In recent years there have been many advances in the production of recombinant proteins in bacterial hosts. A common problem is the sequestration of recombinant protein by the bacterium in an inclusion body which is the general term for a large insoluble protein aggregate. Harsh denaturing conditions are typically needed to solubilize the inclusion body protein with a consequent requirement of refolding to obtain functional protein. Despite these difficulties, inclusion bodies have some attractive features for recombinant protein production including each cell having one inclusion body which contains high purity protein and as much as 50% of the total mass of cellular protein.1 Inclusion bodies are noncrystalline solids and there has been little high-resolution, residue-specific study to ascertain their molecular structure. The most common structural method has been infrared (IR) spectroscopy which provided information about the overall fractions of different types of regular secondary structure. These studies were typically carried out on dehydrated samples and suggested that there was more β sheet structure in the inclusion body protein than in the native fold. It was therefore proposed that the non-native β strands from different proteins associated together to form intermolecular β-sheets in a structure similar to amyloid protein.2–8 Some reports indicate that a fraction of inclusion body protein is natively folded and functional.3, 9
This paper describes application of solid-state NMR to determine residue-specific secondary structure in inclusion body protein. The methods are straightforward and inexpensive and should be broadly applicable to a wide variety of proteins in inclusion bodies. The methods were initially developed to study residue-specific conformation in membrane-reconstituted proteins and were based on the observation that in sequences of proteins of moderate size, a large fraction of the residues are in “unique sequential pairs”, for example, there would only be one instance of a Leu followed by a Leu. In addition, NMR methods such as “rotational-echo double-resonance” (REDOR) can selectively detect the signal of the 13C carbonyl (13CO) nuclei which are directly bonded to 15N nuclei, and there are well-known correlations between the backbone 13CO NMR chemical shift of a residue and its local conformation.10–12 Recombinant protein was therefore produced in E. coli cells in minimal media containing the 1-13C amino acid and 15N-amino acid of the respective N- and C-terminal residues of the unique sequential pair, and the filtered 13CO NMR signal of the N-terminal residue of the pair was used to determine its conformation.12
These methods were first applied to “FHA2” which is a N-terminal 185-residue functional domain of the 221-residue “HA2” subunit of the influenza virus hemagglutinin protein.12 FHA2 contains a ~20-residue N-terminal “fusion peptide” that binds to the host cell membrane and plays a key role in infection. FHA2 is a membrane protein because of the fusion peptide while shorter constructs which lack the fusion peptide are soluble in aqueous solution and have been crystallized.13 There is also a NMR structure of the HA2 fusion peptide in detergent micelles.14
One essential feature of the method was expression in minimal media containing labeled amino acids. However, growth only in minimal media provided <0.5 mg purified FHA2 per L culture and an alternate protocol was developed to first grow to high cell densities in rich media and to then switch to minimal media prior to expression.12, 15, 16 Using this method, the purified yield of FHA2 from the soluble cell lysate was ~10 mg/L culture. This was likely the fraction of FHA2 which was incorporated in cell membranes because the lysis buffer contained N-lauroylsarcosine detergent which is effective at solubilizing membrane proteins.
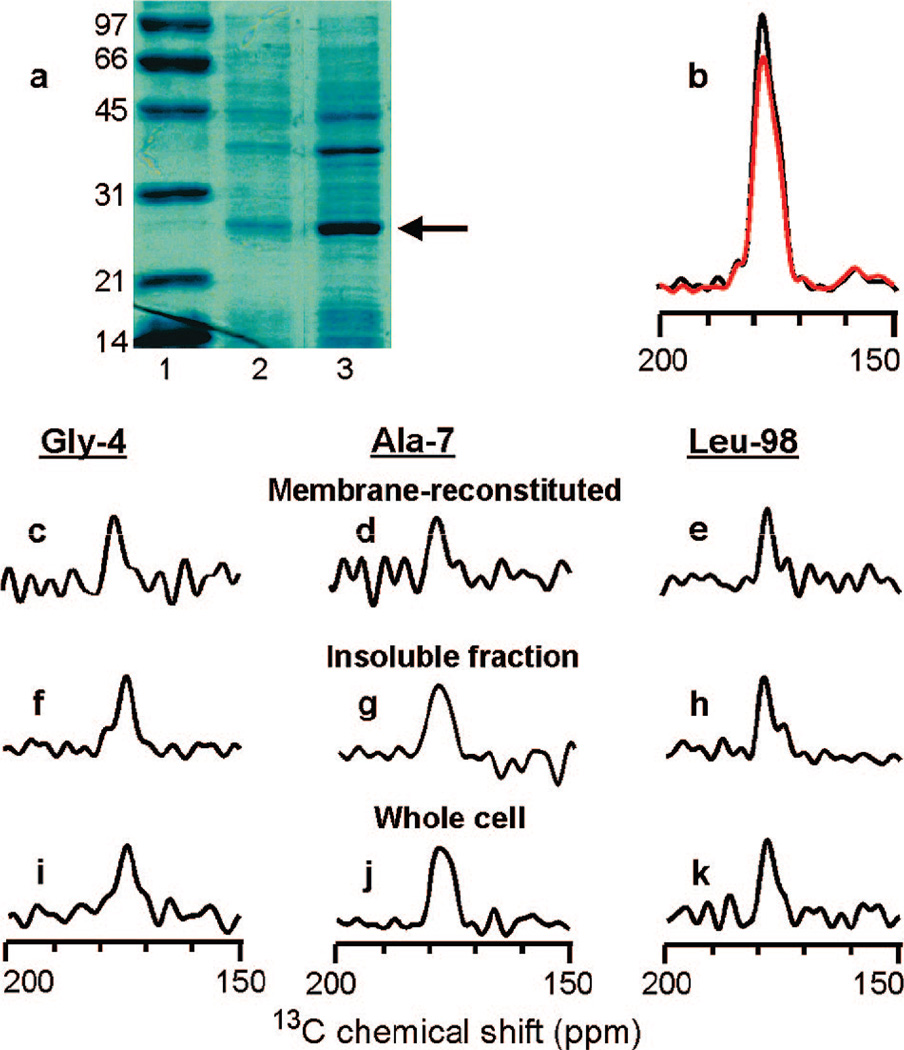
There was a significant amount of FHA2 in inclusion bodies as evidenced by a dominant FHA2 band in the SDS-PAGE gels of the unlysed fully hydrated E. coli cells and in the hydrated pellet formed from centrifugation of the insoluble material in the cell lysate, Figure 1a. In addition, ~10 mg/L more FHA2 was purified after sonication of the aforementioned pellet in 8 M urea, and this denatured FHA2 could be refolded in detergent and reconstituted in membranes.16 The relative band intensities in Figure 1a suggest that FHA2 is at least 10–20% of the total protein mass in the insoluble fraction and in unlysed whole cells and in the present study, the labeling/solid-state NMR method was applied to FHA2 in these two hydrated materials.17, 18 The 13CO NMR signals were dominated by FHA2 as evidenced by comparison of the “S0” (black) and “S1” (red) REDOR NMR spectra of the pellet for which the labeling targeted the Leu-98/Leu-99 unique sequential pair, Figure 1b. The S0 spectrum was composed of all the 13CO signal in the sample whereas the S1 spectrum lacked signal from 13CO nuclei directly bonded to 15N nuclei. The experimental S1/S0 integrated intensity ratio was 0.89 and correlated with the 0.92 ratio expected for fully labeled FHA2 with 13 Leus in the sequence. The experimental and (expected) ratios for labeling which targeted Gly-4 and Ala-7 were 0.93 (0.94) and 0.93 (0.91), respectively and were generally consistent with labeled amino acids being in the media only during the expression period and with FHA2 being the primary protein produced during the expression period.
Figure 1.
(a) SDS-PAGE gel: lane 1, molecular weight standards in kDa; lane 2, whole E. coli cells without lysis; and lane 3, the solid pellet obtained after centrifugation of the cell lysate. Different amounts of total protein were loaded in each lane and the arrow marks FHA2. (b-k) 13C solid-state NMR spectra of FHA2. Panel b is for the insoluble fraction Leu-98 sample and the S0 and S1 REDOR spectra are displayed in black and red, respectively. The remaining panels are S0 - S1 difference spectra. Example expression media contained 1-13C-Gly and 15N-Ala (for Gly-4) or 1-13C,15N-Leu (for Leu-98). The most complete labeling of inclusion body FHA2 was obtained with addition of 30 mg/L of each labeled amino acid at the start of induction and at the 1 and 2 h times during the 3 h induction period. Experimental NMR conditions included 9.4 T spectrometer, 40 µL active sample volume, and sample cooling gas at −20 °C. Each membrane-reconstituted sample contained ~5 mg FHA2 and each c-e spectrum was obtained in ~3 days, while each of the other spectra was obtained in ~1 1/2 days.
The S0 - S1 REDOR difference spectrum primarily represents the filtered 13CO signal of the N-terminal residue of the unique sequential pair and such spectra are presented in Figure 1 for (c, f, i) Gly-4, (d, g, j) Ala-7, or (e, h, k) Leu-98 and for FHA2 (c-e) purified and reconstituted in membranes, (f-h) in inclusion bodies in the insoluble cell lysate, or (i-k) in unlysed whole cells. There is general similarity of the spectra and peak 13CO chemical shifts of the membrane-reconstituted, inclusion body, and whole cell FHA2 protein, Figure 1c-k and Table 1. The similarity of the insoluble fraction and the whole cell spectra correlated with most of the FHA2 being in inclusion bodies. In the existing structures, a N-terminal helix extends to residue 10 in the fusion peptide and there is a helix that extends from residue 38 to residue 105. The peak 13CO shifts for Gly-1, Gly-4, Ala-7, and Leu-98 correlate with these helical conformations as evidenced by better agreement with distributions of backbone 13CO shifts in helical conformation (175.5 ± 1.2, 179.4 ± 1.3, and 178.5 ± 1.3 ppm for Gly, Ala, and Leu, respectively) than with characteristic shifts in β sheet/amyloid samples(170–172, 174–176, and 173–175 ppm, respectively).5, 6, 11, 19, 20 The Ala-7 spectra of the insoluble fraction and whole cell samples are broader than the other spectra which may indicate some conformational heterogeneity at this residue. Overall, the data support a model in which native helical conformation is retained at least for a significant fraction of inclusion body FHA2. It is interesting that dominant β sheet chemical shifts were not observed in the fusion peptide region because this region is hydrophobic and could potentially form amyloid aggregates in the absence of membranes. Although 13CO shifts are not absolutely definitive in determining conformation and are correlated with regions of the Ramachandran plot rather than precise dihedral angles, 13CO shifts do provide significant conformational information particularly when helical shifts at several nearby residues are obtained (as in the fusion peptide region) or when sharp signals (indicating structural order) with very similar helical shifts are obtained in both the natively folded and inclusion body protein (as with Leu-98).11
Table 1.
Peak 13C Carbonyl Chemical Shifts of FHA2 in ppm Units
| sample type | residue | |||
|---|---|---|---|---|
| Gly-1 | Gly-4 | Ala-7 | Leu-98 | |
| membrane-reconstituted | 174.8 | 177.2 | 179.1 | 178.8 |
| insoluble fraction | 174.7 | 174.5 | 178.9 | 178.6 |
| whole cell | 175.2 | 174.7 | 177.9 | 178.4 |
The approach presented in this paper has two significant advantages over the more commonly used IR approach for study of inclusion body structure. First, conformation at individual residues is straightforwardly studied with the NMR method but only overall protein conformation is typically obtained with the IR method. It should therefore be possible to develop more detailed structural models with the NMR data. Second, the inclusion body and whole cell samples are fully hydrated with the NMR method rather than being dehydrated with the IR method. A large number of specifically labeled samples are required for a high-resolution NMR structural model of inclusion body protein but depending on linewidths, it may be possible to obtain structural data for samples with more significant labeling using methods previously applied to NMR structure determination of microcrystalline, amyloid, and membrane-associated proteins.5, 6, 19–21 In summary, this study describes a general approach for residue-specific structural analysis of recombinant protein in inclusion bodies including those in whole E. coli cells as well as evidence for native conformation at some specific residues of a particular inclusion body protein.
Supplementary Material
Acknowledgment
The FHA2 plasmid was provided by Yeon-Kyun Shin and financial support was provided by Michigan State.
Footnotes
Supporting Information Available: FHA2 sequence and descriptions of cell growth and labeling, sample preparation, and NMR methods and analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Baneyx F, Mujacic M. Nat. Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 2.Przybycien TM, Dunn JP, Valax P, Georgiou G. Protein Eng. 1994;7:131–136. doi: 10.1093/protein/7.1.131. [DOI] [PubMed] [Google Scholar]
- 3.Oberg K, Chrunyk BA, Wetzel R, Fink AL. Biochemistry. 1994;33:2628–2634. doi: 10.1021/bi00175a035. [DOI] [PubMed] [Google Scholar]
- 4.Ami D, Natalello A, Gatti-Lafranconi P, Lotti M, Doglia SM. FEBS Lett. 2005;579:3433–3436. doi: 10.1016/j.febslet.2005.04.085. [DOI] [PubMed] [Google Scholar]
- 5.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ritter C, Maddelein ML, Siemer AB, Luhrs T, Ernst M, Meier BH, Saupe SJ, Riek R. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrio M, Gonzalez-Montalban N, Vera A, Villaverde A, Ventura S. J. Mol. Biol. 2005;347:1025–1037. doi: 10.1016/j.jmb.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 8.Umetsu M, Tsumoto K, Ashish K, Nitta S, Tanaka Y, Adschiri T, Kumagai I. FEBS Lett. 2004;557:49–56. doi: 10.1016/s0014-5793(03)01441-8. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Fruitos E, Aris A, Villaverde A. Appl. Environ. Microbiol. 2007;73:289–294. doi: 10.1128/AEM.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gullion T, Schaefer J. J. Magn. Reson. 1989;81:196–200. [Google Scholar]
- 11.Zhang HY, Neal S, Wishart DS. J. Biomol. NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 12.Curtis-Fisk J, Preston C, Zheng ZX, Worden RM, Weliky DP. J. Am. Chem. Soc. 2007;129:11320–11321. doi: 10.1021/ja073644g. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Skehel JJ, Wiley DC. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han X, Bushweller JH, Cafiso DS, Tamm LK. Nat. Struct. Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 15.Murphy OJ, 3rd, Kovacs FA, Sicard EL, Thompson LK. Biochemistry. 2001;40:1358–1366. doi: 10.1021/bi0015109. [DOI] [PubMed] [Google Scholar]
- 16.Curtis-Fisk J, Spencer RM, Weliky DP. Protein Expression Purif. doi: 10.1016/j.pep.2008.06.009. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serber Z, Selenko P, Hansel R, Reckel S, Lohr F, Ferrell JE, Wagner G, Dotsch V. Nat. Protoc. 2006;1:2701–2709. doi: 10.1038/nprot.2006.181. [DOI] [PubMed] [Google Scholar]
- 18.Kim SJ, Cegelski L, Stueber D, Singh M, Dietrich E, Tanaka KSE, Parr TR, Far AR, Schaefer J. J. Mol. Biol. 2008;377:281–293. doi: 10.1016/j.jmb.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siemer AB, Ritter C, Ernst M, Riek R, Meier BH. Angew. Chem. Int. Ed. 2005;44:2441–2444. doi: 10.1002/anie.200462952. [DOI] [PubMed] [Google Scholar]
- 20.Qiang W, Bodner ML, Weliky DP. J. Am. Chem. Soc. 2008;130:5459–5471. doi: 10.1021/ja077302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.