Abstract
Purpose
Acute myeloid leukemia (AML) is the most common acute leukemia in adults and is often resistant to conventional therapies. The MYC oncogene is commonly overexpressed in AML but has remained an elusive target. We aimed to examine the consequences of targeting MYC both directly and indirectly in AML overexpressing MYC/Myc due to trisomy 8/15 (human/mouse), FLT3-ITD mutation, or gene amplification.
Methods
We performed in vivo knockdown of Myc (shRNAs) and both in vitro and in vivo experiments using four drugs with indirect anti-MYC activity: VX-680, GDC-0941, artemisinin, and JQ1.
Results
shRNA knockdown of Myc in mice prolonged survival, regardless of the mechanism underlying MYC overexpression. VX-680, an aurora kinase inhibitor, demonstrated in vitro efficacy against human MYC-overexpressing AMLs regardless of the mechanism of MYC overexpression, but was weakest against a MYC-amplified cell line. GDC-0941, a PI3-kinase inhibitor, demonstrated efficacy against several MYC-overexpressing AMLs, although only in vitro. Artemisinin, an antimalarial, did not demonstrate consistent efficacy against any of the human AMLs tested. JQ1, a bromodomain and extra-terminal bromodomain inhibitor, demonstrated both in vitro and in vivo efficacy against several MYC-overexpressing AMLs. We also confirmed a decrease in MYC levels at growth inhibitory doses for JQ1, and importantly, sensitivity of AML cell lines to JQ1 appeared independent of the mechanism of MYC overexpression.
Conclusions
Our data support growing evidence that JQ1 and related compounds may have clinical efficacy in AML treatment regardless of the genetic abnormalities underlying MYC deregulation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00280-015-2766-z) contains supplementary material, which is available to authorized users.
Keywords: MYC, AML, Myeloid leukemia, JQ1, BRD4
Introduction
Acute myeloid leukemia (AML) accounts for 80 % of adult acute leukemias [1], and the primary treatment for patients remains combination chemotherapy. However, the rate of complete response to induction chemotherapy falls below 50 % for elderly patients [2]. Even in younger patients, 40 % do not survive beyond two years following diagnosis [3] and multidrug resistance is seen in 33–57 % of AML cases [4]. The discovery of all-trans retinoic acid as an effective therapy for acute promyelocytic leukemia (APL) revolutionized the treatment of this unique AML subtype [5], but new molecularly targeted therapies are needed to improve the prognosis and treatment of AML more generally.
MYC is an attractive target for cancer therapeutics due to its regulation by multiple, converging signaling cascades. MYC is a transcription factor of the helix-loop-helix–leucine zipper family that regulates many cellular processes, including proliferation, cell cycle progression, differentiation, and apoptosis [6, 7]. Following dimerization with MAX, MYC binds to target E-box sequences in the regulatory regions of many target genes [8], and appropriate MYC levels are therefore critical to ensure normal cell fate decisions. Deregulation of MYC may result in uncontrolled cell proliferation, immortalization, growth factor independence, genomic instability, and escape from immune surveillance [6]. In addition, MYC has been shown to inhibit myeloid cell differentiation [9] and is found overexpressed or amplified in many hematologic and solid malignancies (reviewed in [6, 7]). In animal models, transduction of unfractionated murine bone marrow (BM) cells with Myc results in AML development [10], and expression of inducible anti-Myc short hairpin RNA (shRNA) in leukemic cells leads to their depletion both in vitro and in vivo [11]. Similarly, expression of a human MYC transgene causes AML in mice, and inactivation of the same transgene causes sustained tumor regression [12].
Various mechanisms can account for MYC overexpression in AML, including trisomy 8 resulting in single copy gain of MYC, MYC amplification, and deregulated expression due to an upstream mutation (for example in FLT3, a gene encoding a receptor tyrosine kinase); 9 % of AMLs are characterized by trisomy 8, making it the most common chromosomal abnormality in human AML [13]. This gain leads to increased MYC levels in such leukemias [14], and importantly, our laboratory showed that MYC contributes to the pathogenic effect of trisomy 8 [14]. MYC amplifications are more occasional, with double minute chromosomes observed in 1 % of cases [15]. Double minute chromosomes in AML most often include MYC [15], and this amplification correlates with higher MYC expression and poorer prognosis [16]. Common mutations upstream of MYC in AML include FLT3 activating mutations, present in 25–30 % of AML patients and associated with a poor prognosis [17]. As FLT3 is found upstream of several leukemogenic pathways including Ras and PI3-kinase [18], such mutations could be anticipated to stimulate MYC mRNA expression or stabilize MYC protein.
Our laboratory isolated transplantable mouse leukemias that arose in the hMRP8-PML/RARA transgenic line [17]. In this model, the human MRP8 promoter controls PML/RARA, the fusion gene hallmark of APL, driving its expression in maturing myeloid progenitors, neutrophils, and monocytes. Using these transgenic mice, we identified and generated leukemias that model MYC overexpression via three different mechanisms: single copy gain (resulting from trisomy 15), retroviral expression of an activating mutation of FLT3, and retroviral overexpression of MYC as a model of gene amplification. Murine trisomy 15, syntenic to trisomy 8 in humans, is the most common recurring abnormality in this model, paralleling the human data [17]. Importantly, retroviral overexpression of MYC suppresses gain of chromosome 15, suggesting that trisomy 15 largely functions to increase MYC levels [17].
Despite its importance in leukemogenesis, MYC has been difficult to target pharmacologically. Most small molecule inhibitors interrupt the MYC:MAX interaction but have shown only mixed results (reviewed in [6]). However, several molecules have demonstrated efficacy in MYC-overexpressing malignancies through indirect targeting of MYC (reviewed in [6, 7]). In this study, we followed a similar strategy to investigate the consequences of targeting MYC indirectly on the growth of AML, using the four compounds described below:
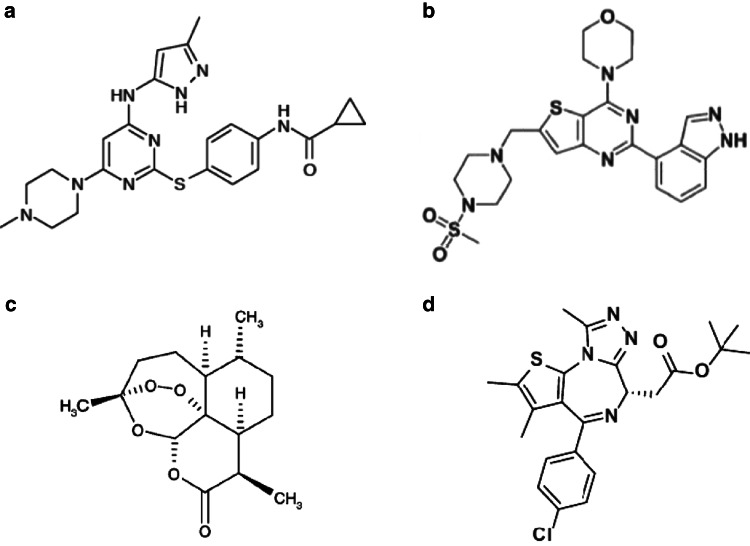
VX-680 (Fig. 1a) belongs to the family of aurora kinase inhibitors [19], which have demonstrated synthetic lethal interactions with overexpressed MYC in prior studies [20, 21]. VX-680 has demonstrated efficacy against AML both in vitro and in vivo [19] and has been recently investigated in a phase I clinical trial for non-AML leukemia patients (Merck, NCT00111683). VX-680 inhibits aurora-A and aurora-B kinases in a p53-independent manner [20], and preliminary data show that it extends survival of mice transplanted with MYC-overexpressing APLs (unpublished data from M. Bishop’s laboratory, University of California San Francisco).
Fig. 1.
Chemical structures of compounds investigated in our study. a VX-680 [19]. b GDC-0941 [22]. c Artemisinin [24]. d JQ1 [26]
GDC-0941 (Fig. 1b) targets and inhibits PI3-kinase [22], a receptor at the top of a key signaling cascade composed of many downstream effectors, including MYC [23]. When the pathway is activated, the inhibitory phosphorylation of MYC by GSK-3β is released, allowing MYC to be stabilized and translate its effects [23]. Illustrating the broad possible applications of such compound, GDC-0941, is currently undergoing a phase I clinical trial in non-Hodgkin lymphoma and solid cancer patients (Genentech, NCT00876122).
Artemisinin (Fig. 1c) has traditionally been used for its antimalarial properties [24], but recent studies suggest that its use could be extended to tumor treatment [25]. Dihydroartemisinin, the principal active metabolite of artemisinin, has been shown to lead to MYC degradation and induce apoptosis of MYC-overexpressing cells [25] in a GSK-3β-dependent manner.
JQ1 (Fig. 1d) is an inhibitor of the bromodomain and extra-terminal bromodomain (BET) protein BRD4 [26, 27]. Interestingly, JQ1 can also attenuate solid tumor growth without affecting MYC levels, indicating additional MYC-independent effects in some settings [28]. The efficacy of JQ1 has been demonstrated against AML in vitro and in vivo [29], but no clinical trials have been initiated to date.
We hypothesized that the mechanism of MYC overexpression would influence the response to the various MYC-targeting compounds described above, as well as the response to direct MYC-targeting shRNAs. Such information would be useful to stratify patients according to their underlying genetic lesions and personalize AML therapy.
Materials and methods
Animals
FVB/n mice were bred and maintained at University of California, San Francisco (UCSF), and their care was in accordance with Institutional Animal Care and Use Committee guidelines.
Retrovirus production
shRNAs against MYC/Myc were previously validated (Lowe laboratory). BOSC23 cells were transfected by gently adding a solution containing CaCl2, HBS (pH 7.05), pCL-Eco (helper plasmid), and the shRNA-containing vectors (see “Materials and Methods” in electronic supplementary material for cloning details). After 7 h at 37 °C 8 % CO2, the transfection mixture was replaced with fresh media and the plate returned to the incubator. After 48 h, the retrovirus-containing supernatant was harvested, filtered (0.2 μm), and frozen. Three distinct anti-Myc and two control (anti-Renilla luciferase and anti-Rpa3) retroviruses were produced.
Retroviral transduction
Two independent cryopreserved leukemias were transplanted retro-orbitally into sublethally irradiated (4.5 Gy) recipients: leukemia 1111 (PML/RARA with trisomy 15, designated as “PR/+15”) and leukemia 1127 (PML/RARA with activated FLT3, designated as “PR/FLT3RV”). Upon euthanasia of sick animals, fresh leukemic bone marrow and spleen cells (in a 1:1 ratio when possible) were harvested, passed through a 70-μm strainer and plated at 1 × 106 cells/well. After spinning, the supernatant was removed, and 1 mL of retrovirus containing 5 % of IL-3 and IL-6 conditioned media and 4 μL of 2 mg/mL polybrene were added/well. The plate was centrifuged at 2500 rpm for 90 min at room temperature and the supernatant replaced with Myelocult M5300 (StemCell Technologies#05300) containing 5 % of IL-3 and IL-6 conditioned media. After 24 h, the transduction procedure was repeated, following which cells were harvested and isolated by flow cytometry.
Fluorescence activated cell sorting (FACS)
Cells were double-sorted on a BD Biosciences FACSAriaIII. Doublets were eliminated, and DAPI− (Invitrogen cat#D3571, 1:60,000) and mCherry+ (and GFP+, if applicable) cells were isolated (purity > 95 %); 7000 transduced cells/animal were transplanted back into sublethally irradiated FVB/n CD45.2 recipients following previously described protocols [30, 31].
Statistics
Survival curves were generated using Prism software (GraphPad) and compared using log-rank analysis. Student’s unpaired t test was used to compare the mean percentage of mCherry+ cells in BM of euthanized recipients.
Cell lines origin, culture conditions, and doubling time measurement
BOSC23 cells were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) containing 10 % FBS; 5637 cells were maintained in RPMI containing 10 % FBS.
MOLM-14, MV4-11, and HL-60 cells were obtained from Dr. Neil Shah’s laboratory at UCSF, which confirmed the presence of FLT3-ITD in MOLM-14 and MV4-11, and NRASQ61 in HL-60. These lines were maintained in RPMI containing 10 % FBS. MUTZ-2, OCI-AML5, AP-1060, and FKH-1 cells were authenticated by and ordered from DSMZ (http://www.dsmz.de/). FKH-1 cells were maintained in RPMI containing 20 % FBS. MUTZ-2 cells were maintained in alpha-MEM containing 20 % FBS and 20 % conditioned medium from cell line 5637. OCI-AML5 cells were maintained in alpha-MEM containing 20 % FBS and 10 % conditioned medium from cell line 5637. AP-1060 cells were maintained in Iscove’s MDM containing 20 % FBS and 10 % conditioned medium from cell line 5637. Penicillin–streptomycin and l-glutamine were added to all the cell line media described above. Cell lines were split 2–3 times per week, using split ratios available at http://www.dsmz.de/, and passaged in the Kogan laboratory for less than 6 months. Cell numbers were plotted against time, and a best-fit exponential equation was used to calculate the doubling time for each cell line.
Drug handling
For in vitro experiments, VX-680 and GDC-0941 (free base) were ordered from LC Laboratories (http://www.lclabs.com/); 99 % of pure artemisinin was ordered from ebiochem (http://www.ebiochem.com/product/artemisinin-99-uv-8157). JQ1 was shipped directly from the laboratory of James Bradner, MD at the Dana-Farber Cancer Institute. Doxorubicin was obtained from the UCSF inpatient pharmacy. For in vivo experiments, GDC-0941 (free base) was obtained directly from the laboratory of Kevin Shannon, MD, and JQ1 was obtained directly from the Bradner laboratory.
In vitro drug assay and GI50 calculation
To measure the effect of drugs on cell growth, MOLM-14, MV4-11, HL-60, OCI-AML5, and FKH-1, cells were plated in 96-well plates at 10,000 cells/well in 100 μL. For MUTZ-2 and AP-1060, cells were first grown in 24-well plates at 500,000 cells/well in 1 mL before being transferred to 96-well plates in a 1:4 dilution. Drugs were prepared at 60 mM in DMSO (or 600 μM in saline for doxorubicin) and further diluted in half-log serial dilutions. Plates were incubated at 37 °C and 5 % CO2, and cell growth measured using the CellTiter-Glo assay (Promega cat#G7570) after two doubling times, per manufacturer’s instructions. Luminescence was measured on a Synergy 2 Multi-Mode Microplate Reader, Biotek model. Relative light units were plotted against drug concentration, and a best-fit logistic curve generated using a four-parameter sigmoidal dose–response model. Data from two independent experiments were averaged to generate the GI50.
To measure the effect of drugs on MYC levels, cells were plated in six well plates at 3 × 106 cells/well and treated as described above for one doubling time. In addition to an untreated control, two drug concentrations were chosen (unless specified in the figure): lower (nearest half-log unit below the calculated GI50) and higher (nearest half-log unit above the calculated GI50).
Western blots
Following one doubling time of exposure, cells were washed three times with PBS and lysed with protease inhibitors-containing RIPA buffer. After measuring protein concentration using the Bio-Rad DC protein assay, 40 ug of protein was loaded with NuPage LDS sample buffer and NuPage reducing agent, onto a 4–12 % Bis–Tris gradient gel. Protein extracts were transferred to a nitrocellulose membrane, which was blocked with a TBST 5 % milk solution. Anti-c-MYC was added at 1:10,000 (Epitomics cat#1472-1, rabbit monoclonal), followed by goat anti-rabbit IgG-HRP (Santa Cruz cat#SC-2054) at 1:10,000. To detect β-actin, anti-actin (Sigma Aldrich cat#A2228, mouse) was added at 1:10,000, followed by a goat anti-mouse IgG-HRP (Santa Cruz cat#SC-2055) at 1:10,000. Details on imaging and calculations are provided in the "Materials and Methods" in the electronic supplementary material.
In vivo GDC-0941 experiments
Vehicle solution was prepared with 0.5 % hydroxypropyl methylcellulose in ddH2O, to which 500× Tween 80 was added. Each day, a 12.5 mg/mL solution of GDC-0941 was prepared in the above vehicle. In vivo experiments were conducted in collaboration with the UCSF Helen Diller Family Comprehensive Cancer Center Preclinical Therapeutics Core. Groups of five FVB/n mice were sublethally irradiated (4.5 Gy) and retro-orbitally injected with the following four murine leukemias: PR/+15 and PR/FLT3RV (see the “Retroviral transduction” section for description), leukemia#478 (resulting from retroviral MYC overexpression, designated as “PR/MYCRV”), and leukemia#214 (constitutively stable MYC control, resulting from retroviral T58A mutant MYC overexpression, designated as “PR/MYCT58ARV”). Mice were treated by oral gavage at 10 μL/g of body weight (125 mg/kg) for 21 days beginning on day 14 post-injection. Pharmacokinetics for GDC-0941 at the dose and administration route utilized in our study has been previously described [22]. Control mice were treated with vehicle. Mice were euthanized upon showing signs of morbidity, and necropsy performed to confirm the presence of leukemia.
In vivo JQ1 experiments
Vehicle solution was prepared with 10 % hydroxypropyl-beta-cyclodextrin. Each day, a 5 mg/mL solution of JQ1 was prepared in the above vehicle. In vivo experiments and euthanasia were performed as described above. Mice were treated by intra-peritoneal injections at 50 mg/kg of animal for 21 days beginning on day 14 post-injection. Pharmacokinetics for JQ1 at the dose and administration route utilized in our study have been previously described [32]. For leukemia 1111 (PR/+15), one animal required euthanasia following a JQ1 intraperitoneal injection and was censored on the date of death. For leukemia 478 (PR/MYCRV), three animals in the placebo-treated group and three animals in the JQ1-treated group developed leukemia and were included in the survival analysis (two animals in each group failed to engraft with leukemic cells).
Results
Myc knockdown in Myc-overexpressing AMLs prolongs survival in recipient mice and prevents AML cells from predominating in the BM
To determine whether Myc knockdown prolongs survival in mice injected with MYC-overexpressing AMLs, leukemic cells isolated from the hMRP8-PML/RARA model were transduced with an anti-Myc shRNA-containing retrovirus [11]. Leukemias 1111 (“PR/+15,” overexpressing MYC through trisomy 15) and 1127 (“PR/FLT3RV,” overexpressing MYC via constitutive FLT3 activation) were passaged into recipient animals. Leukemic BM and spleen were harvested fresh from sick animals and transduced with mCherry-tagged Myc or control shRNAs [11]. mCherry+ cells were double-sorted (mCherry+/GFP+ cells in the case of the PR/FLT3RV leukemia) and injected into sublethally irradiated 45.2 or FVB/n recipients. Mice were euthanized upon showing signs of morbidity, or 15 weeks post-transplant, and BM was assessed for the presence of mCherry+ (and GFP+ if applicable) cells.
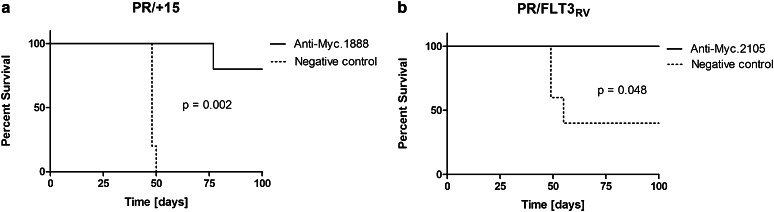
Mice injected with AMLs transduced with anti-Myc shRNA survived longer than mice injected with AMLs transduced with anti-Renilla luciferase shRNA (Fig. 2a, b), consistent with previous data showing that Myc shRNA knockdown could prolong survival in mice injected with murine AML [11]. Furthermore, the effect of MYC knockdown paralleled the survival benefit observed in mice injected with AMLs transduced with anti-Rpa3 shRNA (data not shown), as Rpa3 is an essential gene required for DNA replication [33]. BM from mice injected with anti-Myc-transduced AML contained very few mCherry+ cells at euthanasia, while BM from mice injected with anti-Renilla-transduced AML contained a high percentage of mCherry+ cells (data not shown), indicating that AMLs transduced with anti-Myc shRNA were less able to predominate in vivo. These data suggested that various MYC-overexpressing AMLs may respond to anti-MYC therapy and prompted exploration of MYC inhibition in human AML cell lines.
Fig. 2.
Myc knockdown prolongs survival in mice transplanted with Myc-overexpressing AML. a Kaplan–Meier curve comparing survival of recipient mice injected with leukemia 1111, a PML/RARA leukemia with trisomy 15 (designated as ‘PR/+15’), after shRNA knockdown against Myc (or control) and isolation by flow cytometry. N = 5 mice per group. b, Kaplan–Meier curve comparing survival of recipient mice injected with leukemia 1127, a PML/RARA leukemia obtained by retroviral transduction of PML/RARA BM with activated FLT3 (designated as ‘PR/FLT3RV’), after shRNA knockdown and isolation by flow cytometry. Note that not all animals in the group receiving PR/FLT3RV cells transduced with control vector developed leukemia, likely reflecting the low numbers of cells transplanted. N = 5 mice per group
VX-680, GDC-0941 and JQ1 inhibit growth of MYC-overexpressing human AMLs in vitro, while artemisinin does not
To examine the benefit of personalized therapy for human MYC-overexpressing AMLs, it was important to determine whether various human AMLs would respond differently to putative anti-MYC therapies according to their underlying genetic lesion. We obtained five human AML cell lines displaying deregulated MYC genes, designated herein as “MYC-overexpressing” (OCI-AML5, MUTZ-2, MOLM-14, MV4-11, and HL-60), as well as two control cell lines not known to overexpress MYC (AP-1060 and FKH-1). Mimicking clinical situations, the five AML cell lines had various genetic abnormalities underlying MYC overexpression: single copy gain via trisomy 8 (OCI-AML5 and MUTZ-2), FLT3-ITD mutation and trisomy 8 (MOLM-14 and MV4-11), or MYC amplification (HL-60). Given that anti-MYC shRNA is not currently a viable AML treatment and that clinical utility of direct MYC inhibitors has not been demonstrated, we proceeded to test the effect on AML growth of four molecules with indirect anti-MYC activity (VX-680, GDC-0941, artemisinin, and JQ1, described in the "Introduction"). We exposed the cell lines to varying drug concentrations and measured cell growth (Figure S1 in electronic supplementary material) and MYC protein levels (Fig. 3 and Figures S2b–d in electronic supplementary material). Importantly for each cell line, we standardized the length of exposure to two doubling times of unexposed cells for cell growth measurements and to one doubling time for MYC levels by western blot (Supplementary Methods, data not shown). Growth inhibition of 50 % (GI50) was calculated for each drug–AML pairing (Table S1 and Figures S3a–g in electronic supplementary material).
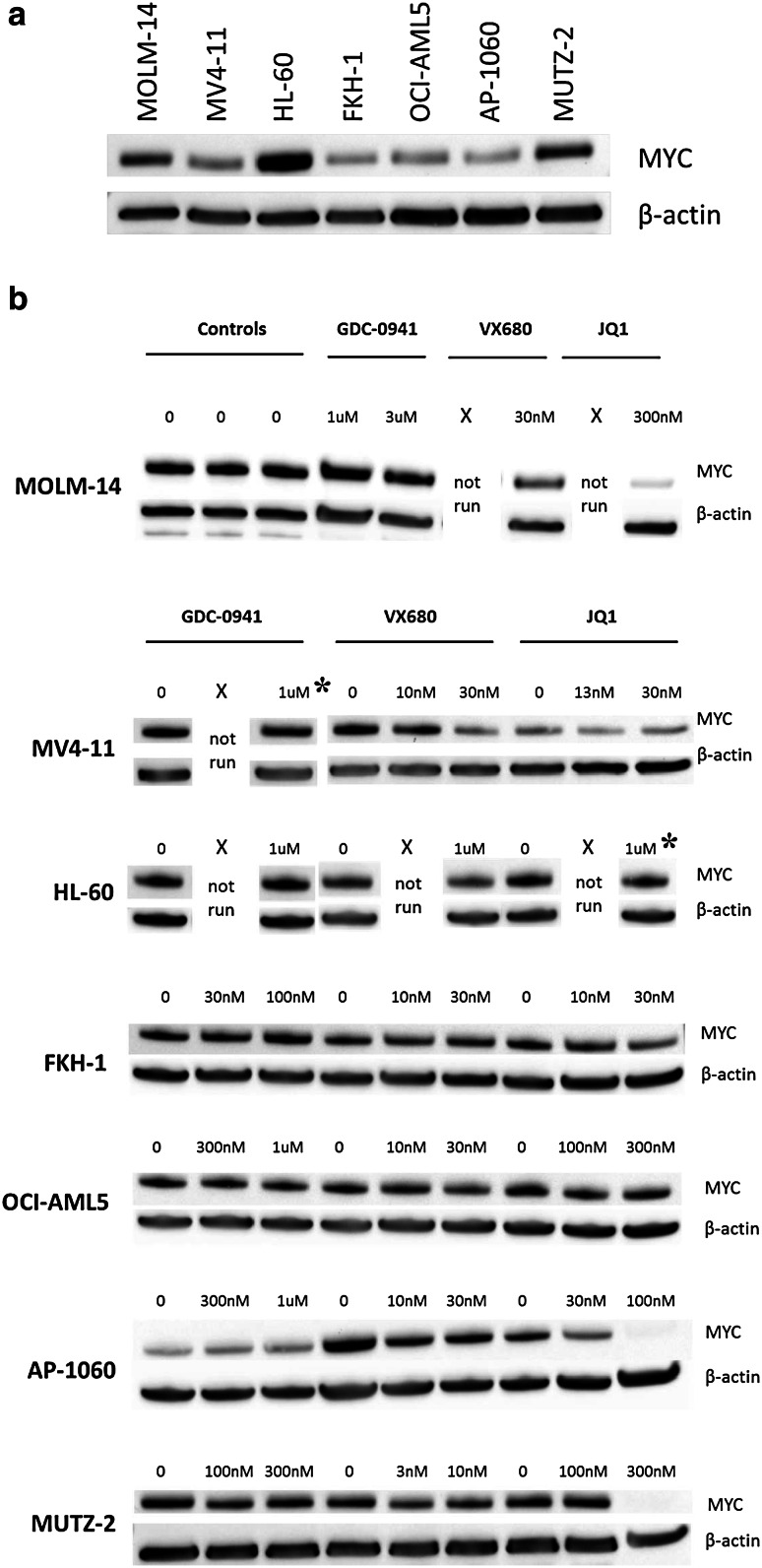
Fig. 3.
Levels of MYC protein observed in the various cell lines, at basal levels and after exposure to GDC-0941, VX-680, or JQ1. a Western blots demonstrating basal MYC levels observed in all cell lines, untreated. Normalized levels to those seen in AP-1060 (lowest expressing cell lines) are shown in Figure S3a in electronic supplementary material. b MYC levels following drug treatment at concentrations spanning the GI50 (unless specified, see asterisk below). Note: in a few instances, only one drug concentration was investigated (above the GI50 in all cases). Asterisk in these two cases, the drug concentration tested is not immediately above the GI50 but rather another half-log unit higher, yet no decrease in MYC levels is observed at these higher concentrations (for MV4-11, GI50 for GDC-0941 was 212.7nM; for HL-60, GI50 for JQ-1 was 250.1nM)
Untreated, the control AP-1060, cell line exhibited the lowest levels of MYC as compared to the other cell lines. The other control cell line FKH-1 showed a nearly twofold increase in MYC levels compared to AP-1060. Further illustrating the variation in baseline MYC levels, OCI-AML5, a cell line with trisomy 8, showed MYC levels slightly higher than AP-1060, but below that of FKH-1. HL-60, a cell line with MYC amplification, had the highest MYC levels of all cell lines investigated (Fig. 3a and Figure S2a in electronic supplementary material). Thus, baseline overexpression of MYC in these cell lines did not appear to correlate with mechanisms of overexpression, apart from the multiple copy gain in HL-60 which associated with the highest MYC protein levels of the cell lines tested.
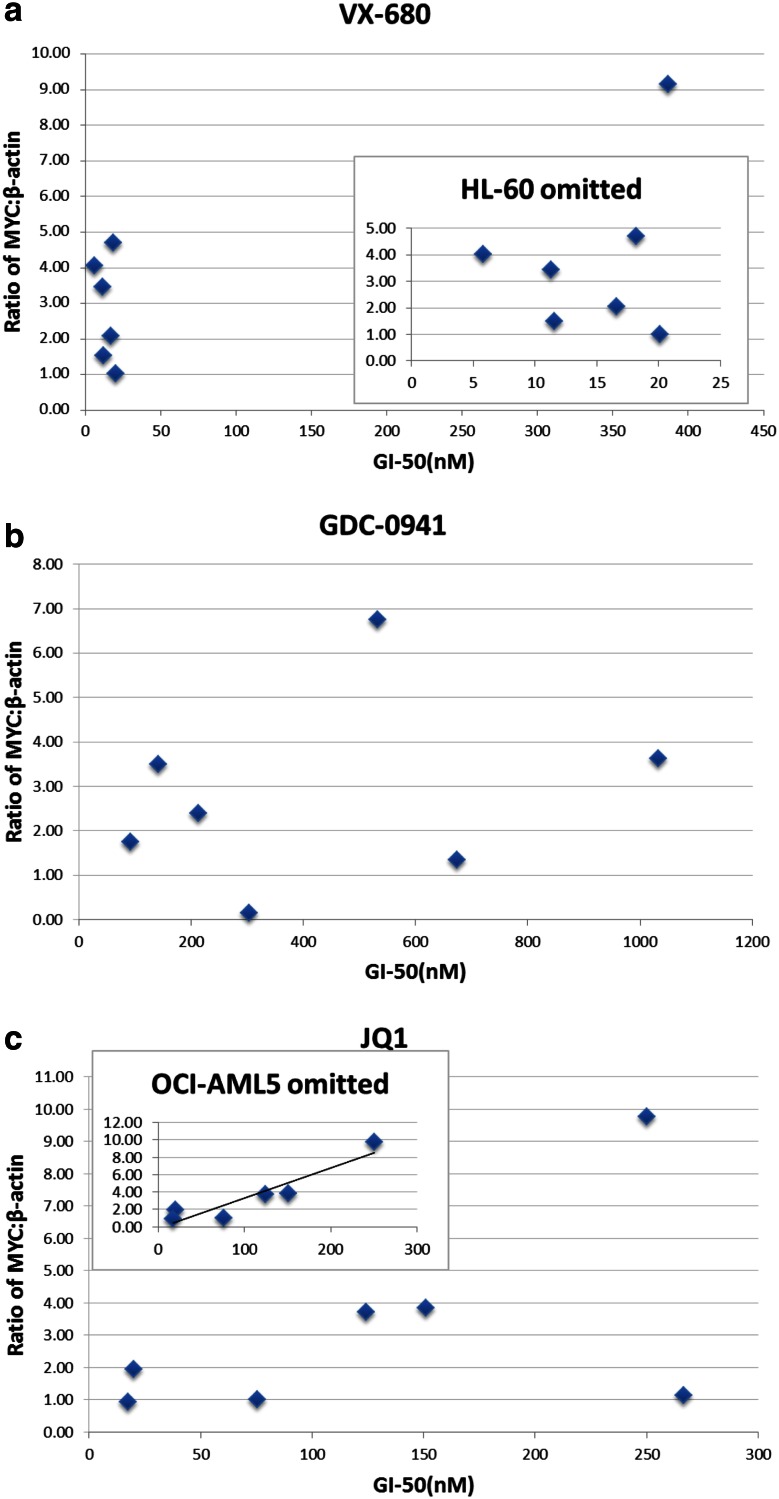
The GI50s calculated for each cell line and drug are shown in Tables S1 and S2 (in electronic supplementary material). GI50s for VX-680 were less than or equal to 20 nM for all cell lines except HL-60, which was less than 400 nM, far below the clinically achievable plasma concentration of 5 μM [34]. Of note, there was no correlation observed between drug efficacy and mechanism of MYC overexpression. Given prior work suggesting that VX-680 exhibits synthetic lethality in the presence of MYC overexpression, we did not anticipate a decrease in MYC protein levels near the GI50s, but in fact did see such a decrease for the two FLT3 positive cell lines, MOLM-14 and MV4-11 (Fig. 3b and Figure S2b in electronic supplementary material), which might reflect FLT3 kinase inhibition by VX-680 (see “Discussion”). Notably, although there was no overall correlation between sensitivity to VX-680 and baseline MYC protein levels (Fig. 4a), HL-60, which had the highest MYC protein level of the cell lines tested, appeared to be the most resistant to VX-680.
Fig. 4.
MYC levels of untreated samples were plotted against calculated GI50s for VX-680, GDC-0941, and JQ1 to assess the correlation between MYC expression level and drug sensitivity. a HL-60 differed markedly from the other cell lines in regard to sensitivity to VX-680. When HL-60 is omitted, no correlation was observed between GI50 and baseline MYC protein level (Inset, R 2 = 0.12). b No correlation was observed between GI50 and baseline MYC protein level when cell lines were treated with GDC-0941 (R 2 = 0.07). c A weak correlation was observed between GI50 and baseline MYC protein level when cell lines were treated with JQ1 (R 2 = 0.29), trending toward higher GI50 with higher baseline MYC level. OCI-AML5 appeared as an outlier, and a marked correlation emerged when omitting this cell line (Inset, R 2 = 0.86)
GI50s for GDC-0941 were less than or equal to 1 μM for all cell lines, also far below the preclinically achievable plasma concentration of 10 μM [22]. A clinical trial assessing oral bioavailability of GDC-0941 in humans has been conducted (NCT01240226), but results are not yet available. Although GDC-0941 demonstrated in vitro efficacy across all cell lines, there was no correlation observed between drug efficacy and mechanism of MYC overexpression (Tables S1 and S2 in electronic supplementary material), and GDC-0941 exposure at levels above the GI50 did not result in a decrease in MYC protein levels in six of the seven cell lines (Fig. 3b and Figure S2c in electronic supplementary material), indicating that the growth inhibitory effects in these six lines are not mediated through decrease in MYC. Further, there was no correlation between sensitivity to GDC-0941 and baseline MYC protein levels (Fig. 4b).
GI50s for artemisinin were less than 2 μM for HL-60 and MV4-11, but above 30 μM for the remaining cell lines, which was the highest drug concentration tested. Importantly, clinically achievable concentrations of artemisinin are approximately of 2 μM (or 530 ng/mL) [35], and given the poor in vitro efficacy observed, western blot data were not generated for artemisinin-treated cell lines.
GI50s for JQ1 were less than 300 nM for all cell lines investigated. Clinically achievable JQ1 concentration has not been verified. As GI50 concentrations were crossed, a decrease in MYC levels was observed, which was >twofold in four of the seven cell lines (Fig. 3b and Figure S2d in electronic supplementary material). Thus, JQ1 demonstrated in vitro efficacy across all cell lines in association with decreasing MYC protein levels. Although there was no correlation observed between drug efficacy and mechanism of MYC overexpression, some correlation was observed between drug efficacy and basal MYC protein level; overall, higher protein level appeared to correlate with decreasing sensitivity to JQ1 (Fig. 4c). OCI-AML5 is an outlier, exhibiting the highest measured GI50 for JQ1 despite having among the lowest measured MYC protein level and showing minimal decrease in MYC level with JQ1 treatment. Removing this datapoint strengthened the correlation between basal MYC protein level and JQ1 resistance (Fig. 4c inset, R2 = 0.86).
We also performed these experiments using doxorubicin, a cytotoxic chemotherapeutic agent with clinically achievable levels of 100 nM [36]. GI50s for all cell lines were less than or equal to 40 nM (Tables S1 and S2 in electronic supplementary material).
Given our results, the termination of clinical trials using VX-680 due to QTc prolongation and a general lack of encouraging clinical results with that drug [37], we chose to proceed with preclinical in vivo studies using GDC-0941 and JQ1.
GDC-0941 does not prolong survival in mice transplanted with MYC-overexpressing AMLs
We wished to determine whether the efficacy of GDC-0941 against MYC-overexpressing AMLs we observed in vitro (Table S1 in electronic supplementary material) was also observed in vivo. Groups of five sublethally irradiated mice were transplanted with one of the four murine AMLs: PR/+15, PR/FLT3RV, PR/MYCRV, and PR/MYCT58ARV (see “Introduction” and “Materials and Methods” for descriptions of these leukemias). Starting 14 days after AML injection, mice were treated with GDC-0941 or vehicle for 21 days. Mice treated with GDC-0941 did not survive longer than mice treated with vehicle for any of the four leukemias (data not shown).
JQ1 prolongs survival in mice transplanted with MYC-overexpressing AMLs
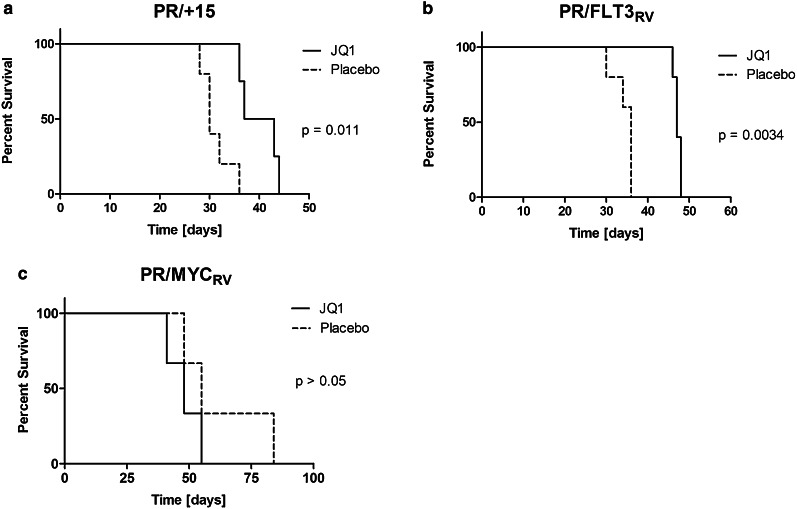
After showing that targeting MYC in murine MYC-overexpressing AMLs prolongs survival (Fig. 2) and that JQ1 demonstrated efficacy against several MYC-overexpressing AML cell lines (Table S1 in electronic supplementary material), we proceeded with preclinical studies using JQ1 in three murine AMLs with mechanisms of MYC overexpression paralleling those seen in human AML. Groups of sublethally irradiated mice were injected with the above AMLs. Starting 14 days after injection, mice were treated with JQ1 or placebo for 21 days.
Mice treated with JQ1 survived significantly longer than mice treated with placebo, whether injected with PR/+15 leukemia (Fig. 5a, P = 0.011) or PR/FLT3RV leukemia (Fig. 5b, P = 0.0034). Of interest, in the group injected with PR/MYCRV, mice treated with JQ1 did not live longer than mice treated with placebo (Fig. 5c, P > 0.05). This result is expected since in the PR/MYCRV leukemia, MYC expression is driven by a heterologous promoter, whereas JQ1 activity relies predominantly on downregulation at the endogenous MYC promoter. The PR/MYCT58ARV leukemia did not engraft sufficiently for study in these JQ1 experiments.
Fig. 5.
JQ1 prolongs survival in mice transplanted with Myc-overexpressing AMLs. a Kaplan–Meier curve comparing survival of FVB/n mice transplanted with PR/+15 leukemia treated for 21 days with JQ1 or placebo (N = 5 per group). b Kaplan–Meier curve comparing survival of FVB/n mice transplanted with PR/FLT3RV leukemia treated for 21 days with JQ1 or placebo (N = 5 per group). c Kaplan–Meier curve comparing survival of FVB/n mice transplanted with PR/MYCRV leukemia treated for 21 days with JQ1 or placebo (N = 3 per group)
Discussion
MYC is commonly overexpressed in AML and has been shown to contribute to leukemogenesis [12, 14–16]. In this report, we compare specifically the effect of targeting MYC across AMLs that overexpress MYC through different mechanisms (trisomy of MYC/Myc-containing chromosomal regions, FLT3-ITD mutation, or gene amplification). Trisomy 8 and FLT3-ITD mutation have strong clinical relevance as they are the two most common mechanisms of MYC overexpression in human AML, and patients with FLT3-ITD AML have a particularly poor prognosis. We showed that shRNA knockdown of Myc in murine AMLs overexpressing MYC by trisomy 15 or FLT3-ITD prolongs survival and inhibits the ability of leukemic cells to predominate in vivo. We showed that VX-680, GDC-0941, and JQ1 significantly inhibit the growth of several human AML cell lines, while artemisinin does not. We further showed that the effect of VX-680, GDC-0941, and JQ1 on MYC levels varies across the AML cell lines tested, but we did not observe a discernible pattern of effect with respect to the mechanism underlying MYC overexpression. Thus, our experiments did not support our initial hypothesis that drug sensitivity would relate to the mechanism of MYC overexpression. JQ1 was observed to decrease MYC protein levels, and for several cell lines, the measured GI50 coincided with a sharp decrease in MYC protein levels. Additionally, JQ1 was generally more efficacious against AML cell lines that contained lower levels of MYC protein. Finally, we showed that JQ1 prolongs survival of mice injected with AMLs when Myc is overexpressed under control of its endogenous promoter, whereas GDC-0941 did not demonstrate efficacy against the MYC-overexpressing AMLs tested in our preclinical studies. Overall, our experiments demonstrated that targeting MYC in various MYC-overexpressing AMLs is an effective strategy that merits further development and that JQ1 appears to be a particularly promising drug in this regard.
In these experiments, we specifically compare drug efficacy across MYC-overexpressing AML cell lines stratified by mechanism of MYC overexpression. VX-680, which has been reported as synthetic lethal toward MYC [20], showed efficacy across all MYC-overexpressing cell lines except HL-60, which overexpresses MYC by gene amplification. There was no correlation between MYC levels and GI50s among the cell lines tested. In fact, HL-60 had the highest level of MYC protein of the cell lines tested and was the least sensitive to VX-680. Previous data suggest that higher MYC levels correlated with synthetic lethality with VX-680 [20], but this association was not observed in our experiments. This difference may have been due to the specific cell lines used, particularly with regard to effects of genes other than MYC that were not explored here. Assessing drug effects relative to doubling time, as we did herein, might also have contributed to our results in comparison with the prior study. Of note, we did observe decreased MYC level after VX-680 exposure in the MV4-11 and MOLM-14 cell lines, which both contain FLT3-ITD mutations. This is possibly due to the ability of VX-680 to inhibit the FLT3 expressed in these cells, which thereby may have contributed to a drop in MYC expression. Since clinical trials utilizing VX-680 have been terminated, testing of alternative aurora kinase inhibitors in MYC-overexpressing AMLs, particularly those with trisomy 8 or FLT3-ITD, may be of interest.
GDC-0941 showed efficacy against all cell lines, but this did not generally correlate with decrease in MYC levels. This lack of correlation suggests that growth inhibition in vitro by GDC-0941 does not reflect decreased MYC levels. Therefore, GDC-0941 does not appear to be a MYC-specific therapy in this setting. Furthermore, GDC-0941 was not efficacious in vivo against MYC-overexpressing leukemias. Given that GDC-0941 does not appear to be specific for MYC and did not demonstrate efficacy in our preclinical studies, these experiments do not support the continued exploration of this particular drug in the treatment of MYC-overexpressing AML. Of note, recent analysis suggests that overexpression of MYC may be a mechanism of resistance to PI3-kinase inhibitors [38].
In general, artemisinin showed minimal effect. GI50 was achieved for HL-60 and MV4-11. However, the relatively low GI50 against HL-60 may have been due to an early inflection point in the curve and may not reflect true efficacy. Nevertheless, the result may warrant further trials of artemisinin in the rare cases of AML with amplified MYC. In vivo testing was not undertaken due to limited drug effect in vitro. Overall, our experiments do not support the continued exploration of artemisinin as drug therapy against MYC-overexpressing AML.
JQ1 demonstrated strong efficacy across all cell lines. Among the drugs tested, JQ1 was the only compound effective at reducing MYC levels regardless of the mechanism of overexpression, and JQ1 doses near the GI50s were associated with a decrease in MYC levels. HL-60, the cell line with the highest basal levels of MYC, had the highest GI50, and we observed a positive correlation between GI50 and MYC level, suggesting that JQ1 may be particularly efficacious in AMLs with low to moderate MYC expression. Lower MYC levels may lead to increased susceptibility to growth inhibition by JQ1. In our preclinical in vivo studies, JQ1 demonstrated efficacy against leukemias with deregulated MYC, and this effect was lost when MYC was expressed under a heterologous promoter, indicating that downregulation at the Myc promoter is a major mechanism mediating JQ1 activity in these experiments. Together, these data support the role of JQ1 as an agent that lowers MYC protein levels and is thus able to inhibit growth of MYC-dependent leukemias, with an efficacy inversely correlated with MYC expression levels.
Previous work showed that JQ1 inhibits MYC transcription in a dose- and time-dependent manner, leading to the depletion of the c-MYC oncoprotein and selective downregulation of downstream transcriptional targets [26]. In addition, JQ1 may limit the activity of c-MYC by disrupting P-TEFb and transcription elongation [39, 40]. JQ1 has demonstrated antileukemic effects in vitro against a variety of human AML cell lines and primary patient samples [29, 41, 42], as well as in vivo against human AML xenografts [41] and murine MLL/AF9; KrasG12D transplants [29]. These effects include growth inhibition [42], cell cycle arrest, and apoptosis [29], among others. Of additional interest, isocitrate dehydrogenase (IDH) gene mutations have been shown to initiate the development of AML by cooperating with oncogenic FLT3 or NRAS. IDH mutant AMLs showed great sensitivity to JQ1, and western blots of IDH mutant leukemias treated with JQ1 showed a decrease in MYC levels [43]. JQ1 has also demonstrated efficacy against B-ALL cell lines with high-risk cytogenetics [44]. Another bromodomain inhibitor, I-BET, has shown efficacy against several MLL-fusion leukemias, including cell lines, murine leukemias, and primary patient samples [45].
Our data support the idea that targeting MYC may be beneficial for the treatment of most AMLs, regardless of mechanism of MYC overexpression, and that bromodomain inhibitors may be useful for this purpose until a direct MYC inhibitor can be developed. These experiments support future investigation of both direct and indirect MYC-targeting AML therapies and further establish MYC as an oncogene of central importance to AML pathogenesis and treatment.
Electronic supplementary material
Acknowledgments
Sam Brondfield was a Howard Hughes Medical Institute Research Fellow. We thank James Bradner for providing JQ1. We thank Kevin Shannon for reagents and helpful discussion. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA095274 (to S.C.K) and R01CA136717 (to A.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional support was received from: the Leukemia and Lymphoma Society (Specialized Center of Research to S.W.L and Scholar Award to A.G.); the Howard Hughes Medical Institute (to S.B.); and the Austrian Science Fund (FWF) SFB project F4710-B20 (to J.Z).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted.
References
- 1.Yamamoto JF, Goodman MT. Patterns of leukemia incidence in the United States by subtype and demographic characteristics, 1997–2002. Cancer Causes Control. 2008;19(4):379–390. doi: 10.1007/s10552-007-9097-2. [DOI] [PubMed] [Google Scholar]
- 2.Estey E. Acute myeloid leukemia and myelodysplastic syndromes in older patients. J Clin Oncol. 2007;25(14):1908–1915. doi: 10.1200/JCO.2006.10.2731. [DOI] [PubMed] [Google Scholar]
- 3.Juliusson G, Antunovic P, Derolf A, Lehmann S, Mollgard L, Stockelberg D, Tidefelt U, Wahlin A, Hoglund M. Age and acute myeloid leukemia: real world data on decision to treat and outcomes from the Swedish Acute Leukemia Registry. Blood. 2009;113(18):4179–4187. doi: 10.1182/blood-2008-07-172007. [DOI] [PubMed] [Google Scholar]
- 4.Appelbaum FR, Gundacker H, Head DR, Slovak ML, Willman CL, Godwin JE, Anderson JE, Petersdorf SH. Age and acute myeloid leukemia. Blood. 2006;107(9):3481–3485. doi: 10.1182/blood-2005-09-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang ME, Ye YC, Chen SR, Chai JR, Lu JX, Zhoa L, Gu LJ, Wang ZY. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood. 1988;72(2):567–572. [PubMed] [Google Scholar]
- 6.Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16(4):318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 7.Horiuchi D, Anderton B, Goga A (2014) Taking on challenging targets: making MYC druggable. Am Soc Clin Oncol Educ Book e497–502. doi:10.14694/EdBook_AM.2014.34.e497 [DOI] [PMC free article] [PubMed]
- 8.Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16(4):253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 9.Leon J, Ferrandiz N, Acosta JC, Delgado MD. Inhibition of cell differentiation: a critical mechanism for MYC-mediated carcinogenesis? Cell Cycle. 2009;8(8):1148–1157. doi: 10.4161/cc.8.8.8126. [DOI] [PubMed] [Google Scholar]
- 10.Luo H, Li Q, O’Neal J, Kreisel F, Le Beau MM, Tomasson MH. c-Myc rapidly induces acute myeloid leukemia in mice without evidence of lymphoma-associated antiapoptotic mutations. Blood. 2005;106(7):2452–2461. doi: 10.1182/blood-2005-02-0734. [DOI] [PubMed] [Google Scholar]
- 11.Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, Shi J, Weissmueller S, Fellmann C, Taylor MJ, Weissenboeck M, Graeber TG, Kogan SC, Vakoc CR, Lowe SW. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011;25(15):1628–1640. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4(2):199–207. doi: 10.1016/S1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 13.Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A, Goldstone A. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood. 1998;92(7):2322–2333. [PubMed] [Google Scholar]
- 14.Jones L, Wei G, Sevcikova S, Phan V, Jain S, Shieh A, Wong JC, Li M, Dubansky J, Maunakea ML, Ochoa R, Zhu G, Tennant TR, Shannon KM, Lowe SW, Le Beau MM, Kogan SC. Gain of MYC underlies recurrent trisomy of the MYC chromosome in acute promyelocytic leukemia. J Exp Med. 2010;207(12):2581–2594. doi: 10.1084/jem.20091071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothlisberger B, Heizmann M, Bargetzi MJ, Huber AR. TRIB1 overexpression in acute myeloid leukemia. Cancer Genet Cytogenet. 2007;176(1):58–60. doi: 10.1016/j.cancergencyto.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Rayeroux KC, Campbell LJ. Gene amplification in myeloid leukemias elucidated by fluorescence in situ hybridization. Cancer Genet Cytogenet. 2009;193(1):44–53. doi: 10.1016/j.cancergencyto.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Sohal J, Phan VT, Chan PV, Davis EM, Patel B, Kelly LM, Abrams TJ, O’Farrell AM, Gilliland DG, Le Beau MM, Kogan SC. A model of APL with FLT3 mutation is responsive to retinoic acid and a receptor tyrosine kinase inhibitor, SU11657. Blood. 2003;101(8):3188–3197. doi: 10.1182/blood-2002-06-1800. [DOI] [PubMed] [Google Scholar]
- 18.Zheng R, Small D. Mutant FLT3 signaling contributes to a block in myeloid differentiation. Leuk Lymphoma. 2005;46(12):1679–1687. doi: 10.1080/10428190500261740. [DOI] [PubMed] [Google Scholar]
- 19.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, Su M, Golec JM, Miller KM. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10(3):262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 20.Yang D, Liu H, Goga A, Kim S, Yuneva M, Bishop JM. Therapeutic potential of a synthetic lethal interaction between the MYC proto-oncogene and inhibition of aurora-B kinase. Proc Natl Acad Sci USA. 2010;107(31):13836–13841. doi: 10.1073/pnas.1008366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.den Hollander J, Rimpi S, Doherty JR, Rudelius M, Buck A, Hoellein A, Kremer M, Graf N, Scheerer M, Hall MA, Goga A, von Bubnoff N, Duyster J, Peschel C, Cleveland JL, Nilsson JA, Keller U. Aurora kinases A and B are up-regulated by Myc and are essential for maintenance of the malignant state. Blood. 2010;116(9):1498–1505. doi: 10.1182/blood-2009-11-251074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raynaud FI, Eccles SA, Patel S, Alix S, Box G, Chuckowree I, Folkes A, Gowan S, De Haven Brandon A, Di Stefano F, Hayes A, Henley AT, Lensun L, Pergl-Wilson G, Robson A, Saghir N, Zhyvoloup A, McDonald E, Sheldrake P, Shuttleworth S, Valenti M, Wan NC, Clarke PA, Workman P. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol Cancer Ther. 2009;8(7):1725–1738. doi: 10.1158/1535-7163.MCT-08-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asano T, Yao Y, Zhu J, Li D, Abbruzzese JL, Reddy SA. The PI 3-kinase/Akt signaling pathway is activated due to aberrant Pten expression and targets transcription factors NF-kappaB and c-Myc in pancreatic cancer cells. Oncogene. 2004;23(53):8571–8580. doi: 10.1038/sj.onc.1207902. [DOI] [PubMed] [Google Scholar]
- 24.Cui L, Su XZ. Discovery, mechanisms of action and combination therapy of artemisinin. Expert Rev Anti Infect Ther. 2009;7(8):999–1013. doi: 10.1586/eri.09.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu JJ, Meng LH, Shankavaram UT, Zhu CH, Tong LJ, Chen G, Lin LP, Weinstein JN, Ding J. Dihydroartemisinin accelerates c-MYC oncoprotein degradation and induces apoptosis in c-MYC-overexpressing tumor cells. Biochem Pharmacol. 2010;80(1):22–30. doi: 10.1016/j.bcp.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci USA. 2012;109(47):19408–19413. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Truong BT, Lee YJ, Lodie TA, Park DJ, Perrotti D, Watanabe N, Koeffler HP, Nakajima H, Tenen DG, Kogan SC. CCAAT/Enhancer binding proteins repress the leukemic phenotype of acute myeloid leukemia. Blood. 2003;101(3):1141–1148. doi: 10.1182/blood-2002-05-1374. [DOI] [PubMed] [Google Scholar]
- 31.Lee YJ, Jones LC, Timchenko NA, Perrotti D, Tenen DG, Kogan SC. CCAAT/enhancer binding proteins alpha and epsilon cooperate with all-trans retinoic acid in therapy but differ in their antileukemic activities. Blood. 2006;108(7):2416–2419. doi: 10.1182/blood-2006-02-003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, Lemieux ME, Picaud S, Yu RN, Qi J, Knapp S, Bradner JE. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150(4):673–684. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McJunkin K, Mazurek A, Premsrirut PK, Zuber J, Dow LE, Simon J, Stillman B, Lowe SW. Reversible suppression of an essential gene in adult mice using transgenic RNA interference. Proc Natl Acad Sci USA. 2011;108(17):7113–7118. doi: 10.1073/pnas.1104097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fei F, Stoddart S, Groffen J, Heisterkamp N. Activity of the Aurora kinase inhibitor VX-680 against Bcr/Abl-positive acute lymphoblastic leukemias. Mol Cancer Ther. 2010;9(5):1318–1327. doi: 10.1158/1535-7163.MCT-10-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rath K, Taxis K, Walz G, Gleiter CH, Li SM, Heide L. Pharmacokinetic study of artemisinin after oral intake of a traditional preparation of Artemisia annua L. (annual wormwood) Am J Trop Med Hyg. 2004;70(2):128–132. [PubMed] [Google Scholar]
- 36.Gajewski E, Gaur S, Akman SA, Matsumoto L, van Balgooy JN, Doroshow JH. Oxidative DNA base damage in MCF-10A breast epithelial cells at clinically achievable concentrations of doxorubicin. Biochem Pharmacol. 2007;73(12):1947–1956. doi: 10.1016/j.bcp.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boss DS, Beijnen JH, Schellens JH. Clinical experience with aurora kinase inhibitors: a review. Oncologist. 2009;14(8):780–793. doi: 10.1634/theoncologist.2009-0019. [DOI] [PubMed] [Google Scholar]
- 38. Martins MM, Zhou AY, Corella A, Horiuchi D, Yau C, Rakshandehroo T, Gordan JD, Levin RS, Johnson J, Jascur J, Shales M, Sorrentino A, Cheah J, Clemons PA, Shamji AF, Schreiber SL, Krogan NJ, M Shokat KM, McCormick F, Goga A, Bandyopadhyay S. Linking tumor mutations to drug responses via a quantitative chemical-genetic interaction map. Cance. Cancer Discov. 2014;5(2):154–167. doi: 10.1158/2159-8290.CD-14-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141(3):432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang CH, Lujambio A, Zuber J, Tschaharganeh DF, Doran MG, Evans MJ, Kitzing T, Zhu N, de Stanchina E, Sawyers CL, Armstrong SA, Lewis JS, Sherr CJ, Lowe SW. CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev. 2014;28(16):1800–1814. doi: 10.1101/gad.244368.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ., III Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108(40):16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herrmann H, Blatt K, Shi J, Gleixner KV, Cerny-Reiterer S, Mullauer L, Vakoc CR, Sperr WR, Horny HP, Bradner JE, Zuber J, Valent P. Small-molecule inhibition of BRD4 as a new potent approach to eliminate leukemic stem- and progenitor cells in acute myeloid leukemia AML. Oncotarget. 2012;3:1588–1599. doi: 10.18632/oncotarget.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C, Liu Y, Lu C, Cross JR, Morris JP, IV, Shroff AS, Ward PS, Bradner JE, Thompson C, Lowe SW. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes Dev. 2013;27(18):1974–1985. doi: 10.1101/gad.226613.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, Rodig SJ, Kung AL, Bradner JE, Weinstock DM. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120(14):2843–2852. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, Huthmacher C, Gudgin E, Lugo D, Beinke S, Chapman TD, Roberts EJ, Soden PE, Auger KR, Mirguet O, Doehner K, Delwel R, Burnett AK, Jeffrey P, Drewes G, Lee K, Huntly BJ, Kouzarides T. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.