytc variants expressed from yeast (i.e., tmK72), AcH73 (this variant has H26N, H33N and H39Q and K73H mutations as well as two mutations near the N-terminus which lead to N-terminal acetylation in vivo [9]) and K73H/K79A [29], exhibit the presence of an intermediate phase from pH 5 to 9 with low amplitude and observed rate constants of a comparable magnitude to values reported here for kobs,2. This phase has previously been attributed to a conformational change involving an acid state below pH 7, or a high-spin species above pH 7 [9, 26, 29].
Two slow phases, on a 10 – 100 s timescale are observed. Based on previous work on K73H variants of iso-1-Cytc, these phases likely correspond to formation of a Lys79-heme ligated conformer as well as isomerization of proline in the His73-heme alkaline conformer [29, 30, 42]. The observed rate constant for the first slow phase, kobs,3 (Fig. S3), ranges from 0.1 to 0.2 s−1, consistent with the rate of proline isomerization reported for yeast-expressed K73H/K79A iso-1-Cytc (i.e., tmK72) in the same pH regime [29, 42]. The amplitude of this phase shows a modest increase from pH 6 to 8, consistent with the slow phase associated with proline isomerization linked to the His73-heme alkaline transition observed with K73H/K79A iso-1-Cytc [42].
The slowest kinetic phase (Fig. S4) yields an observed rate constant, kobs,4, near 0.03 s−1 from pH 5 to 8. Above pH 8, kobs,4 begins to increase. Corresponding amplitude data for this slow phase also begins to increase near pH 8. The increase in amplitude and observed rate constant for this phase is consistent with thermodynamic data which show that the Lys79-heme ligated conformer populates from pH 8 to 10 (Fig. 3).
Direct measurement of microscopic rate constants for the alkaline transition by gated ET
Gated ET methods were used to directly measure the microscopic rate constants corresponding to Ω-loop D opening and replacement of the native Met80-heme ligand with either histidine or lysine. As previously described [4, 9, 21, 27, 29, 44, 52], oxidized protein with the Met80-heme ligand will undergo direct reduction by hexammineruthenium(II), a6Ru2+ (Fig. 4), whereas the His73-heme alkaline conformer is unable to undergo reduction due to a lower reduction potential [53]. Thus, a conformational change must occur to achieve the native Met80-heme ligated conformer before reduction can occur (Path B in Fig. 4). If the conformational change occurs on a much slower timescale than direct ET, a conformational ET gate exists.
Fig. 4.
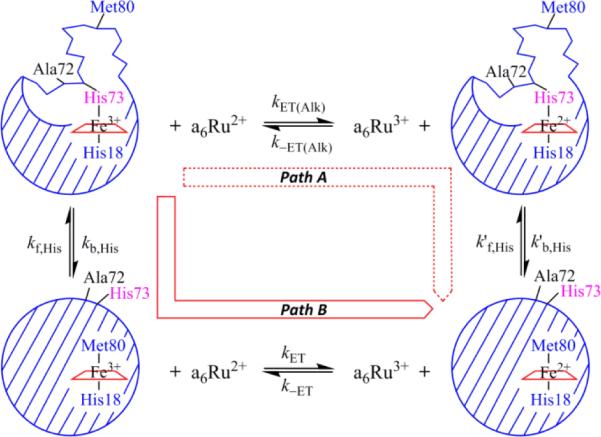
Gated ET square scheme for the WT*/K73H variant showing paths A and B.
Short time scale data acquisitions at pH 5 and 6 were fit to a two-exponential equation (Fig. S5). These fits are consistent with equilibrium population of both the native Met80-heme ligated conformer and the His73-heme ligated alkaline conformer at these pH values, as illustrated in Fig. 4. Above pH 7, a quadruple exponential equation was needed to fit the data (Fig. S5) indicating the presence of additional conformers with alternate heme ligands at equilibrium in solution that must convert to the native Met80-heme conformer before being reduced by a6Ru2+. Amplitude and rate constant data for each phase are given in Tables S7-S10. To distinguish them from the observed rate constants from pH jump experiments, we designate the observed rate constants from the gated ET experiments, kgET1, kgET2, kgET3, and kgET4, from fastest to slowest kinetic phase, and the associated amplitudes as, AgET1, AgET2, AgET3 and AgET4. Amplitude data from gated ET experiments are plotted against pH in Fig. 5. When kET[a6Ru2+] is much greater than kf,His73 (see Fig. 4, or kf,L's, rate constants for formation of conformers with other ligands, L, replacing Met80), individual amplitudes are directly proportional to the population of a given iso-1-Cytc conformer present at each pH because reduction is fast compared to the rate of the conformational change that returns the alternatively ligated conformer to the native state. Values of amplitudes from reduction of WT*/K73H iso-1-Cytc with a6Ru2+ concentrations between 2 and 4 mM were optimal for this purpose. At higher a6Ru2+ concentration, the direct electron transfer rates are fast enough relative to the dead time of our stopped-flow instrument that the associated amplitude may be underestimated (see Fig. S5). Conversely, at lower a6Ru2+ concentrations, kET[a6Ru2+] begins to approach kf,His73, such that Met80-ligated WT*/K73H can convert to the His73-heme bound conformer before it is reduced, leading to an apparent increase in the amplitude resulting from the His73-heme alkaline conformer (see Table S8).
Fig. 5.
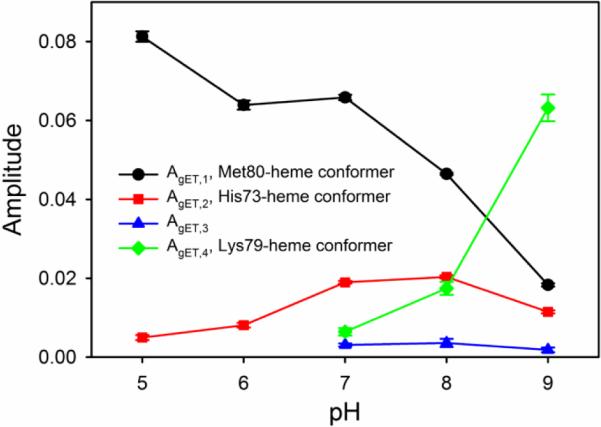
Amplitude data vs pH from gated ET experiments on WT*/K73H. Amplitude values correspond to the change in absorbance at 550 nm. Concentrations of a6Ru2+ for AgET,1 (black circles and line) and AgET,2 (red squares and line) for pH 5, 6, 7, 8 and 9 were 2.8, 3.0, 3.7, 3.9 and 2.8 mM, respectively. AgET,1 and AgET,2 are both from short (1 to 5 s) data acquisitions, Concentrations of a6Ru2+ for AgET,3 (blue triangles and line) and AgET,4 (green diamonds and line) for pH 7, 8 and 9 were 1.9, 2.24 and 1.78 mM, respectively. AgET,3 and AgET,4 are both from long timescale (150 to 300 s) data acquisitions. Error bars are the standard deviation of the average.
In Fig. 5, AgET,1 shows a decrease in magnitude as pH increases. AgET,1 decreases slowly from pH 5 to 7 and then more rapidly above pH 7. This behavior mimics the loss of absorbance at 695 nm due to Fe3+-Met80 ligation as pH is increased in the equilibrium pH titration data in Fig. 3. Thus, the fastest kinetic phase is assigned to WT*/K73H in the native conformer with Met80-heme ligation. AgET,2 initially increases as pH is increased above pH 5, maximally populates near pH 7-8, and then decreases. This behavior is consistent with the His73-heme conformer, which should increase in population as pH is increased reaching a maximum when His73 is fully deprotonated near pH 7.5. AgET,3 remains relatively invariant from pH 7-9. There is some uncertainty regarding the species corresponding to AgET,3, but it may correspond to iso-1-Cytc with a high spin heme as has been observed with other K73H variants of iso-1-Cytc [9, 26, 29]. The equilibrium population of this species is clearly small (Fig. 5), so, absorbance features near 650 nm, typical of a high spin heme, are not evident in equilibrium pH titration data (not shown). AgET,4, grows from a minor phase at pH 7 to the largest amplitude at pH 9. The increase in AgET,4 as AgET,1 and AgET,2 decrease is consistent with the behavior expected for the Lys79-heme conformer. Lys79 is expected to displace both Met80 and His73 as the heme ligand at high pH. The pH dependence of amplitude data from gated ET experiments observed here is similar to that previously reported for AcH73 iso-1-Cytc [9].
The magnitude of the rate constant for the fastest phase of the gated ET experiment, kgET,1, is strongly dependent on the concentration of a6Ru2+ consistent with direct ET with bimolecular rate constant, kET (Fig. 4), to the native state of WT*/K73H iso-1-Cytc (Table S7 and Fig. S6). The other rate constants from the gated ET experiments show little dependence on the concentration of a6Ru2+ (Fig. S7 and Tables S8 to S10). Thus, these rate constants reflect reduction of WT*/K73H iso-1-Cytc gated by a conformational change.
Discussion
Effects of the tmK72A mutation on the global and local stability of yeast iso-1-Cytc
The crystal structure of yeast iso-1-Cytc shows that tmK72 lies across Ω-loop D, potentially sterically hindering opening of the heme crevice (Fig. 1 and Fig. 8, below). Previously, we have shown that for variants of iso-1-Cytc carrying a K79H mutation, similar stability parameters are obtained from GdnHCl denaturation regardless of whether trimethyllysine or alanine is present at residue 72 [26]. Here, within error, we also report only a modest effect of the tmK72A mutation on the stability of a K73H variant of iso-1-Cytc (Table 1). Similarly, the tmK72A mutation does not affect the global stability of iso-1-Cytc (yWT versus WT* in Table 1)
Global unfolding experiments were performed at pH 7.5 where WT*/K73H partially populates the His73-heme conformer as is apparent from Figs. 3 and 5. The decrease in the denaturant m-value of WT*/K73H iso-1-Cytc relative to WT* iso-1-Cytc (see Table 1) is consistent with global unfolding monitored by CD occurring primarily from the partially unfolded His73-heme alkaline conformer as for other K73H variants of iso-1-Cytc [21, 33, 42].
pH titration studies demonstrate that pKapp is similar for WT* iso-1-Cytc and yWT iso-1-Cytc indicating that the tmK72A mutation has little effect on the Lys-mediated alkaline conformational transition. When the K73H mutation is introduced, within error pKC2(Lys), which corresponds to the Lys79-mediated alkaline transition, is also unaffected by the tmK72A mutation (Table 2). However, pKC1(His), which corresponds to the His73-mediated alkaline transition is about twice the positive magnitude for WT*/K73H compared to yWT/K73H (Table 2). Thus, the tmK72A mutation destabilizes the His73-heme alkaline conformer relative to the native conformer (ΔΔG of 0.53 ± 0.09 kcal/mol). By contrast, data in Table 2 for previously reported K79H variants of iso-1-Cytc, show that tmK72A mutation has little effect on the stability of the His79-heme alkaline conformer relative to the native conformer [26]. Interestingly, the tmK72A mutation makes pKC2(Lys) for the Lys73-heme alkaline conformer less favorable relative to the native conformer (ΔΔG of 0.45 ± 0.31 kcal/mol) for K79H variants. Thus, the tmK72A mutation appears to have less effect on the stability of alkaline conformers relative to the native state when the alkaline state ligand is from position 79, whereas it appears to destabilize the alkaline conformer relative to the native conformer by about 0.5 kcal/mol when the ligand is from position 73. Thus, the Ω-loop D substructure (Red in Fig. 1) clearly responds differently to the tmK72A mutation when deformed from position 73 versus position 79.
Histone binding proteins interact with trimethyllysine residues on histone tails using aromatic cage motifs, which are rich in aromatic residues (Tyr, Phe, Trp) and methionine [54, 55]. Trimethyllysine binding is stabilized by π-cation interactions. In the NMR structure of the Lys73-heme alkaline conformer [10], Met80 and Tyr67 are proximal to the side chain of position 72 providing a potential aromatic cage-like motif to bind tmK72 that could stabilize alkaline conformers with His/Lys73-heme ligation (see Fig. 8 below).
The tmK72A mutation affects the dynamics of the His73-mediated alkaline transition at both low and high pH
Fig. 6 compares the pH dependence of kobs,1 from pH jump experiments for the His73-heme alkaline conformational transition of WT*/K73H to previously reported data for yWT/K73H [30]. Below pH 6, kobs,1 remains constant for WT*/K73H whereas it increases for yWT/K73H [30]. Between pH 6 and 8, kobs,1 is identical for the two variants. From pH 8 to 10, kobs,1 initially increases more slowly with increasing pH for WT*/K73H compared to yWT/K73H. Thus, except for near neutral pH the tmK72A mutation significantly affects kobs,1. This behavior contrasts sharply with the effect of the tmK72A mutation on the dynamics of the His79-heme alkaline transition. In this case, the change in dynamics caused by the mutation is more pronounced near neutral pH and less pronounced near pH 5 and 10 [26, 27]. The mechanism of the alkaline transition is different when mediated by His73 than when mediated by His79. In general, three ionizable groups are required to fit kinetic data for the His73-mediated alkaline transition [9, 29, 30, 42], while only two are required to fit the kinetics of the His79-mediated alkaline transition [23, 26, 27]. Thus, it is perhaps not surprising that the two alkaline transitions respond differently to the tmK72A mutation.
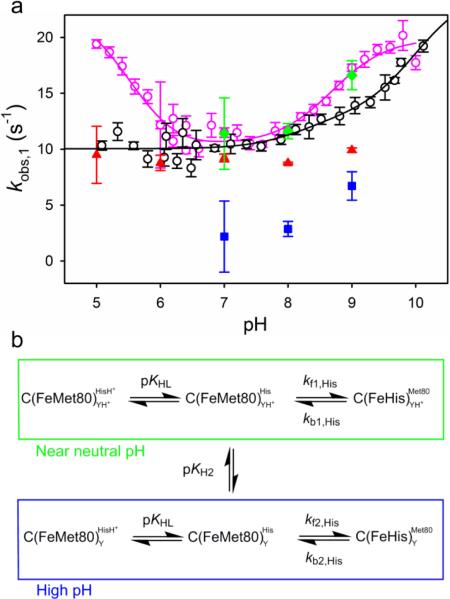
Fig. 6.
a Overlay of kobs,1 vs pH from pH jump experiments for WT*/K73H (open black circles) and yWT/K73H (open pink circles, data are from ref. [30]) iso-1-Cytc. All data were collected at 25 °C in 10 mM buffer containing 0.1 M NaCl. The black solid line is a fit of the WT*/K73H data to Eq. 3 from the Discussion and the pink solid line is a fit of the yWT/K73H data to Eq. 5 from ref. [30]. The microscopic rate constants, kb,His (red triangles) and kf,His (blue squares), extracted from gated ET experiments are shown for comparison. kobs,1 (green diamonds) determined from gated ET experiments is also shown, where kobs,1 is given by the sum of kf,His and kb,His. b Kinetic scheme for the His73-mediated alkaline transition involving two ionizations used for the derivation of Eqs. 3 and 4 [23, 30]. The YH+/Y ionization in the scheme corresponds to pKH2.
Normally the His73-mediated alkaline conformational transition is best fit to a model involving three ionizable groups. Since the ionizable group with a pKa near 5.5 (pKH1 in ref. [30]) is not evident for WT*/K73H (Fig. 6) we fit the kobs,1 versus pH data to a model involving two ionizable groups (Fig. 6b) [23, 30].
This model yields Eq. 3 for the pH dependence of kobs,1.
| (3) |
In Eq. 3, the methionine-heme to histidine-heme ligand conformational switch corresponds to forward rate constant, kf1,His, near neutral pH and the reverse rate constant for the conformational switch is kb1,His. KHL (or pKHL) is the ionization constant for the ligand (His73 in this case) which replaces Met80. As a result of the second ionization event (KH2 or pKH2), the rate constants change to kf2,His and kb2,His. In this model, kb1,His is equal to kobs,1 at low pH where the native conformer is fully populated. kb2,His cannot be determined directly from the kobs,1 versus pH data. From amplitude data, we can estimate the KC1(His) at pH values above pKH2, KC1(His)Hi, to be 0.94 (see Electronic supplementary material). With this value for KC1(His)Hi as a constraint, kf2,His and kb2,His can be obtained from the fit of Eq. 3 to the kobs,1 versus pH data for WT*/K73H iso-1-Cytc in Fig. 6a. The parameters from the fit of the kobs,1 versus pH data for WT*/K73H in Fig. 6a are summarized in Table 3. Compared to yWT/K73H the forward rate constant, kf1,His, is similar but, kb1,His is larger for WT*/K73H, consistent with the destabilizing effect of the tmK72A mutation on the His73-heme alkaline conformer observed in our thermodynamic data (Table 2).
Table 3.
Rate and ionization constants for the His73-mediated alkaline transition of K73H variants of iso-1-Cytc at 25 °C in 10 mM buffer, 0.1 M NaCl
| Variant |
||
|---|---|---|
| Parameter | WT*/K73H | yWT/K73Ha |
| k f1,His | 3 ± 2 | 3.5 ± 0.2 |
| k b1,His | 10.0 ± 0.2 | 7.0 ± 0.4 |
| k f2,His | 12 ± 3 | 6.6 ± 0.2 |
| k b2,His | 13 ± 9 | 13.2 ± 0.4 |
| pKHL | 8.2 ± 0.6 (6.8 ± 0.1)b | 6.4 ± 0.5 |
| pKH2 | 10.0 ± 0.6 (9.9 ± 0.2)b | 8.7 ± 0.2 |
Parameters are from ref. [30]
Values in brackets are from fits of the two ionization mechanism to amplitude versus pH data.
The behavior of the K73H variants is quite opposite of that of the K79H variants. kb1,His is unaffected by the tmK72A mutation with the K79H variants whereas kf1,His increases by 1.5- to 2.5-fold [26, 27]. It is also noteworthy that following the second ionization event for K79H variants, the magnitude of kobs,1 decreases [23, 26, 27], opposite of what occurs with K73H variants [9, 29, 30, 42].
The pKHL from fitting the kobs,1 versus pH data for WT*/K73H iso-1-Cytc in Fig. 6a has a higher value than the pKa(His) of about 6.7, reported in Table 2 from pH titration data. The amplitude versus pH data for the His73-heme alkaline transition shows two phases (Fig. 7). Thus, we can also fit the amplitude versus pH data to obtain values for pKHL and pKH2 using Eq. 4 [23, 30], which describes the behavior of the amplitude of the His73-heme alkaline transition
| (4) |
expected for the two ionization mechanism (Fig. 6b). In Eq. 4, CT is the amplitude if the conformational transition goes to completion. In fitting the data, the microscopic rate constants (kf1,His, kb1,His, kf2,His and kb2,His) from the fit of the kobs,1 versus pH data in Fig. 6a (Table 3) were used as constants in Eq. 4. The fit of the data yields a value for pKHL of 6.8 ± 0.1 which is in good agreement with the pKa(His) obtained from thermodynamic data (Table 2). This value for pKHL is likely more reliable than the one obtained from the kobs,1 versus pH data, given the size of errors bars and the scatter in the kobs,1 data below pH 7. Within error the value for pKH2 did not change whether fitting kobs,1 or amplitude versus pH data (Table 3). It is possible that a third ionization event may occur at a lower pH, however, within the error of the reported data points we are unable to reliably detect this ionization, which is clearly present for yWT/K73H iso-1-Cytc (Fig. 6). pKHL is similar for both WT*/K73H and yWT/K73H [30]. However, pKH2 increases by about 1 unit for WT*/K73H compared to yWT/K73H (Table 3). By contrast, the tmK72A mutation does not affect pKH2 for the His79-heme alkaline transition. We previously speculated that the outer heme propionate (also referred to as heme propionate-7 and heme propionate D), which is believed to have a pKa greater than 9 [2, 56, 57], could be responsible for the pKH2 ionization [23]. If our earlier speculation is correct, the tmK72A mutation increases the pKa of the outer heme propionate in the context of the K73H mutation, but not in the context of the K79H mutation [26]. In summary, our kinetic data also show that the effect of the tmK72A mutation depends on whether Ω-loop D is deformed from position 73 or 79.
Fig. 7.
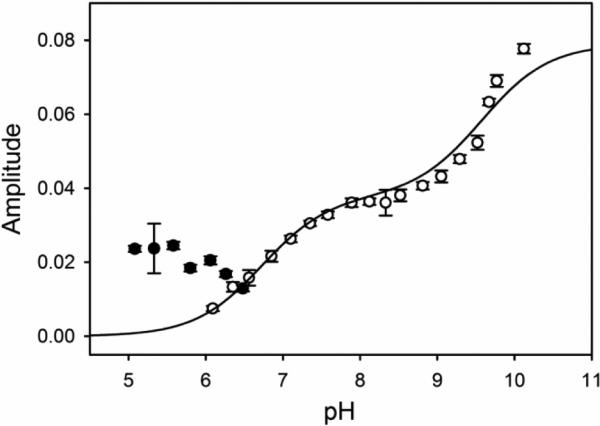
Amplitude versus pH for the fast phase (kobs,1) from pH jump stopped-flow experiments. Amplitudes correspond to the change in absorbance at 406 nm. Upward (open circles) pH jump amplitudes are fit to Eq. 4 in the Discussion, derived from the kinetic scheme in Fig. 6b [23, 30]. The fit yields CT equal to 0.164 ± 0.009. Downward pH jump amplitudes (filled black circles) are not used in the fit. Error bars on data points are the standard deviation of the average.
Determining forward and reverse rate constants of the alkaline conformational transition
The individual rate constants from the gated ET experiments were plotted against a6Ru2+ concentration. kgET,1, which corresponds to bimolecular ET from a6Ru2+ to the native state, shows a linear dependence on a6Ru2+ concentration (Fig. S6) as expected for a bimolecular reaction under pseudo-first-order conditions (see Fig. 4). Fitting the kgET,1 versus a6Ru2+ concentration data to Eq. 5, yields the bimolecular rate constant kET as the slope. In Eq. 5, kuni
| (5) |
represents a summation of all unimolecular rate constants (kf,His, see Fig. 4, and kf,L's for species corresponding to AgET,3 and AgET,4, see Fig. 5) that lead to the disappearance of the native conformer. The kET values as a function of pH are listed in Table 4 and plotted in Fig. S6. kET values decrease with increasing pH. A similar pH dependence for kET is observed for the AcH73 variant of iso-1-Cytc [9], as well as, for a yWT/K73H/K79A variant of iso-1-Cytc [29] both of which contain the native tmK72 residue. This observation suggests that the mechanism of direct ET is similar, regardless of whether Ω-loop D is sterically hindered by tmK72 or sterics have been relieved by the tmK72A mutation.
Table 4.
Rate constants obtained from reduction of oxidized WT*/K73H by hexaammineruthenium(II) chloride at 25 °C in 10 mM buffer, 0.1 M NaCl.a
| pH | kET (mM−1, s−1)b | kb,His (s−1)c | kf,His (s−1)d |
|---|---|---|---|
| 5 | 44 ± 2 | 9 ± 2 | - |
| 6 | 47 ± 1 | 8.8 ± 0.7 | - |
| 7 | 42.4 ± 0.5 | 9.2 ± 0.4 | 2 ± 3 |
| 8 | 34.6 ± 0.7 | 8.76 ± 0.08 | 2.8 ± 0.7 |
| 9 | 31.6 ± 0.2 | 9.9 ± 0.2 | 7 ± 1 |
Errors are standard errors reported by SigmaPlot unless otherwise noted
kET is obtained from fits to Eq. 5, as described in the text. The intercepts, kuni, of the fits to Eq. 5 were close to zero within error
At pH 5 and 6, kb,His73 is taken as the average value of kgET,2 across all concentrations of a6Ru2+. Error is the standard deviation of the average. At pH 7 to 9, kb,His is from fits to Eq. 6, as described in the text
From fits to Eq. 6, as described in the text
By contrast, kET for reduction of WT*/K79H increases as pH increases [27]. The relative proximity of His73 versus His79 to the heme may account for these differences. At higher pH, deprotonation of His79, which is close to the heme edge (see Fig. 1), will make it easier for a6Ru2+ to form a precursor complex [58], which has a6Ru2+ close to the heme of iso-1-Cytc. Thus, the observed increase in kET as pH increases is reasonable. His73 is located above Met80 far removed from the heme edge (see Fig. 1). Deprotonation of His 73 would thus increase the probability that a6Ru2+ would form a precursor complex with Cytc at a greater distance from the heme edge. At this location, ET is expected be less favorable, leading to the observed drop in kET at higher pH.
At all pH values, the magnitude of kgET,2, which corresponds to reduction of the His73-heme conformer, is independent of a6Ru2+ concentration at high concentrations of a6Ru2+. Evidently at high a6Ru2+ concentration, kET[a6Ru2+] is much greater than kf,His, such that the rate of reduction is controlled by the unimolecular conformational change back to the native state (i.e., kgET,2 is approximately equal to kb,His, see Eq 6 and Fig. 4). At pH 8 and 9, there is some decrease in kgET,2 at the lowest concentrations of a6Ru2+. Thus, it is possible to fit the dependence of kgET,2 on a6Ru2+ concentration to the full form of Eq. 6, which is derived assuming a steady-state approximation [59-61], so that both rate constants, kb,His and kf,His, associated with the
| (6) |
conformational transition can be obtained (Fig. S7). At pH 7 and below, no clear decrease in kgET,2 is observed at the lowest a6Ru2+ concentrations. The data at pH 7 were fit to Eq. 6, to obtain kb,His. The fit also provides kf,His, but the error in its magnitude is large (Table 4) and at best it provides an upper limit for kf,His at pH 7. At pH 5 and 6, the errors in the magnitudes of kgET,2 are larger, so, we have used the average of kgET,2 at all concentrations of a6Ru2+ as an estimate for the magnitude of kb,His. All values of kf,His and kb,His obtained from gated ET experiments are reported in Table 4.
In Fig. 6a, the microscopic rate constants, kf,His and kb,His, obtained from the gated ET experiments along with their sum, which should be equal to kobs,1 obtained from pH jump experiments, are plotted against kobs,1 for the His73-heme alkaline transition obtained from pH jump experiments. Across the pH regime studied, values for kb,His are invariant within error for WT*/K73H, except at pH 9 where kb,His appears to increase by about 10% (Table 4). For yWT/K73H and other variants with K73H mutations, an ionizable group with a pKa near 5.5 is observed (see Fig. 6 and refs. [9, 29]). If there is an ionization affecting the His73-heme alkaline transition in this pH regime, its effect on the magnitude of kb,His is within the error of our measurements. As the pH is increased, the rate constant associated with opening of the heme crevice and replacing the methionine-heme ligand with histidine, kf,His, increases. Below pH 7, kf,His is too small to evaluate. At pH 5 and 6, kb,His is within error of the magnitude of kobs,1 from pH jump experiments (Fig. 6a), indicating that kf,His is close to zero. At pH 7 and above, the sum of kf,His and kb,His matches kobs,1 from pH jump experiments reasonably well (Fig. 6a). At pH 8, where the His73-heme alkaline conformer is maximally populated (Fig. 5), the ratio of kf,His to kb,His should be close to KC1(His) obtained from equilibrium measurements. At pH 8, KC1(His) (obtained as kf,His divided by kb,His) equals 0.32 ± 0.08, which is close to KC1(His) of 0.21 ± 0.02 from equilibrium data for WT*/K73H (derived from pKC1(His) in Table 2). At pH 9, kf,His and kb,His yield 0.7 ± 0.1 for KC1(His). This value approaches our estimate of the increase from KC1(His) near 0.21 to KC1(His)Hi of about 0.94 due to the ionization of the group with pKH2 of 10 (Fig. 7 and Table 3). Interestingly, the pKH2 ionization also increases the stability of the His73-heme conformer relative to the native conformer for the AcH73 variant of iso-1-Cytc [29].
Effect of the tmK72A mutation on the free energy landscape of K73H variants of iso-1-Cytc
Fig. 8 shows the effect of the tmK72A mutation on the free energy landscape of the His73-mediated alkaline transition of iso-1-Cytc using thermodynamic and kinetic data presented here for WT*/K73H and reported previously for yWT/K73H [20, 30]. In Fig. 8, we have assumed that the tmK72A mutation does not affect the stability of the native state. This choice is partially dictated by the small decrease in the stability of the His73-heme conformer based on GdnHCl unfolding data in Table 1 (given that global unfolding of K73H variants proceeds primarily from the His73-heme conformer, ref. [20]), which is similar in magnitude to the effect of the tmK72A mutation on the relative stability of the native conformer versus the His73-heme alkaline conformer (Table 2). As discussed above, the NMR structure of the Lys73-heme alkaline conformer suggests that tmK72 could interact with an aromatic cage-like motif [54, 55], derived from Tyr67 and Met80 in the Lys/His73-heme alkaline conformer [10, 26]. Truncation of tmK72 to an Ala would eliminate this interaction [10] (see Fig. 8, lower right), supporting the contention that the increase in the positive magnitude of pKC1(His) (less favorable equilibrium) results primarily from a destabilization of the His73-heme conformer with little effect on the free energy of the native state (Fig. 8). The lesser effect of the tmK72A mutation on the transition state (TS, see Fig. 8) would be consistent with partial formation of the aromatic cage-like motif in the TS. Assigning effects of mutations to one ground state or another is always uncertain. The global stability of the yWT/K73H is sensitive to how the global unfolding data are fit and could be somewhat lower than for the WT*/K73H variant [62]. Also, the overlap between partial unfolding to the His73-heme alkaline conformer by GdnHCl and global unfolding is likely greater for the WT*/K73H variant than for the yWT/K73H variant because the His73-heme alkaline conformer is destabilized by the tmK72A mutation (Table 2). Thus, the estimation of the free energy of the His73-heme alkaline conformer for WT*/K73H iso-1-Cytc versus yWT/K73H iso-1-Cytc has some uncertainty. The discussion here is supported by structural data for the Lys73-heme alkaline conformer (Fig. 8, lower right) [10]. However, the factors affecting the stabilities of the two ground states as a result of the tmK72A mutation may be more complex. What is clear is that tmK72A mutation decreases the barrier from the His73-heme alkaline conformer to the TS and may cause a slight increase in the barrier from the native conformer to the TS (although the errors in kf,His for WT*/K73H and yWT/K73H are overlapping, see Fig. 8).
Fig. 8.

Effect of the tmK72A mutation on the free energy landscape of the His73-heme mediated alkaline conformational transition of iso-1-Cytc. yWT/K73H has tmK72 and WT*/K73H carries the tmK72A mutation. The range given for the relative stabilities of the native conformer and His73-heme alkaline conformer for WT*/K73H is based on kinetic (0.7 kcal/mol; using kf,His and kb,His at pH 8 in Table 4 to obtained KC1(His)) and thermodynamic data (0.9 kcal/mol, using pKC1(His) in Table 2). The range in the magnitude of KC1(His) produces a similar range for the relative stabilities of the His73-heme alkaline conformers of WT*/K73H and yWT/K73H. The change in the height of the barrier is calculated using the Eyring equation, yielding a decrease in the barrier for return to the native conformer by about 0.15 kcal/mol with the tmK72A mutation (kb,His increases from 7 to 8.8 s−1). The range in the ΔG of the TS for WT*/K73H versus yWT/K73H results from the range in the ΔG of the His73-heme conformers of WT*/K73H and yWT/K73H. Lower left: structure of native iso-1-Cytc (pdb code: 2YCC with Ω-loop D colored salmon shown as a space-filling model. Lower right: structure of the Lys73-heme alkaline conformer (pdb code: 1LMS) with Ω-loop D colored salmon. Met80, Tyr67, Pro71 and Ala/Lys72 are shown as space filling models. Ala72 in this structure has been converted to Lys using the mutate function of PyMol (added carbons are shown in gray).
Thus, the tmK72A mutation has very different effects on the free energy landscape of Ω-loop D with respect to formation of a His73-heme versus a His79-heme alkaline conformer. For the His79-heme alkaline transition, the tmK72A mutation decreases the barrier from the native conformer to the TS allowing faster formation of the His79-heme alkaline conformer [26, 27]. For the His73-heme alkaline transition, the tmK72A mutation decreases the barrier from the His73-heme alkaline conformer to the TS speeding formation of the native conformer. This observation has interesting general implications for the effects of mutations on the tier 0 dynamics of protein substructures involved in function. In particular, our results show that the dynamic accessibility of different higher energy states of a protein substructure can be affected differently by mutation, which could change the partitioning between different functions or modulate the specificity of a function. In the case of iso-1-Cytc, peroxidase activity is enhanced by the tmK72A mutation [25], suggesting that the tier 0 dynamics required to access the higher energy state of Ω-loop D that mediates peroxidase activity may be similar to the tier 0 dynamics required to access the His79-heme alkaline conformer but not those to access His73-heme alkaline conformer. Given the high conservation of Ω-loop D between yeast and mammals, it is possible that the residue at position 72 has similar effects on the tier 0 dynamics required for peroxidase activity early in apoptosis for mammalian Cytc.
Conclusion
The primary observation of the present work is that the effect of the tmK72A mutation on the His73-heme alkaline transition is entirely opposite from that observed for the His79-heme alkaline transition. Near neutral pH, this mutation destabilizes the His73-heme alkaline conformer relative to the native conformer, whereas it destabilizes the native conformer relative to the His79-heme alkaline conformer. This mutation also enhances the rate of formation of the His79-heme alkaline conformer near neutral pH while enhancing the rate of formation of the native conformer from the His73-heme alkaline conformer. Therefore, the effect of the tmK72A mutation on the dynamics of Ω-loop D clearly depends of the nature of the deformation of this substructure. Peroxidase activity assays on yWT versus WT* iso-1-Cytc show that kcat is 2-fold larger near pH 8 for WT* iso-1-Cytc, which carries the tmK72A mutation [25]. Given the high degree of conservation of Ω-loop D from yeast to mammals, and the current results on the effect of the tmK72A mutation on the dynamics of the His73-heme alkaline transition, our previous peroxidase activity results suggest that the dynamics of heme crevice opening required for peroxidase activity early in apoptosis may be similar to the dynamics of Ω-loop D which lead to formation of the His79-heme alkaline conformer.
Supplementary Material
Acknowledgments
This research was supported by NSF grants CHE-0910166 and CHE-1306903 to B.E.B. The Bruker microflex MALDI-ToF mass spectrometer was acquired with support from an NSF Major Research Instrumentation Grant CHE-1039814. B.E.B. also acknowledges support from CoBRE grant P20GM103546 funded by the NIGMS.
Abbreviations
- ET
electron transfer
- gated ET
Conformationally-gated electron transfer
- GdnHCl
guanidine hydrochloride
- iso-1-Cytc
iso-1-cytochrome c
- tmK
trimethyllysine
References
- 1.Henzler-Wildman K, Kern D. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 2.Moore GR, Pettigrew GW. Cytochromes c: Evolutionary, Structural and Physicochemical Aspects. Springer-Verlag; New York: 1990. [Google Scholar]
- 3.Kagan VE, Bayir HA, Belikova NA, Kapralov O, Tyurina YY, Tyurin VA, Jiang J, Stoyanovsky DA, Wipf P, Kochanek PM, Greenberger JS, Pitt B, Shvedova AA, Borisenko G. Free Radical Biol. Med. 2009;46:1439–1453. doi: 10.1016/j.freeradbiomed.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherney MM, Bowler BE. Coord. Chem. Rev. 2011;255:664–677. [Google Scholar]
- 5.Ansari A, Berendzen J, Bowne SF, Frauenfelder H, Iben IET, Sauke TB, Shyamsunder E, Young RD. Proc. Natl. Acad. Sci. U.S.A. 1985;82:5000–5004. doi: 10.1073/pnas.82.15.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banci L, Bertini I, Rosato A, Varani G. J. Biol. Inorg. Chem. 1999;4:824–837. doi: 10.1007/s007750050356. [DOI] [PubMed] [Google Scholar]
- 7.Berghuis AM, Brayer GD. J. Mol. Biol. 1992;223:959–976. doi: 10.1016/0022-2836(92)90255-i. [DOI] [PubMed] [Google Scholar]
- 8.Barker PD, Mauk AG. J. Am. Chem. Soc. 1992;114:3619–3624. [Google Scholar]
- 9.Bandi S, Bowler BE. Biochemistry. 2011;50:10027–10040. doi: 10.1021/bi201082h. [DOI] [PubMed] [Google Scholar]
- 10.Assfalg M, Bertini I, Dolfi A, Turano P, Mauk AG, Rosell FI, Gray HB. J. Am. Chem. Soc. 2003;125:2913–2922. doi: 10.1021/ja027180s. [DOI] [PubMed] [Google Scholar]
- 11.Döpner S, Hildebrandt P, Rosell FI, Mauk AG, von Walter M, Buse G, Soulimane T. Eur. J. Biochem. 1999;261:379–391. doi: 10.1046/j.1432-1327.1999.00249.x. [DOI] [PubMed] [Google Scholar]
- 12.Döpner S, Hildebrandt P, Rosell FI, Mauk AG. J. Am. Chem. Soc. 1998;120:11246–11255. [Google Scholar]
- 13.Krishna MM, Maity H, Rumbley JN, Lin Y, Englander SW. J. Mol. Biol. 2006;359:1410–1419. doi: 10.1016/j.jmb.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 14.Krishna MM, Lin Y, Rumbley JN, Englander SW. J. Mol. Biol. 2003;331:29–36. doi: 10.1016/s0022-2836(03)00697-1. [DOI] [PubMed] [Google Scholar]
- 15.Bai Y, Sosnick TR, Mayne L, Englander SW. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laun P, Buettner S, Rinnerthaler M, Burhans WC, Breitenbach M. In: Subcellular Biochemistry: Aging Research in Yeast. Breitenbach M, Jazwinski SM, Laun P, editors. Springer; Netherlands: 2012. pp. 207–232. [Google Scholar]
- 17.Ferrer JC, Guillemette JG, Bogumil R, Inglis SC, Smith M, Mauk AG. J. Am. Chem. Soc. 1993;115:7507–7508. [Google Scholar]
- 18.Rosell FI, Ferrer JC, Mauk AG. J. Am. Chem. Soc. 1998;120:11234–11245. [Google Scholar]
- 19.Blouin C, Guillemette JG, Wallace CJA. Biophys. J. 2001;81:2331–2338. doi: 10.1016/S0006-3495(01)75879-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson CJ, Bowler BE. Biochemistry. 2000;39:13584–13594. doi: 10.1021/bi0017778. [DOI] [PubMed] [Google Scholar]
- 21.Bandi S, Bowler BE. J. Am. Chem. Soc. 2008;130:7540–7541. doi: 10.1021/ja801941r. [DOI] [PubMed] [Google Scholar]
- 22.Kristinsson R, Bowler BE. Biochemistry. 2005;44:2349–2359. doi: 10.1021/bi048141r. [DOI] [PubMed] [Google Scholar]
- 23.Bandi S, Baddam S, Bowler BE. Biochemistry. 2007;46:10643–10654. doi: 10.1021/bi700992y. [DOI] [PubMed] [Google Scholar]
- 24.Pollock WB, Rosell FI, Twitchett MB, Dumont ME, Mauk AG. Biochemistry. 1998;37:6124–6131. doi: 10.1021/bi972188d. [DOI] [PubMed] [Google Scholar]
- 25.McClelland LJ, Mou T-C, Jeakins-Cooley ME, Sprang SR, Bowler BE. Proc. Natl. Acad. Sci. U.S.A. 2014;111:6648–6653. doi: 10.1073/pnas.1323828111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherney MM, Junior C, Bowler BE. Biochemistry. 2013;52:837–846. doi: 10.1021/bi301599g. [DOI] [PubMed] [Google Scholar]
- 27.Cherney MM, Junior CC, Bergquist BB, Bowler BE. J. Am. Chem. Soc. 2013;135:12772–12782. doi: 10.1021/ja405725f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kostrzewa A, Páli T, Wojciech F, Marsh D. Biochemistry. 2000;39:6066–6074. doi: 10.1021/bi992559l. [DOI] [PubMed] [Google Scholar]
- 29.Bandi S, Bowler BE. Biopolymers (Pept Sci) 2013;100:114–124. doi: 10.1002/bip.22164. [DOI] [PubMed] [Google Scholar]
- 30.Martinez RE, Bowler BE. J. Am. Chem. Soc. 2004;126:6751–6758. doi: 10.1021/ja0494454. [DOI] [PubMed] [Google Scholar]
- 31.Myers JK, Pace CN, Scholtz JM. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schellman JA. Biopolymers. 1978;17:1305–1322. doi: 10.1002/bip.1978.360170510. [DOI] [PubMed] [Google Scholar]
- 33.Godbole S, Dong A, Garbin K, Bowler BE. Biochemistry. 1997;36:119–126. doi: 10.1021/bi961915m. [DOI] [PubMed] [Google Scholar]
- 34.Duncan MG, Williams MD, Bowler BE. Protein Sci. 2009;18:1155–1164. doi: 10.1002/pro.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosell FI, Mauk AG. Biochemistry. 2002;41:7811–7818. doi: 10.1021/bi016060e. [DOI] [PubMed] [Google Scholar]
- 36.Rumbley JN, Hoang L, Englander SW. Biochemistry. 2002;41:13894–13901. doi: 10.1021/bi026543y. [DOI] [PubMed] [Google Scholar]
- 37.Deng WP, Nickoloff JA. Anal. Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 38.Redzic JS, Bowler BE. Biochemistry. 2005;44:2900–2908. doi: 10.1021/bi048218b. [DOI] [PubMed] [Google Scholar]
- 39.Wandschneider E, Hammack BN, Bowler BE. Biochemistry. 2003;42:10659–10666. doi: 10.1021/bi034958t. [DOI] [PubMed] [Google Scholar]
- 40.Pace CN. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 41.Hammack BN, Smith CR, Bowler BE. J. Mol. Biol. 2001;311:1091–1104. doi: 10.1006/jmbi.2001.4909. [DOI] [PubMed] [Google Scholar]
- 42.Baddam S, Bowler BE. Biochemistry. 2005;44:14956–14968. doi: 10.1021/bi0515873. [DOI] [PubMed] [Google Scholar]
- 43.Margoliash E, Frohwirt N. Biochem. J. 1959;71:570–572. doi: 10.1042/bj0710570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baddam S, Bowler BE. J. Am. Chem. Soc. 2005;127:9702–9703. doi: 10.1021/ja0527368. [DOI] [PubMed] [Google Scholar]
- 45.Fergusson JE, Love JL. Inorganic Synthesis. 1972;13:208–213. [Google Scholar]
- 46.Allen AD, Senoff CV. Can. J. Chem. 1967;45:1337–1341. [Google Scholar]
- 47.Matsubara T, Ford PC. Inorg. Chem. 1978;17:1747–1752. [Google Scholar]
- 48.Meyer TJ, Taube H. Inorg. Chem. 1968;7:2369–2370. [Google Scholar]
- 49.Godbole S, Hammack B, Bowler BE. J. Mol. Biol. 2000;296:217–228. doi: 10.1006/jmbi.1999.3454. [DOI] [PubMed] [Google Scholar]
- 50.Eaton WA, Hochstrasser RM. J. Chem. Phys. 1967;46:2533–2539. doi: 10.1063/1.1841081. [DOI] [PubMed] [Google Scholar]
- 51.Dragomir I, Hagarman A, Wallace C, Schweitzer-Stenner R. Biophys. J. 2007;92:989–998. doi: 10.1529/biophysj.106.095976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baddam S, Bowler BE. Inorg. Chem. 2006;45:6338–6346. doi: 10.1021/ic0603712. [DOI] [PubMed] [Google Scholar]
- 53.Bortolotti CA, Paltrinieri L, Monari S, Ranieri A, Borsari M, Battistuzzi G, Sola M. Chem Sci. 2012;3:807–810. [Google Scholar]
- 54.Sanchez R, Zhou M-M. Trends Biochem. Sci. 2011;36:364–372. doi: 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daze KD, Hof F. Acc. Chem. Res. 2013;46:937–945. doi: 10.1021/ar300072g. [DOI] [PubMed] [Google Scholar]
- 56.Hartshorn RT, Moore GR. Biochem. J. 1989;258:595–598. doi: 10.1042/bj2580595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moore GR. FEBS Lett. 1983;161:171–175. doi: 10.1016/0014-5793(83)81001-1. [DOI] [PubMed] [Google Scholar]
- 58.Cusanovich MA, Tollin G. In: Cytochrome c: A Multidisciplinary Approach. Scott RA, Mauk AG, editors. University Science Books; Sausalito, CA: 1996. pp. 489–513. [Google Scholar]
- 59.Wijetunge P, Kulatilleke CP, Dressel LT, Heeg MJ, Ochrymowycz LA, Rorabacher DB. Inorg. Chem. 2000;39:2897–2905. doi: 10.1021/ic0000909. [DOI] [PubMed] [Google Scholar]
- 60.Meagher NE, Juntunen KL, Salhi CA, Ochrymowycz LA, Rorabacher DB. J. Am. Chem. Soc. 1992;114:10411–10420. [Google Scholar]
- 61.Rorabacher DB. Chem. Rev. 2004;104:651–697. doi: 10.1021/cr020630e. [DOI] [PubMed] [Google Scholar]
- 62.Godbole S, Bowler BE. Biochemistry. 1999;38:487–495. doi: 10.1021/bi981698k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.