Abstract
Bicuspid aortic valve (BAV) is the most common adult congenital heart defect and is found in 0.5% to 2.0% of the general population. The term “BAV” refers to a heterogeneous group of disorders characterized by diverse aortic valve malformations with associated aortopathy, congenital heart defects, and genetic syndromes. Even after decades of investigation, the genetic determinants of BAV and its complications remain largely undefined. Just as BAV phenotypes are highly variable, the genetic etiologies of BAV are equally diverse and vary from complex inheritance in families to sporadic cases without any evidence of inheritance. In this paper, the authors discuss current concepts in BAV genetics and propose a roadmap for unraveling unanswered questions about BAV through the integrated analysis of genetic and clinical data.
Keywords: bicuspid, consortium, roadmap, valve
The most common adult congenital heart defect, a bicuspid aortic valve (BAV), is found in 0.5% to 2.0% of the general population (1,2), with a higher prevalence in men. BAVs result from abnormal aortic cusp formation during valvulogenesis, in which adjacent cusps fuse into a single cusp that is bigger than its counterpart but smaller than 2 normal cusps combined (2). Overall, “BAV” refers to a heterogeneous group of disorders distinguished by aortic valve malformations with associated aortopathy, congenital heart defects, and genetic syndromes. Despite years of investigation, researchers have yet to define genetic determinants of BAV and its complications, although most patients with BAV will develop complications requiring treatment. Just as BAV phenotypes are highly variable, the genetic etiologies of BAV are equally diverse and vary from complex inheritance in families to sporadic cases without evidence of inheritance (3).
CURRENT KNOWLEDGE OF THE GENETICS OF BICUSPID AORTIC VALVE
GENETIC SYNDROMES
Although most patients with BAV have no extracardiac abnormalities, BAV also occurs as a component of known genetic syndromes (Table 1). The highest penetrance of BAV for a genetic syndrome occurs in women with Turner syndrome, which is caused by partial or complete absence of one X chromosome in women. BAV appears in >30% of women with Turner syndrome, and the prevalence of associated coarctation, aortic aneurysms, and acute aortic dissections exceeds that in sporadic BAV cases (4). In the general population, BAV is more prevalent in men (1% to 2%) than women (0.5%) (5), suggesting that the loss of genes on the X chromosome may predispose to BAV formation. The genes responsible for this effect have not been identified.
TABLE 1.
Human Genetic Syndromes That Include Bicuspid Aortic Valve
| Syndrome | Genetic Defect | Clinical Features | OMIM No. |
|---|---|---|---|
| Turner | Monosomy X | Short stature, infertility, coarctation | — |
| Loeys-Dietz | TGFBR1, TGFBR2 mutations | TAAD, craniosynostosis, bifid uvula, skeletal defects | 609192, 608967 (type 1) |
| 610168, 610380 (type 2) | |||
| DiGeorge | 22q11.2 deletion | Truncus arteriosus, tetralogy of Fallot, craniofacial defects | 188400 (DiGeorge) |
| 192430 (VCF) | |||
| Familial TAAD | ACTA2 mutations (10%–15%) | TAAD, Premature coronary artery disease, cerebral aneurysms | 607086 (AAT1) |
| 607087 (AAT2) | |||
| 132900 (AAT4) | |||
| 608967 (AAT5) | |||
| 611788 (AAT6) | |||
| 613780 (AAT7) | |||
| 614816 (LDS type 4) | |||
| Andersen-Tawil | KCNJ2 mutations (60%) | Dysmorphic features, cardiac arrhythmias, periodic paralysis | 170390 (LQTS 7) |
| Larsen | FLNB mutations | Craniofacial and skeletal defects | 150250 |
| Kabuki | KMT2D, KDM6A mutations (70%) | Mental retardation, hearing loss, coarctation | 147920 (type 1) |
| 300867 (type 2) | |||
AAT = aortic aneurysm, familial thoracic; LDS = Loeys-Dietz syndrome; LQTS = long-QT syndrome; OMIM = Online Mendelian Inheritance in Man; TAAD = thoracic aortic aneurysms and dissections; VCF = velocardiofacial syndrome.
CONGENITAL HEART DEFECTS
Although the genetic links remain unknown, BAV is also seen with some congenital heart disorders involving the left ventricular outflow tract (LVOT), such as hypoplastic left heart syndrome, coarctation of the aorta, and some ventricular septal defects (6–8). Hypoplastic left heart syndrome and coarctation are significantly enriched among primary relatives of patients with BAV, and according to linkage analysis, mutations in the same genes probably caused some cases of hypoplastic left heart syndrome and BAV (9). One possible interpretation of these observations is that LVOT morphology and obstruction may also influence aortic valve development because of alterations in blood flow or shear stress (10).
FAMILIAL INHERITANCE
BAV occasionally demonstrates complex inheritance in large families without syndromic features. Autosomal-dominant transmission of BAV was observed in some 3-generation pedigrees, but no single-gene model clearly explains BAV inheritance (11–13). The prevalence of BAV stands nearly 10-fold higher in primary relatives of patients with BAV than in the general population (1). Inheritance is observed in >50% of families if associated nonvalvular complications are involved, such as aortic coarctation, thoracic aortic aneurysms, and mitral valve or ventricular septal defects, which are each found in 10% of family members (14). The heritability of BAV has been estimated to be as high as 0.89 in these studies, and multiple genetic alleles can interact to cause BAV or different congenital heart defects without BAV in the same family. Genetic, epigenetic, and environmental modifiers may be responsible for the variable penetrance and phenotypic expression.
Through linkage analysis of BAV pedigrees, investigators have uncovered multiple genetic loci for BAV. The most significantly associated genes were located on chromosomes 3p22 (TGFBR2), 5q15-21, 9q22.33 (TGFBR1), 9q34-35 (NOTCH1), 10q23.3 (ACTA2), 13q33-qter, 15q25-q26.1, 17q24 (KCNJ2), and 18q, with a maximal logarithm of odds score of 3.8 on chromosome 18q22.1 (13,15,16). Most families were identified because of thoracic aortic aneurysms and dissections (TAAD); BAV was found incidentally in some patients.
These studies, along with the diverse genetic syndromes associated with BAV, highlight BAV’s substantial genetic heterogeneity and implicate many discrete genes in its formation. Importantly, BAV may not have been detected on the echocardiograms of index patients. Without definitive phenotypic classification of affected patients, linkage analysis offers substantially diminished power and has not generally succeeded in identifying genes that cause BAV without aortic or extravascular manifestations. NOTCH1 remains the only gene for isolated BAV identified using linkage analysis and positional cloning strategies, but it is unlikely to cause more than a small proportion of familial cases (17,18).
In families with inherited BAV, the mutated gene often correlates with specific features and prognosis. ACTA2 mutations cause a syndrome that may include BAV as well as thoracic aortic aneurysms, premature coronary artery disease, and cerebrovascular disease (19). NOTCH1 mutations predispose to BAV with calcific aortic stenosis but are not associated with aortic disease or other extracardiac abnormalities. BAV occurs in 20% of cases of Loeys-Dietz syndrome, associated with craniofacial defects and connective tissue fragility with vulnerability to acute arterial dissections (20). Therefore, knowledge of the particular mutation in a family can direct clinicians to screen for additional clinical features in the proband and family members. As more BAV genes are discovered, genetic information will increasingly influence clinical decisions about diagnostic tests and therapies for patients with BAV.
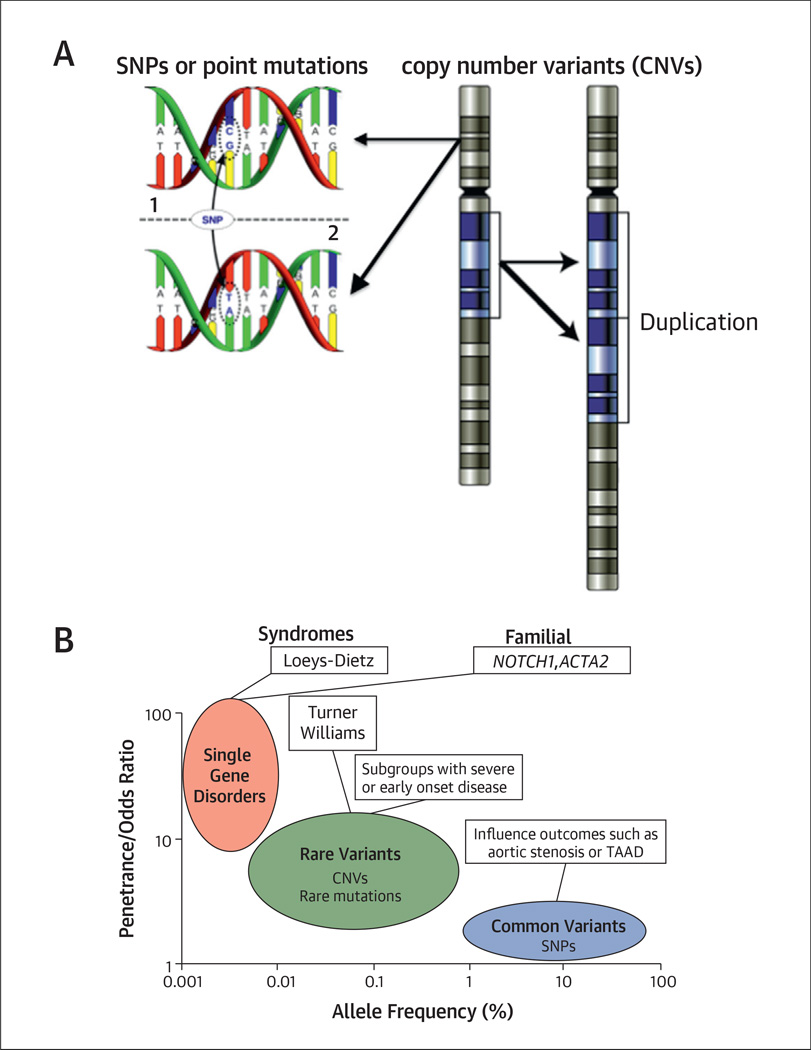
Studies involving large cohorts of patients with BAV and thoracic aortic disease indicate that many types of genetic variation contribute to the risk for this complex disease, from common single-nucleotide polymorphisms to rare copy-number variants and chromosomal abnormalities (Fig. 1A). Mutations in the same gene appear to cause TAAD across an allelic spectrum ranging from recognizable genetic syndromes to sporadic disease. For example, some mutations inMYH11 cause autosomal-dominant inheritance of TAAD associated with patent ductus arteriosus, but rare duplications involving the same gene predispose to isolated, nonfamilial cases of aortic dissection (21–23). In the majority of these patients, the total burden of genetic variants and environmental factors, rather than a single variant or class of variants, contributes to TAAD risk.
FIGURE 1. Genetic Variants on the Spectrum.
(A) The diversity of genetic variants in the human genome. (Left) A single-nucleotide variant at a specific location in the genome in 2 different individuals. Single-nucleotide variants are classified as single-nucleotide polymorphisms (SNPs), which are found in 1% or more of the general population, or rare variants, including disease-causing mutations. (Right) A chromosomal duplication that results in a copy gain of a chromosomal segment. Structural variants in the human genome include copy-number variants (CNVs) such as duplications and deletions, as well as copy-neutral events such as inversions and translocations. (B) The genetic spectrum of bicuspid aortic valve (BAV) disease. The allele frequency equals the proportion of subjects harboring a genetic variant. Single-gene disorders with high penetrance tend to cause genetic syndromes with BAV or familial forms of BAV. Rare mutations such as CNVs are expected to cause early onset forms of BAV with severe manifestations but may not be recognizable as genetic syndromes. Common variation in genes that regulate aortic valve calcification or connective tissue integrity can modify the phenotype of BAV disease progression but are unlikely to cause disease on their own. Any combination of these variants may predispose to BAV in individual cases.
TAAD = thoracic aortic aneurysms and dissections. Figure 1B is adapted from McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 2008;9:356–69.
Because BAV and TAAD frequently occur together, the genetic architecture of BAV may also consist of many different genetic variants that interact in an additive manner to increase risk for BAV and its complications (Fig. 1B). Therefore, successful gene discovery in BAV will require a combination of different methods, such as linkage analysis, positional cloning, copy-number analysis, genome-wide association (GWA) studies, and ultimately whole-genome sequencing as its cost continues to decrease.
FEATURES OF BICUSPID AORTIC VALVE THAT INFLUENCE GENE DISCOVERY
CLINICAL HETEROGENEITY
Reduced penetrance and variable expressivity present major impediments to identifying novel BAV genes in families (24). Thus, some subjects who transmit BAV to their offspring who are assumed to harbor causative genetic mutations may not have BAV themselves or may have other cardiovascular abnormalities. BAV disease also displays remarkable variation in the age of onset of complications, such as aortic valve disease or TAAD. Although BAV formation is genetically determined, the rate of progression to clinically evident disease or the development of aortopathy may be driven by interactions between genetic factors causing BAV, genetic factors associated with the development of aortic stenosis in tricuspid aortic valves, and well-established cardiovascular risk factors such as smoking, hypertension, and dyslipidemia (25,26). These factors heavily influence valve calcification leading to aortic stenosis or regurgitation (the most common indications for valve replacement surgery in patients with BAV) (27–30).
The interaction between genes and environment in influencing aortic valve complications is also apparent in mice, in which the development of calcific tricuspid aortic valve stenosis required both dietary modifications and specific genetic mutations (31). A recent GWA study demonstrated that an intronic genetic variant at the lipoprotein(a) locus promotes human aortic valve calcification (25). Distinct genetic variants and environmental hazards, independent of the factors that cause BAV formation, may influence the complications and outcomes of BAV disease and thus the recognition of BAV in an individual patient.
ANIMAL MODELS
The paucity of animal studies erects another obstacle to understanding BAV’s etiology. It develops spontaneously in an inbred strain of Syrian hamsters but without valve or aortic complications typical of human BAV, and the underlying genetic defect is unknown (32,33). Targeted deletions of fibroblast growth factor, bone morphogenetic protein, and Notch pathway genes in mice produced BAV with incomplete penetrance, as well as various malformations of the LVOT and aorta (Table 2). This approach, if sufficiently scaled to accommodate the large numbers of BAV candidate genes and pathways, can help prioritize candidate genes for further analysis.
TABLE 2.
Transgenic Mouse Models That Include Bicuspid Aortic Valve
| Allele | Penetrance | Other Phenotypes | Ref. # |
|---|---|---|---|
| Gata5tm1.1Nemr | 7/28 | LVH | (47) |
| Tg(GATA5-cre)1Krc/Acvr1tm1Vk | 3/22 | Perimembranous VSDs | (59) |
| Nos3tm1Unc | 5/12 | None | (60) |
| Hoxa1tm3Mrc | 4/17 | IAA, VSD | (61) |
| Tbx1tm1Pa | 5/34 | IAA, TOF, coronary anomalies | (62) |
| Hoxa3tm1(cre)Moon/Fgf8tm2Moon | 7/33 | IAA, coronary anomalies | (63) |
| Nkx2–5tm1(cre)Rjs/Frs2tm1Fwan | 20% | OA, DORV | (64) |
| Nkx2–5tm1Wehi* Nkx2–5tm4Rph* | 11/100 2/98 | Conduction defects, PFO | (65) |
Allele information is taken from Mouse Genome Informatics (http://www.informatics.jax.org).
Heterozygotes.
DORV = double-outlet right ventricle; IAA = interrupted aortic arch; LVH = left ventricular hypertrophy; OA = overriding aorta; PFO = patent foramen ovale; TOF = tetralogy of Fallot; VSD = ventricular septal defect.
However, with rare exceptions, mutations in genes that cause BAV when targeted in animals have not been found in humans with BAV. Therefore, typical animal models with single gene mutations may not be sufficient, and incorporation of multiple mutations or gene-environment interactions will likely be required to understand the complexity of human BAV disease.
CLINICAL IMPLICATIONS
Because BAV formation and disease involve many genes, developing clinical genetic tests to identify patients at risk for BAV-related complications, such as aortic stenosis or TAAD, will require considerable time and research. Until then, early identification and careful surveillance of at-risk family members will likely remain primary clinical strategies. The 2010 American College of Cardiology and American Heart Association guidelines for thoracic aortic disease provide a class I indication to image first-degree relatives of patients with BAV and TAAD to detect valvular and extravalvular complications (34,35). Affected relatives can then receive interventions to reduce traditional cardiovascular risk factors and, perhaps, emerging therapies targeted to prevent these complications. It is also recommended that every newly diagnosed BAV patient undergo a comprehensive clinical evaluation that includes complete imaging, at a minimum, of the heart and thoracic aorta to rule out other defects and additional imaging as warranted by examination or family history. Genetic tests are offered to patients with BAV when features of single-gene disorders or syndromes are present, and findings are confirmed in first-degree relatives whenever possible.
BICUSPID AORTIC VALVE GENETICS: ROADMAP TO DISCOVERY
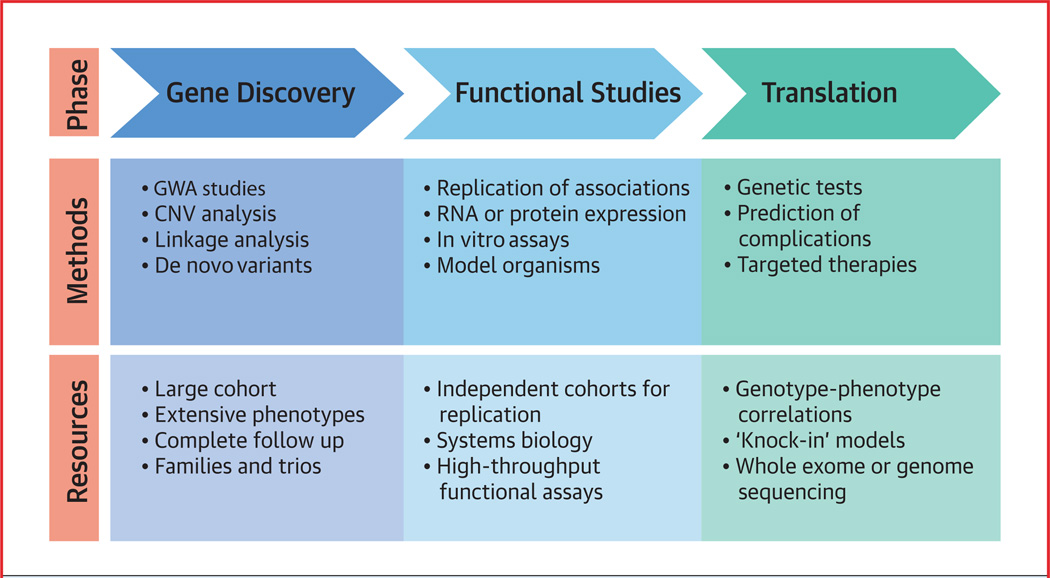
With so many issues to be resolved, how can we unravel the genetic complexity of BAV? A proposed roadmap for translational applications of genetic discoveries is illustrated in the Central Illustration.
CENTRAL ILLUSTRATION. Roadmap to Translate Genetic Discoveries Into Clinical Applications.
“Knock-out” models include tissue-specific and whole-organism deletion of genes. “Knock-in” models are generated by introducing a specific human sequence variant into an orthologous gene. In real time, these processes would most likely occur simultaneously. CNV = central nervous system; GWA = genome-wide analysis.
The International Bicuspid Aortic Valve Consortium (BAVCon) (Online Appendix) was recently founded to translate genetic discoveries into clinically useful diagnostic tests and therapies, as well as address broader challenges related to knowledge gaps in the pathophysiology and natural history of BAV disease. Designed as a registry for long-term surveillance, BAVCon is conceptually similar to the National Registry of Genetically Triggered Thoracic Aortic Aneurysms and Cardiovascular Conditions (GenTAC), which enrolled subjects with diverse forms of thoracic aortic disease, including BAV. The BAVCon Registry consists of independently arbitrated echocardiographic and computed tomographic images from more than 4,000 patients with BAV and tricuspid controls retrospectively enrolled at 16 centers in North America and Europe. Prospective collection of images and tissue samples from additional patients, and the eventual incorporation of GenTAC patients with BAV into BAVCon, will create a deeply phenotyped and collaborative resource for clinical, genetic, and molecular studies available to all BAVCon investigators.
The enormous diversity of BAV with respect to anatomic variation, coexisting abnormalities, and natural history poses a significant challenge to researchers whose goal is to identify genes that cause BAV or predict BAV-associated complications. To sort through this genetic complexity, large and ethnically diverse cohorts that are well characterized in terms of BAV anatomy, associated complications, and cardiovascular and noncardiovascular risk factors are needed. BAVCon will permit adequately powered GWA studies, which are particularly important for understanding and predicting endpoints of BAV disease, such as valve calcification and acute aortic dissection.
Similarly, well-characterized families predisposed to BAV and its complications inherited in a Mendelian manner are needed to identify genes with mutations that confer high disease risk. Investigators may need to follow this cohort for a decade or longer to capture outcomes, such as aortic valve stenosis, that are slow to develop. Designed with these challenges in mind, BAVCon represents the first worldwide registry of ethnically diverse patients with BAV characterized by experts in the field using stringent, uniform criteria and standardized measurements. Valve morphology and hemodynamic data will be precisely assessed using state-of-the-art imaging methods and will be correlated with these outcomes. Because different genetic variants may contribute to outcomes, we envision BAVCon as a rich and continuous source of future genetic studies.
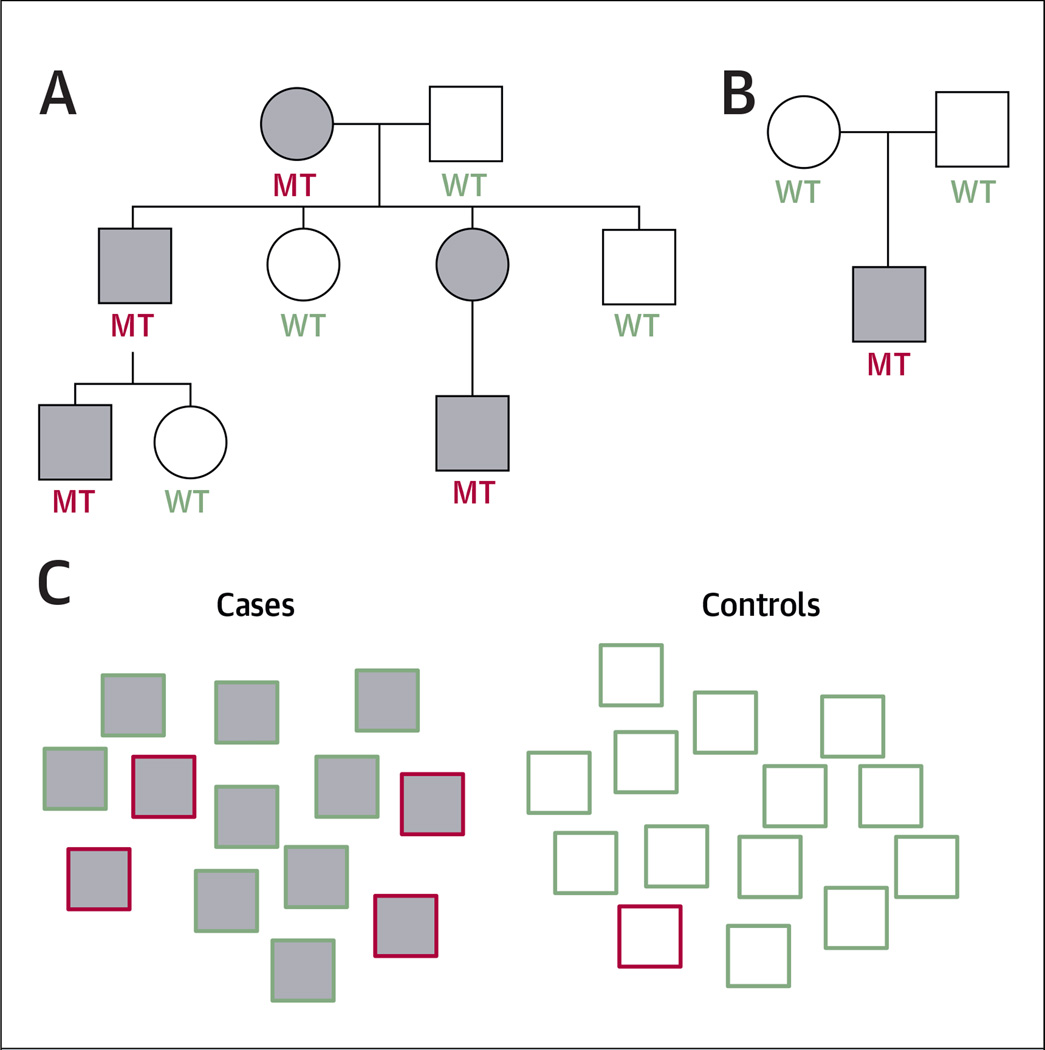
Large families with inherited predisposition to BAV present the most promising opportunities for gene discovery, because family based studies optimize the statistical power to identify causative genetic variants. When BAV is inherited as an autosomal-dominant trait, the mutant gene can be traced from parents to offspring using segregation analysis, and the causative mutations can then be identified by sequencing the genes of distantly related subjects (Fig. 2A). The ability to sequence entire exomes or genomes of multiple family members has greatly reduced the time and labor required for gene discovery and has led to the discovery of several novel genes for TAAD (21,36–39).
FIGURE 2. Experimental Designs to Identify Disease-Causing and Susceptibility Genetic Variants.
(A) Example of a 3-generation pedigree with autosomal-dominant inheritance. Affected patients are gray. Mutation carriers (MT) and subjects without mutations (WT) are highlighted in red and green, respectively. In this instance, the penetrance of the mutation is complete, and the mutant gene can be mapped to a segment of the genome using linkage analysis. (B) Example of a trio. De novo mutations in the affected offspring that are not found in the parents may be prioritized for analysis in independent groups of cases. In this situation, the penetrance of the phenotype in the parents is uncertain and may confound the results. (C) Example of a case-control association study. The genotypes of large groups of cases (gray) and controls (white) are compared at sites across the genome. The red border identifies subjects who carry the risk allele that is being tested for association. Variants that are enriched in cases compared with controls are considered to be associated with the disease. In this case, there is a 4-fold enrichment of the variant among the cases compared with controls.
Even if BAV inheritance is not observed, genetic tests of family members may still be informative. Genetic variants present in affected children but not their parents are referred to as de novo variants (Fig. 2B), which are generally prioritized because they tend to be more deleterious than inherited variations (40). Recent trio-based exome sequencing studies identified causal de novo mutations in several rare syndromes that are not inherited in families (41,42). Because dozens of de novo variants arise stochastically in each generation, candidate genes with de novo variants need to be validated by performing mutation analysis of independent groups of cases.
In contrast to families with inherited BAV disease, >80% of patients with BAV have no known affected relatives and are regarded as sporadic cases. Because linkage analysis is unfeasible to investigate single genetic variants in unrelated patients, case-control association methods such as GWA studies must be used (Fig. 2C). GWA studies typically require thousands of affected individuals and controls to generate meaningful results and are measured by indirect associations between the genuine risk variants and genotyped single-nucleotide polymorphisms, rather than by direct identification of the causal variants. Thus, GWA studies are most likely to identify genetic variants that increase susceptibility to BAV or its complications. The size of the BAVCon cohort will provide sufficient power to discover these common but low-impact BAV-associated variants.
Among sporadic BAV cases, patients at the severe end of the BAV phenotypic spectrum are most likely to have highly penetrant mutations in single genes because they resemble patients with genetic syndromes or inherited BAV. This includes patients with unicuspid aortic valves, those with additional congenital malformations, and those who require early interventions for valve disease or TAAD manifesting in childhood or adolescence. Genomic comparisons between these subgroups and older patients with BAV without significant valve or aortic complications can uncover genetic variants that predispose to BAV-associated complications. In normal populations, these variants may only weakly influence the likelihood of developing disease and may be difficult to detect, but the effects of these variants are likely to be amplified in individuals with BAV, who tend to develop more pronounced aortic disease at younger ages. In other situations, this strategy of sampling at the phenotypic extremes led to the discovery of individually rare variants that would otherwise be missed by traditional GWA studies (43–46).
Genetic studies may lead to the discovery of novel genetic variants whose biological relevance to BAV disease is unknown. Leveraging systems biology to identify data patterns can help prioritize candidate genes. Systems analyses test associations of biological pathways, rather than single genes, with disease. For example, the discovery of NOTCH1 mutations in BAV families led to the systematic investigation of Notch pathway genes in aortic valve development and calcification (47–49). De novo variants of genes that regulate histone modification were found to be enriched in patients with severe congenital heart disease (50). These pathways and others with relevance to BAV outcomes include genes involved in aortic valve calcification, connective tissue integrity, and aortic disease. Using systems-based approaches, BAV candidate genes can be ranked according to their biological functions; interacting genes in the same pathways can then be screened for mutations in BAV cases by sequencing or copy-number analysis.
Because BAV displays incomplete penetrance, a substantial proportion of BAV-associated genetic variants in sporadic cases may also be present in apparently unaffected subjects. Therefore, independent observations of genetic discoveries in separate cohorts of patients with BAV will be essential for the prioritization of candidate genes. With an enrollment of several thousand patients, BAVCon will provide an important platform for these types of validation studies.
Despite significant limitations in BAV animal models, targeted gene deletions in mice, zebrafish, and cultured aortic valve interstitial cells have provided useful insights into candidate gene involvement in LVOT or aortic valve development (3,51,52). Some of these methods can be developed into high-throughput assays to screen candidate genes for roles in BAV formation or disease progression. To investigate therapies for BAV-associated complications, improved genetic models are needed. “Knock-in” experiments, in which human mutations are introduced into homologous mouse or zebrafish genes, may result in more faithful models of human BAV phenotypes than current models with null mutations (53). Pathobiological studies may complement these strategies by validating candidate genes’ biological functions or identifying pathways that can be explored using systems methods. These approaches already have led to the discovery of new drug targets and genetic tests for inherited cardiomyopathies, and hold tremendous promise for BAV (54–56).
BACK TO THE BEDSIDE OF THE PATIENT WITH BICUSPID AORTIC VALVE
How then do we translate these discoveries into clinically meaningful tools for patients with BAV and their families? One approach is to develop genetic tests or biomarkers that identify patients with BAV who are at high risk for adverse outcomes such as aortic valve stenosis or TAAD. Selected patients with BAV can then be enrolled into clinical trials to assess novel preventative strategies or drug therapies on the basis of their individualized genetic risk profiles. Clinical trials will determine if selectively interfering with these pathways using this targeted approach can retard or prevent disease progression.
Similarly, genetic risk profiles may influence surgical decisions. For example, young patients with TAAD with normally functioning or mildly dysfunctional BAVs frequently receive valve-sparing aortic grafts, but we do not know which of them may require future valve replacements or more distal aortic repair (57,58). Genetic profiles may dictate when valve replacements are indicated during aortic repair and, most important, which aortas need to be repaired at the time of valve replacement, a subject of significant current controversy. These are only 2 of many potential examples that illustrate the impact of genetic discoveries on BAV diagnosis and therapies. BAVCon will provide us with unprecedented detail about subtypes of BAV disease through its large and well-phenotyped cohort of patients, making this type of comparative analysis possible for the first time.
Supplementary Material
ABBREVIATIONS AND ACRONYMS
- BAV
bicuspid aortic valve
- BAVCon
International Bicuspid Aortic Valve Consortium
- GWA
genome-wide association
- LVOT
left ventricular outflow tract
- TAAD
thoracic aortic aneurysms and dissections
APPENDIX
For a list of the BAVCon sites and investigators, please see the online version of this article.
Footnotes
The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
REFERENCES
- 1.Siu SC, Silversides CK. Bicuspid aortic valve disease. J Am Coll Cardiol. 2010;55:2789–2800. doi: 10.1016/j.jacc.2009.12.068. [DOI] [PubMed] [Google Scholar]
- 2.Fedak PW, Verma S, David TE, Leask RL, Weisel RD, Butany J. Clinical and pathophysiological implications of a bicuspid aortic valve. Circulation. 2002;106:900–904. doi: 10.1161/01.cir.0000027905.26586.e8. [DOI] [PubMed] [Google Scholar]
- 3.Laforest B, Nemer M. Genetic insights into bicuspid aortic valve formation. Cardiol Res Pract. 2012;2012:180297. doi: 10.1155/2012/180297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson M, Silberbach M. Dissection of the aorta in Turner syndrome: two cases and review of 85 cases in the literature. BMJ Case Rep. 2009;2009:bcr0620091998. doi: 10.1136/bcr.06.2009.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner JI, Kuehl K. Hypoplastic left heart syndrome and other left heart disease: evolution of understanding from population-based analysis to molecular biology and back again—a brief overview. Cardiol Young. 2011;21(Suppl 2):23–27. doi: 10.1017/S1047951111001399. [DOI] [PubMed] [Google Scholar]
- 6.John AS, McDonald-McGinn DM, Zackai EH, Goldmuntz E. Aortic root dilation in patients with 22q11.2 deletion syndrome. Am J Med Genet A. 2009;149A:939–942. doi: 10.1002/ajmg.a.32770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duran AC, Frescura C, Sans-Coma V, Angelini A, Basso C, Thiene G. Bicuspid aortic valves in hearts with other congenital heart disease. J Heart Valve Dis. 1995;4:581–590. [PubMed] [Google Scholar]
- 8.Hor KN, Border WL, Cripe LH, Benson DW, Hinton RB. The presence of bicuspid aortic valve does not predict ventricular septal defect type. Am J Med Genet A. 2008;146A:3202–3205. doi: 10.1002/ajmg.a.32609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinton RB, Martin LJ, Rame-Gowda S, Tabangin ME, Cripe LH, Benson DW. Hypoplastic left heart syndrome links to chromosomes 10q and 6q and is genetically related to bicuspid aortic valve. J Am Coll Cardiol. 2009;53:1065–1071. doi: 10.1016/j.jacc.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hickey EJ, Caldarone CA, McCrindle BW. Left ventricular hypoplasia: a spectrum of disease involving the left ventricular outflow tract, aortic valve, and aorta. J Am Coll Cardiol. 2012;59:S43–S54. doi: 10.1016/j.jacc.2011.04.046. [DOI] [PubMed] [Google Scholar]
- 11.Calloway TJ, Martin LJ, Zhang X, Tandon A, Benson DW, Hinton RB. Risk factors for aortic valve disease in bicuspid aortic valve: a family-based study. Am J Med Genet A. 2011;155A:1015–1020. doi: 10.1002/ajmg.a.33974. [DOI] [PubMed] [Google Scholar]
- 12.Huntington K, Hunter AG, Chan KL. A prospective study to assess the frequency of familial clustering of congenital bicuspid aortic valve. J Am Coll Cardiol. 1997;30:1809–1812. doi: 10.1016/s0735-1097(97)00372-0. [DOI] [PubMed] [Google Scholar]
- 13.Martin LJ, Ramachandran V, Cripe LH, et al. Evidence in favor of linkage to human chromosomal regions 18q, 5q and 13q for bicuspid aortic valve and associated cardiovascular malformations. Hum Genet. 2007;121:275–284. doi: 10.1007/s00439-006-0316-9. [DOI] [PubMed] [Google Scholar]
- 14.Cripe L, Andelfinger G, Martin LJ, Shooner K, Benson DW. Bicuspid aortic valve is heritable. J Am Coll Cardiol. 2004;44:138–143. doi: 10.1016/j.jacc.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 15.McBride KL, Pignatelli R, Lewin M. Inheritance analysis of congenital left ventricular outflow tract obstruction malformations: segregation, multiplex relative risk, and heritability. Am J Med Genet A. 2005;134A:180–186. doi: 10.1002/ajmg.a.30602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goh DL, Han LF, Judge DP, et al. Linkage of familial bicuspid aortic valve with aortic aneurysm to chromosome 15q. Presented at: Annual Meeting of the American Society of Human Genetics; October 2002; Baltimore, MD. [Google Scholar]
- 17.Garg V, Muth AN, Ransom JF, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 18.Ellison JW, Yagubyan M, Majumdar R, et al. Evidence of genetic locus heterogeneity for familial bicuspid aortic valve. J Surg Res. 2007;142:28–31. doi: 10.1016/j.jss.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 19.Guo DC, Papke CL, Tran-Fadulu V, et al. Mutations in smooth muscle alpha-actin (ACTA2) cause coronary artery disease, stroke, and moyamoya disease, along with thoracic aortic disease. Am J Hum Genet. 2009;84:617–627. doi: 10.1016/j.ajhg.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loeys BL, Schwarze U, Holm T, et al. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N Engl J Med. 2006;355:788–798. doi: 10.1056/NEJMoa055695. [DOI] [PubMed] [Google Scholar]
- 21.Pannu H, Tran-Fadulu V, Papke CL, et al. MYH11 mutations result in a distinct vascular pathology driven by insulin-like growth factor 1 and angiotensin II. Hum Mol Genet. 2007;16:2453–2462. doi: 10.1093/hmg/ddm201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu L, Vranckx R, Khau Van Kien P, et al. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat Genet. 2006;38:343–349. doi: 10.1038/ng1721. [DOI] [PubMed] [Google Scholar]
- 23.Kuang SQ, Guo DC, Prakash SK, et al. Recurrent chromosome 16p13.1 duplications are a risk factor for aortic dissections. PLoS Genet. 2011;7:e1002118. doi: 10.1371/journal.pgen.1002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent KC, Crenshaw ML, Goh DL, Dietz HC. Genotype-phenotype correlation in patients with bicuspid aortic valve and aneurysm. J Thorac Cardiovasc Surg. 2013;146:158–165.e1. doi: 10.1016/j.jtcvs.2012.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thanassoulis G, Campbell CY, Owens DS, et al. Genetic associations with valvular calcification and aortic stenosis. N Engl J Med. 2013;368:503–512. doi: 10.1056/NEJMoa1109034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dweck MR, Boon NA, Newby DE. Calcific aortic stenosis: a disease of the valve and the myocardium. J Am Coll Cardiol. 2012;60:1854–1863. doi: 10.1016/j.jacc.2012.02.093. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed S, Honos GN, Walling AD, et al. Clinical outcome and echocardiographic predictors of aortic valve replacement in patients with bicuspid aortic valve. J Am Soc Echocardiogr. 2007;20:998–1003. doi: 10.1016/j.echo.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Roberts WC, Ko JM. Frequency by decades of unicuspid, bicuspid, and tricuspid aortic valves in adults having isolated aortic valve replacement for aortic stenosis, with or without associated aortic regurgitation. Circulation. 2005;111:920–925. doi: 10.1161/01.CIR.0000155623.48408.C5. [DOI] [PubMed] [Google Scholar]
- 29.Tzemos N, Therrien J, Yip J, et al. Outcomes in adults with bicuspid aortic valves. JAMA. 2008;300:1317–1325. doi: 10.1001/jama.300.11.1317. [DOI] [PubMed] [Google Scholar]
- 30.Michelena HI, Desjardins VA, Avierinos JF, et al. Natural history of asymptomatic patients with normally functioning or minimally dysfunctional bicuspid aortic valve in the community. Circulation. 2008;117:2776–2784. doi: 10.1161/CIRCULATIONAHA.107.740878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nus M, Macgrogan D, Martinez-Poveda B, et al. Diet-induced aortic valve disease in mice haploinsufficient for the Notch pathway effector RBPJK/CSL. Arterioscler Thromb Vasc Biol. 2011;31:1580–1588. doi: 10.1161/ATVBAHA.111.227561. [DOI] [PubMed] [Google Scholar]
- 32.Fernandez B, Duran AC, Fernandez-Gallego T, et al. Bicuspid aortic valves with different spatial orientations of the leaflets are distinct etiological entities. J Am Coll Cardiol. 2009;54:2312–2318. doi: 10.1016/j.jacc.2009.07.044. [DOI] [PubMed] [Google Scholar]
- 33.Sans-Coma V, Carmen Fernandez M, Fernandez B, Duran AC, Anderson RH, Arque JM. Genetically alike Syrian hamsters display both bifoliate and trifoliate aortic valves. J Anat. 2012;220:92–101. doi: 10.1111/j.1469-7580.2011.01440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guntheroth WG. A critical review of the American College of Cardiology/American Heart Association practice guidelines on bicuspid aortic valve with dilated ascending aorta. Am J Cardiol. 2008;102:107–110. doi: 10.1016/j.amjcard.2008.02.106. [DOI] [PubMed] [Google Scholar]
- 35.Hiratzka LF, Bakris GL, Beckman JA, et al. 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with thoracic aortic disease. J Am Coll Cardiol. 2010;55:e27–e129. doi: 10.1016/j.jacc.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 36.Boileau C, Guo DC, Hanna N, et al. TGFB2 mutations cause familial thoracic aortic aneurysms and dissections associated with mild systemic features of Marfan syndrome. Nat Genet. 2012;44:916–921. doi: 10.1038/ng.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo DC, Pannu H, Tran-Fadulu V, et al. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat Genet. 2007;39:1488–1493. doi: 10.1038/ng.2007.6. [DOI] [PubMed] [Google Scholar]
- 38.Guo DC, Regalado ES, Minn C, et al. Familial thoracic aortic aneurysms and dissections: identification of a novel locus for stable aneurysms with a low risk for progression to aortic dissection. Circ Cardiovasc Genet. 2011;4:36–42. doi: 10.1161/CIRCGENETICS.110.958066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Regalado ES, Guo DC, Villamizar C, et al. Exome sequencing identifies SMAD3 mutations as a cause of familial thoracic aortic aneurysm and dissection with intracranial and other arterial aneurysms. Circ Res. 2011;109:680–686. doi: 10.1161/CIRCRESAHA.111.248161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 41.Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindhurst MJ, Sapp JC, Teer JK, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khetarpal SA, Edmondson AC, Raghavan A, et al. Mining the LIPG allelic spectrum reveals the contribution of rare and common regulatory variants to HDL cholesterol. PLoS Genet. 2011;7:e1002393. doi: 10.1371/journal.pgen.1002393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li D, Lewinger JP, Gauderman WJ, Murcray CE, Conti D. Using extreme phenotype sampling to identify the rare causal variants of quantitative traits in association studies. Genet Epidemiol. 2011;35:790–799. doi: 10.1002/gepi.20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fahmi S, Yang C, Esmail S, Hobbs HH, Cohen JC. Functional characterization of genetic variants in NPC1L1 supports the sequencing extremes strategy to identify complex trait genes. Hum Mol Genet. 2008;17:2101–2107. doi: 10.1093/hmg/ddn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guey LT, Kravic J, Melander O, et al. Power in the phenotypic extremes: a simulation study of power in discovery and replication of rare variants. Genet Epidemiol. 2011;35:236–246. doi: 10.1002/gepi.20572. [DOI] [PubMed] [Google Scholar]
- 47.Laforest B, Andelfinger G, Nemer M. Loss of Gata5 in mice leads to bicuspid aortic valve. J Clin Invest. 2011;121:2876–2887. doi: 10.1172/JCI44555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Acharya A, Hans CP, Koenig SN, et al. Inhibitory role of Notch1 in calcific aortic valve disease. PLoS One. 2011;6:e27743. doi: 10.1371/journal.pone.0027743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z, Feng L, Wang CM, et al. Deletion of RBP-J in adult mice leads to the onset of aortic valve degenerative diseases. Mol Biol Rep. 2012;39:3837–3845. doi: 10.1007/s11033-011-1162-y. [DOI] [PubMed] [Google Scholar]
- 50.Zaidi S, Choi M, Wakimoto H, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staudt D, Stainier D. Uncovering the molecular and cellular mechanisms of heart development using the zebrafish. Annu Rev Genet. 2012;46:397–418. doi: 10.1146/annurev-genet-110711-155646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nigam V, Srivastava D. Notch1 represses osteogenic pathways in aortic valve cells. J Mol Cell Cardiol. 2009;47:828–834. doi: 10.1016/j.yjmcc.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doyle A, McGarry MP, Lee NA, Lee JJ. The construction of transgenic and gene knockout/knockin mouse models of human disease. Transgenic Res. 2012;21:327–349. doi: 10.1007/s11248-011-9537-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bick AG, Flannick J, Ito K, et al. Burden of rare sarcomere gene variants in the Framingham and Jackson Heart Study cohorts. Am J Hum Genet. 2012;91:513–519. doi: 10.1016/j.ajhg.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greenway SC, Pereira AC, Lin JC, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41:931–935. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang J, Wakimoto H, Seidman JG, Seidman CE. Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science. 2013;342:111–114. doi: 10.1126/science.1236921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park CB, Greason KL, Suri RM, Michelena HI, Schaff HV, Sundt TM. Fate of nonreplaced sinuses of Valsalva in bicuspid aortic valve disease. J Thorac Cardiovasc Surg. 2011;142:278–284. doi: 10.1016/j.jtcvs.2010.08.055. [DOI] [PubMed] [Google Scholar]
- 58.Schafers HJ, Kunihara T, Fries P, Brittner B, Aicher D. Valve-preserving root replacement in bicuspid aortic valves. J Thorac Cardiovasc Surg. 2010;140:S36–S40. doi: 10.1016/j.jtcvs.2010.07.057. [DOI] [PubMed] [Google Scholar]
- 59.Thomas PS, Sridurongrit S, Ruiz-Lozano P, Kaartinen V. Deficient signaling via Alk2 (Acvr1) leads to bicuspid aortic valve development. PLoS One. 2012;7:e35539. doi: 10.1371/journal.pone.0035539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee TC, Zhao YD, Courtman DW, Stewart DJ. Abnormal aortic valve development in mice lacking endothelial nitric oxide synthase. Circulation. 2000;101:2345–2348. doi: 10.1161/01.cir.101.20.2345. [DOI] [PubMed] [Google Scholar]
- 61.Makki N, Capecchi MR. Cardiovascular defects in a mouse model of HOXA1 syndrome. Hum Mol Genet. 2012;21:26–31. doi: 10.1093/hmg/ddr434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 63.Macatee TL, Hammond BP, Arenkiel BR, Francis L, Frank DU, Moon AM. Ablation of specific expression domains reveals discrete functions of ectoderm-and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development. 2003;130:6361–6374. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J, Lin Y, Zhang Y, et al. Frs2alpha-deficiency in cardiac progenitors disrupts a subset of FGF signals required for outflow tract morphogenesis. Development. 2008;135:3611–3622. doi: 10.1242/dev.025361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Biben C, Weber R, Kesteven S, et al. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2–5. Circ Res. 2000;87:888–895. doi: 10.1161/01.res.87.10.888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.