Abstract
The protein-protein docking server ClusPro is used by thousands of laboratories, and models built by the server have been reported in over 300 publications. Although the structures generated by the docking include near-native ones for many proteins, selecting the best model is difficult due to the uncertainty in scoring. Small Angle X-ray Scattering (SAXS) is an experimental technique for obtaining low resolution structural information in solution. While not sufficient on its own to uniquely predict complex structures, accounting for SAXS data improves the ranking of models and facilitates the identification of the most accurate structure. Although SAXS profiles are currently available only for a small number of complexes, due to its simplicity the method is becoming increasingly popular. Since combining SAXS experiments will provide a viable strategy for fairly high-throughput determination of protein complex structures, the option of using SAXS restraints is added to the ClusPro server.
Keywords: protein complex, structure prediction, docking method, scoring function, SAXS restraints
Introduction
Determination of the three dimensional structures of protein complexes is frequently crucial for the mechanistic understanding of cell function. Since it is often more difficult to obtain complex structures experimentally than the structures of the component proteins, computational protein docking is an important alternative method. The quality of models generated by docking methods is continuously monitored by CAPRI (Critical Assessment of Predicted Interactions), the ongoing blind prediction experiment.1 The results of CAPRI indicate substantial recent progress in methodology, including improved performance of automated docking methods.2–4 According to CAPRI evaluations,2–4 our docking server ClusPro has been the most accurate docking server since 2007, and in the most recent rounds of CAPRI its performance was comparable to that of the best human predictor groups.4,5 ClusPro is heavily used: by December 2013 we registered close to 12,000 unique user IPs, and the server completed over 96,000 docking calculations, currently adding about 3,500 per month. Models built by ClusPro have been reported in over 300 publications.
In spite of the recent progress, computational methods still have uncertainties in structure determination. Although docking programs, including ClusPro, generate a number of near-native structures for a large fraction of interacting proteins, current scoring functions are not reliable enough for selecting the best models. It was shown that using ClusPro it may be necessary to retain up to the 30 of lowest energy models in order to assure that the set includes a near-native structure.7 Thus, additional information can be very useful for correct structure determination. Many users of ClusPro are aware of this limitation, and combine computational docking with information from a variety of experimental techniques, including site-directed mutagenesis, cross-linking, and radiolytic protein footprinting with mass spectrometry.5
Small Angle X-ray Scattering (SAXS) is emerging as an effective approach to obtaining low-resolution structural information that can increase the reliability of docking results.8 The basic idea of the method is observing the X-ray scattering of a macromolecule in solution as a function of the scattering angle. The results of the experiment are encoded in a one-dimensional scattering profile determined from the spherical averaging of random orientations that a biomolecule can adopt in aqueous solution, and contains information about the shape and size of the macromolecule.9 Without the need for obtaining protein crystals or for labeling the protein, obtaining data using SAXS is relatively easy, and thus very appealing. SAXS experiments can be performed under a wide variety of solution conditions, including near physiological conditions, and usually take only a few minutes on a well-equipped synchrotron beam line. However, the information content from scattering is much lower than the one that can be obtained by X-ray crystallography, which makes docking a natural complement to SAXS for the determination of complex structures.
Recently, several groups reported combinations of SAXS with protein docking approaches. Pons et al.10 ranked docked structures by weighted docking energy and SAXS fit score as the combined scoring function. In the method developed by Sali and co-workers11 rigid body solutions were filtered by a coarse SAXS fit score, clustered, and ranked by a combined scoring function. Thus, both methods used combinations of docking and SAXS fit to facilitate model selection. Here we take a slightly different approach, and combine the docking method implemented in the ClusPro server with SAXS experimental data without modifying the scoring function. This is achieved by generating a very large number (at least 70,000) of docked structures by global sampling of the conformational space on a dense grid, and retaining a smaller but still large number (at least 2000) configurations that best agree with the observed SAXS profile. These structures are then ranked by the scoring function that was shown to perform well in ClusPro, clustered, and the centers of a number of the largest clusters are considered as models of the complex, as ordinarily done in ClusPro. The main motivation for this approach is that it is based on a well established docking method that for many proteins provides good accuracy docked models without the use of any additional information.5,12,13 Accounting for the SAXS data we just focus on the regions of the configurational space containing the structures that are most compatible with the scattering results, but otherwise perform the docking as usual. This approach has the advantage that we avoid “overfitting” to the SAXS data, and hence the docking results will not get worse even in cases where the SAXS experiment provides very limited additional information. In fact, the information content of SAXS profiles substantially depends on the shape of the complex considered, and it is generally higher for elongated complexes than for ones with more spherical shapes. The parameters of the method, primarily the number of structures that should be retained after SAXS filtering, will be selected by considering a training set of protein-protein interactions with simulated SAXS data, and the resulting algorithm will be applied to a validation set of proteins with experimental SAXS information available.
Currently results of SAXS experiments can be found only for a few protein-protein complexes. Although the application of the method is simple, the main problem is that unless the binding is very strong, an experimental SAXS profile for a complex may be a mixture of values for the complex and the unbound component proteins, thus complicating the analysis. However, due to recent developments in the methodology, particularly the ability of obtaining more homogeneous samples using size exclusion chromatography, we expect that the popularity of SAXS for determining protein complex structures will substantially increase. Therefore, we think that expanding the already well-tested docking server ClusPro by enabling it to account for SAXS data will be useful in the near future. The use of the server is free for academic and governmental research.
Methods
Docking with SAXS restraints
The method presented here addresses the docking problem restrained by a SAXS profile. Thus, given two structures of molecules (referred to as a receptor and a ligand) and the SAXS profile of their complex, we use ClusPro to find the complex structure. We assume at most moderate conformational changes, primarily in the side chains and backbones that can accounted for by using a smooth scoring function and by performing local energy minimization. The docking protocol involves three steps as follows.
Step 1: Generating docked structures.14
PIPER, the docking program implemented in ClusPro, is based on the fast Fourier transform (FFT) correlation approach, and uses a pairwise interaction potential as part of its scoring function E=Eattr+w1Erep+w2Eelec+w3Epair.14 Here Eattr and Erep denote the attractive and repulsive contributions to the van der Waals interaction energy Evdw, Eelec is an electrostatic energy term, and the pairwise term Epair represents the desolvation contributions.14 The repulsive term is designed to not penalize small conformational clashes, thus resulting in a “smooth” scoring function. The coefficients w1, w2, and w3 specify the weights of the corresponding terms, and are optimally selected for different types of docking problems.5 Unless specified otherwise, ClusPro simultaneously generates four types of models using the scoring schemes called (1) balanced, (2) electrostatic-favored, (3) hydrophobic-favored, and (4) van der Waals + electrostatics. The balanced option works generally well for enzyme-inhibitor complexes, whereas options (2) and (3) are suggested for complexes where the association is primarily driven by electrostatic and hydrophobic interactions, respectively. The fourth option, van der Waals + electrostatics, means that w3=0, i.e., the pairwise potential Epair is not used. For each parameter set, ClusPro explores 70,000 rotations of the ligand on a translational grid with 1 Å spacing, and retains the best (i.e., lowest energy) translation for each rotation, thus resulting in 70,000 structures. In addition to the above modes, the “other mode” can be selected as an advanced option for the so-called “other” type of complexes that primarily occur in signal transduction pathways,6 and generally have substantially less perfect shape and electrostatic complementarity than the enzyme-inhibitor complexes. Due to the diverse nature implied by the “other” classification, this mode uses three different sets of weighting coefficients, generating 70,000 structures for each.5
Step 2: Calculation of the SAXS profile and SAXS based filtering of docked structures
We calculate the theoretical SAXS profile using the Debye formula15
where the scattering intensity I is a function of the momentum transfer q = (4π sin θ)/λ at the scattering angle, and I is computed by summing over all pairs of atoms. The quantities fi(q) and dij are the scattering factor of atom i and the distance between atoms i and j respectively. The scattering form factor is a function of the atom, as well as the displaced solvent and hydration layer, f(q) = fv(q) − c1fs(q) + c2sfw(q), where fv is the form factor in vacuo, fs is the form factor of a dummy atom of solvent, s is the fraction of solvent accessible surface area, and fw is the form factor of water. The two constants c1 and c2 adjust the volume of the dummy atom and the difference in density between the hydration layer and bulk water respectively. The default values of these parameters are c1 = 1.0 and c2 = 0, and since the deviations from these values are small, they are fixed at the default values to reduce computational efforts as proposed by Sali and co-workers.16 This simplification can be used here, because we utilize the approximate SAXS profile only to select the region of conformational space, and do not directly incorporate SAXS values into the scoring function. The SAXS profile I(q) is calculated for each structure generated in Step 1, and the difference between the this and the experimental profile Iexp(q) is measured in terms of the χ score, defined by
where M is the number of points, and σ(q) is the error of the experimental profile. As described in Step 1, unless the “other mode” is used, 70,000 structures are saved for each of the four parameter sets used by ClusPro. The structures in each result file are ranked based on the χ score, and the 2000 structures that have the best fit to the experimental SAXS profile are retained. When the “other mode” is used, the structures are ranked based on the χ score in each of the three result files, resulting in three times 2,000 structures.
Step 3. Rescoring and clustering
Unless the “other mode” is used, we have four result files from Step 2 for the different parameter sets, each containing 2000 structures. In each file the structures are re-ranked based on the PIPER energy, and the 1000 lowest energy structures are clustered as described previously.5 The standard ClusPro output shows the centers and populations of the 10 largest clusters for each of the four different parameter sets.5 In contrast, using the “other mode” we re-rank the 2,000 structures in each of the three result files, and select the 500 lowest energy structures from each file. The retained 1500 structures are merged and clustered, and the centers and populations of the 10 largest clusters are shown. Model selection based on filtering by χ values, followed by the selection and clustering of a number of low energy structures has two advantages relative to methods that seek structures with the lowest values of scoring functions combining an energy score and a SAXS fit score. First, retaining many structures that fit well the SAXS profile eliminates too heavy dependence on this type of measurements that may carry very limited information for roughly spherical protein complexes. Second, retaining the largest clusters of low energy structures rather than the ones with the lowest scores makes our results less sensitive both to the inherent errors in the SAXS data and to the conformational variation in the structures generated by docking
Training Data Set
The method was trained using simulated profiles generated from crystal structures of 49 "other type" complexes in the protein docking benchmark.6 The “other type” complexes, including cell surface receptors and signal transduction proteins, were chosen since they generally are the most challenging for docking. Simulated SAXS profiles were generated using c1 = 1.0 and c2 = 0, for a range of the q parameter between 0.0 and 0.3, with a step size of 0.05 using the method for computing theoretical SAXS profiles as described in Step 2. As will be described, the main goal of training is the selection of the number of structures with good fit to the experimental SAXS profile that should be retained in order to optimally account for the information provided by the SAXS data.
Experimental SAXS Data
The impact of accounting for SAXS information was demonstrated by applying the method to experimental data for a lysozyme-inhibitor complex, where the Protein Data Bank (PDB) code for the X-ray crystal structure of the complex is 4G9S, and for the inhibitor structure it is 4DY3.17 Merged SAXS data are provided in the supplement. SAXS data for three homodimers suitable for use as tests cases were taken from the Bioisis database (http://bioisis.net) and from the SASBDB database (http://www.sasbdb.org/) (Table 1). (Table 1). The two dimers from Bioisis are a superoxide dismutase (Bioisis ID: APSODP) and the protein PYR1 (Bioisis ID: 1PYR1P). The dimer from SASBDB is a myomesin dimer (SASBDB ID: SASDAK5).
Table 1.
The four validation cases using experimental data. The database ID can be used to find the SAXS data from the Bioisis or SASBDB databases. The template structures were used to build homology models of the ligand for the PliG-Lysozyme case, and of the monomer in the dimer cases. The ranks shown are the rank of the near native cluster as predicted by our method.
| Experimental Case | Database ID | Template PDB ID |
Sequence Identity |
Original Rank |
Final Rank |
|---|---|---|---|---|---|
| PliG-Lysozyme | N/A | 1GBS | 57.75% | 6 | 3 |
| Superoxide dismutase dimer | APSODP (Bioisis) | 3F7K | 62% | 3 | 2 |
| PYR1 dimer | 1PYR1P (Bioisis) | 3K3K | 100% | 3 | 3 |
| Myomesin-1 dimer | SASDAK5 (SASBDB) | 2RL5 | 99% | N/A | 2 |
Homology modeling
Models were built using Modeller v9.0 (Sali and Blundell 1993), using the templates shown in Table 1. Lys side chains that were not present in the template were not modeled since they have uncertain localization. Aromatic residues (Tyr, Phe, and Trp) that were not present in the template were placed in the most probable non-clashing rotamer positions.
Results and Discussion
Results for the training set
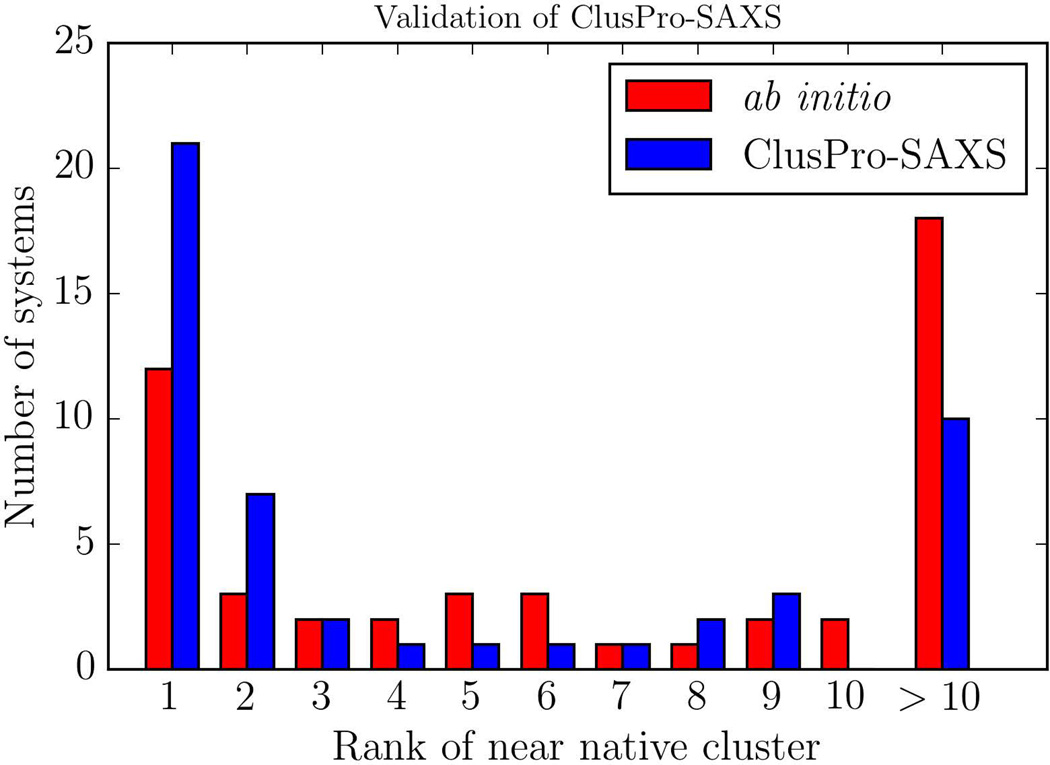
Figure 1 shows the histogram of docking performance, as compared to the ab initio docking approach, for the 49 test complexes with simulated SAXS data in the training set. According to this result, accounting for SAXS profiles almost doubles the number of systems (from 12 to 21) that have a near-native structure in the first (largest) cluster. The top 10 clusters include near-native structures for 39 of the 49 systems if we use the SAXS-based filtering, but only for 30 if no SAXS data are taken into account. We have studied the performance of the method depending on the number of structures retained in the SAXS filtering step (Figure S1, Table S1). As shown, the best performance occurs if around 2000 structures with the best fit to the SAXS profile are selected. The detailed results show that in almost all cases, both the rank and the RMSD of the near-native structure is improved. In a few cases, we do not find any predictions within 10 Å RMSD from the native pose. However, in these cases the ab-initio prediction is also relatively far from the native pose, thus these predictions would have been filtered out during the SAXS filtering step. Retaining fewer structures, and thus putting more emphasis on SAXS data, results in worse performance for a number of complexes. The reason is that we use cluster size for model discrimination. Clustering requires a large number of near-native structures that are close to each other in terms of the pairwise interface root mean square deviation (RMSD). However, not all such structures have very low SAXS scores, and thus we should retain enough structures within a SAXS score range for reliable clustering. On the other extreme, retaining too many structures in the SAXS filtering would yield results that are similar to those obtained by docking without considering the SAXS data. However, the results remain fairly similar within the range of 500 to 5000 structures retained, demonstrating the robustness of the protocol.
Figure 1.
Validation using 49 complexes from the protein docking benchmark.6 Distribution of ranks of near-native models for ab initio docking shown in red, and SAXS docking in blue.
Results for Complexes with Experimental SAXS Data
In spite of the potential of combining protein-protein docking with SAXS, experimental SAXS data on protein complexes remains scarce. However, as mentioned, recent methodology development such as size exclusion chromatography (SEC) SAXS, which allows for obtaining much more homogenous samples, should increase usage of SAXS for complex structure determination. Here we demonstrate the approach on one case of protein complex and 3 dimer test cases with experimental data (Table 1.)
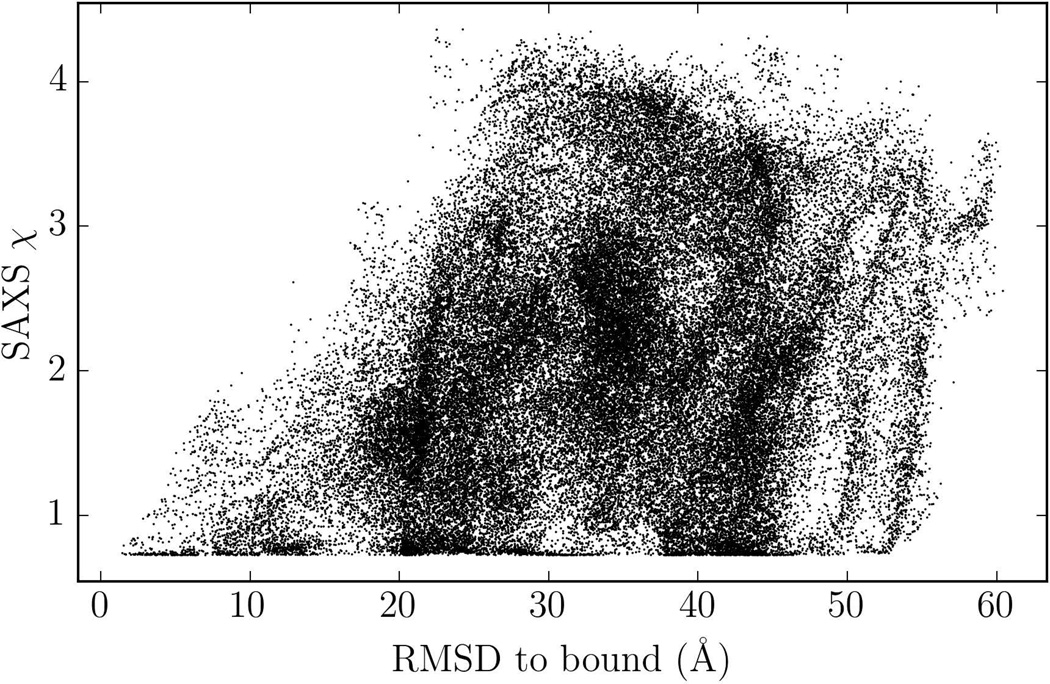
To get insight on how the approach works, we show SAXS fit score versus the RMSD values in Figure 2 for the systematic docking of E. coli PliG with the model of Atlantic salmon g-type lysozyme, where SAXS experimental data was available.17 The merged SAXS data for this complex are provided as Supporting Information. We note that this complex was one of the targets in CAPRI docking experiment, but at that time ClusPro did not have the SAXS filtering option.
Figure 2.
RMSD versus SAXS fit score for all docked conformations of PliG-lysozyme complex. Many conformations have a low SAXS score but a large RMSD.
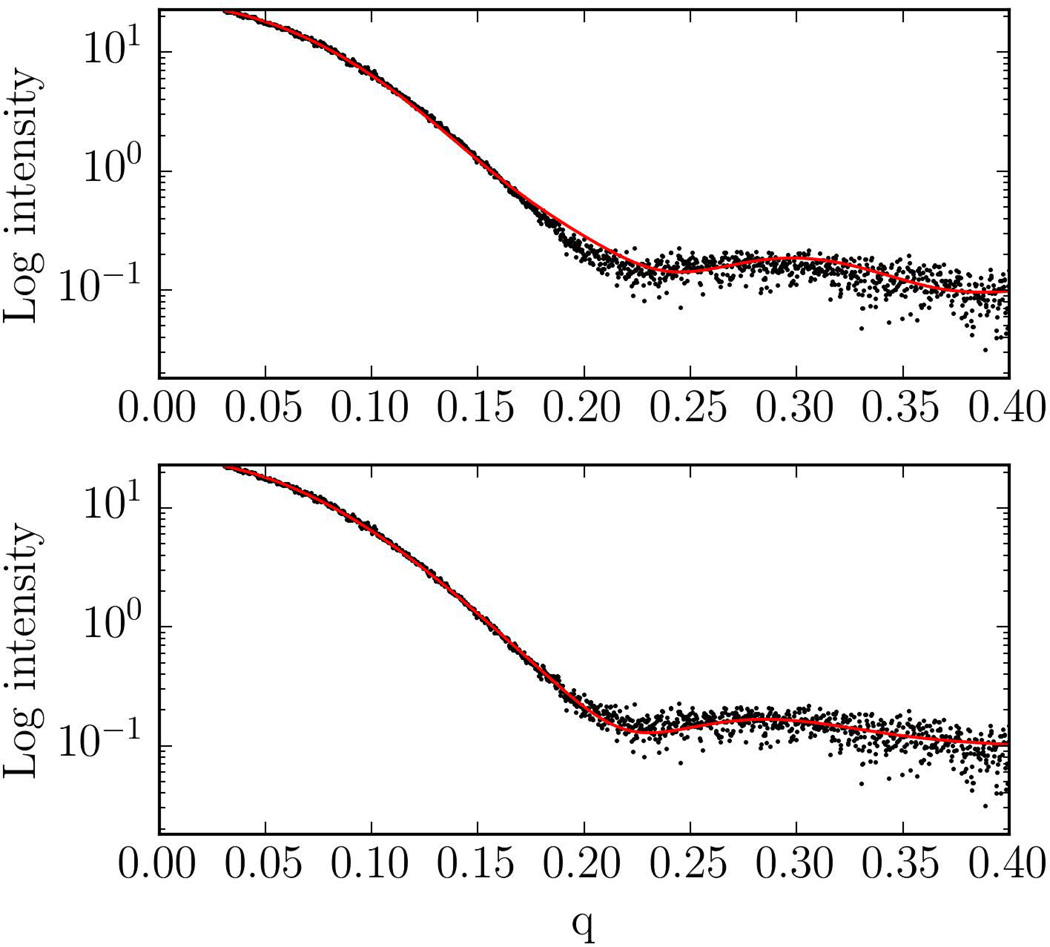
Due to spherical averaging, the SAXS data frequently provide limited information for protein docking. In fact, two conformations can have equally low SAXS fit scores but very different RMSDs from the native structure. Plots for the other experimental cases are shown on the Figure S2. Similar to the lysozyme case, discrimination of the near-native conformations by SAXS chi-score is limited for the globular system PYR1. However, when the geometry of the complex is more elongated (myomesin-1 and superoxide dismutase), the SAXS chi-score becomes more discriminative and we can see sharper funnels in a neighborhood of the native structure (with 10Å RMSD for the myomesin-1 dimer and 7Å RMSD for the superoxide dismutase dimer). In Figure 3 we show the SAXS profile of an incorrect model with a relatively low SAXS fit score, compared to near-native model to demonstrate that they both satisfy the SAXS constraints. Nevertheless, if we dock the PliG protein to lysozyme without the SAXS filtering step, the near native model is ranked 6th, whereas it is ranked 3rd if the SAXS data are taken into account. Improvement was also observed for two of the three dimers in Table 1. Although the improvement may be moderate, the docking did not yield any near-native structure for Myomesin-1 dimer without the SAXS constraints, and thus accounting for the additional information was crucial.
Figure 3.
Top: SAXS profile of non-near native top ranked model predicted by SAXS docking protocol (SAXS fit score χ = 0.87). Bottom: SAXS profile of near-native model (SAXS Fit score χ = 0.78).
Conclusions
In summary we have combined a global systematic docking approach with SAXS data for improving the ranking of docking solutions. The method was trained using simulated data for 49 complexes, and then applied to PliG-Lysozyme dimer cases. Our approach uses SAXS profile only for selecting the region of the conformational space that includes structures with good fit to the SAXS data. This allows us to utilize the additional information provided by SAXS, but also yields fairly good docking results if the information is limited, which is frequently the case. The method is implemented as an advanced option of the ClusPro docking server, which is free for academic and governmental research. Our SAXS profile calculation implementation is also available at https://github.com/StructuralBioinformaticsLab/libsaxs.
Supplementary Material
Acknowledgments
This research was partially supported by NIH/NIGMS under grants GM093147 and GM061867, and by NSF under grants DBI-1147082 and DBI-1458509. SL and SVS were supported by KU Leuven and the Research Foundation Flanders (FWO).
References
- 1.Janin J, Henrick K, Moult J, Eyck LT, Sternberg MJ, Vajda S, Vakser I, Wodak SJ. Proteins. 2003;52:2. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 2.Lensink MF, Mendez R, Wodak SJ. Proteins. 2007;69:704. doi: 10.1002/prot.21804. [DOI] [PubMed] [Google Scholar]
- 3.Lensink MF, Wodak SJ. Proteins. 2010;78:3073. doi: 10.1002/prot.22818. [DOI] [PubMed] [Google Scholar]
- 4.Lensink MF, Wodak SJ. Proteins. 2013;81:2082. doi: 10.1002/prot.24428. [DOI] [PubMed] [Google Scholar]
- 5.Kozakov D, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, Vajda S. Proteins. 2013;81:2159. doi: 10.1002/prot.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen R, Mintseris J, Janin J, Weng Z. Proteins. 2003;52:88. doi: 10.1002/prot.10390. [DOI] [PubMed] [Google Scholar]
- 7.Vajda S, Kozakov D. Curr Opin Struct Biol. 2009;19:164. doi: 10.1016/j.sbi.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graewert MA, Svergun DI. Curr Opin Struct Biol. 2013;23:748. doi: 10.1016/j.sbi.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Yang S. Advanced materials. 2014;26:7902. doi: 10.1002/adma.201304475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pons C, D'Abramo M, Svergun DI, Orozco M, Bernado P, Fernandez-Recio J. J Mol Biol. 2010;403:217. doi: 10.1016/j.jmb.2010.08.029. [DOI] [PubMed] [Google Scholar]
- 11.Schneidman-Duhovny D, Hammel M, Sali A. J Struct Biol. 2011;173:461. doi: 10.1016/j.jsb.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Comeau SR, Kozakov D, Brenke R, Shen Y, Beglov D, Vajda S. Proteins. 2007;69:781. doi: 10.1002/prot.21795. [DOI] [PubMed] [Google Scholar]
- 13.Kozakov D, Hall DR, Beglov D, Brenke R, Comeau SR, Shen Y, Li K, Zheng J, Vakili P, Paschalidis I, Vajda S. Proteins. 2010;78:3124. doi: 10.1002/prot.22835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozakov D, Brenke R, Comeau SR, Vajda S. Proteins. 2006;65:392. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 15.Debye P. Annalen der Physik. 1915;351:809. [Google Scholar]
- 16.Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. Biophys J. 2013;105:962. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leysen S, Vanderkelen L, Weeks SD, Michiels CW, Strelkov SV. Cell Mol Life Sci. 2013;70:1113. doi: 10.1007/s00018-012-1184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.