Abstract
A characteristic immunopathology of human cancers is the induction of tumor antigen-specific T lymphocyte responses within solid tumor tissues. Current strategies for immune monitoring focus on the quantification of the density and differentiation status of tumor-infiltrating T lymphocytes; however, properties of the TCR repertoire ‒ including antigen specificity, clonality, as well as its prognostic significance ‒ remain elusive. In this study, we enrolled 28 gastric cancer patients and collected tumor tissues, adjacent normal mucosal tissues, and peripheral blood samples to study the landscape and compartmentalization of these patients’ TCR β repertoire by deep sequencing analyses. Our results illustrated antigen-driven expansion within the tumor compartment and the contracted size of shared clonotypes in mucosa and peripheral blood. Most importantly, the diversity of mucosal T lymphocytes could independently predict prognosis, which strongly underscores critical roles of resident mucosal T-cells in executing post-surgery immunosurveillance against tumor relapse.
Keywords: gastric cancer, mucosal-residence, prognosis, repertoire sequencing, T-cell repertoire
Abbreviation: TCR, T-cell receptor
Introduction
Cancer is a major public health problem worldwide. At present, surgery remains the primary form of therapy for solid tumors. The pathological assessment of the resected specimen reflects the anatomical extent of the tumor. The tumor-node-metastasis (TNM) stage classification is widely used to estimate the postoperative outcome and evaluate the rationale for adjuvant therapy. Despite the gross prognostic value of the TNM staging system,1 predicting individual patient outcome based on tumor parameters remains imprecise.2 Complementarily, the majority of human solid tumors are infiltrated by lymphocytes and it has been long suspected that the immune responses can be utilized as a benchmark to predict tumor progression or the efficacy of therapy.3 However, the complexities of tumor-elicited immune responses had rendered in situ immune monitoring impractical and inconclusive, until the development of systems biology and methods for high throughput objective measurement.4 The success in colorectal carcinoma4 and the urgent clinical need triggered a new wave of studies on immune biomarker discovery. Nevertheless, the reliability and predictivity of most of these new biomarkers for tumor prognosis remain to be determined.5
The infiltration of T lymphocytes in human tumors is a universal clinical observation and is considered as a major benchmark for immune surveillance.6 Correspondingly, “avoiding immune destruction” is an emerging hallmark of cancer progression, which is achieved by the immunosuppressive tumor microenvironment and selection pressures for tumor-associated antigens (TAAs).7 A major enabling characteristic of tumors is genetic mutations 7 that allow tumor cells to escape from T-cell antigen recognition – termed immunoediting.8 However, this enabling characteristic also leads to the generation of tumor-specific antigens (TSAs) and the accumulation of an aberrant spectrum of TSAs or TAAs.9 During an antitumor response, naïve T-cells are primed by presented TSA/TAAs and differentiate in peripheral lymphoid organs. Mostly by route of the circulation, differentiated effector T-cells enter the tumor to execute their antitumor functions. A small portion of these effector T-cells will survive to form long-lived memory T-cells (TMs). It has been hypothesized that TAA-specific TM cells play a central role in post-surgical immunosurveillance against tumor recurrence.4,10
Studies on infectious immunity have provided convincing evidence that, a specific tissue “niche” is necessary for the maintenance and function of TMs , especially CD8+ cytotoxic TM cells.11 Detached from the niche, circulating effector TMs numbers decline over time12; after entering the circulation, it becomes difficult for TMs to return to their original effector sites, e.g. the epithelial compartment13; and, when tissue resident TMs were forced into circulation by transplantation, their capacity to control localized peripheral infection becomes significantly dampened.13,14 Transcriptome analysis also indicated that tissue-resident memory T-cells (TRMs) represent a uniquely programmed TM population.15 Taken together, TRM cells colonize non-lymphoid tissue niches and form the first line of defense against local pathogen reoccurrence.16 However, the surveillance capacity of TRM cells against tumor relapse remains elusive.
During their brief encounters with antigen presenting cells (APC), T-cells scan through the presented peptide matrix and react with remarkable sensitivity and specificity.17 The specificity of T-cell responses to antigens are largely dictated by the affinity of TCR:peptide-MHC (pMHC) complexes.18 In response to the immense amount of foreign antigens, it is critical to develop and maintain a highly diversified TCR repertoire.19 Ultimately, the diversity of TCR repertoire is generated by somatic recombination of the TCRA and TCRB loci during early development in the thymus.20 During the rearrangement process, arrays of variable (V), diversity (D: only for the Beta chain of T-cell receptor) and joining (J) gene segments are reorganized and assembled by random and non-templated ligation to generate an antigen receptor.21 Besides the combinatorial diversity encoded in the genome, the junctional diversity generated by nonhomologous end joining of V to DJ/J segments22 is the major contributor in shaping the spectrum of TCR antigen recognition.18 The DNA sequence of this joint region between segments encodes a protein domain called the complementarity determining region 3 (CDR3), which is the hot spot for peptide contact23 and the determinant for the specificity and affinity of antigen recognition.18 Therefore, the central benchmarks of T-cell-mediated antitumor response – the local expansion of TAA-specific TCR clones, and the consequent landscape shift in the intratumoral TCR repertoire – are reflected by CDR3 clonal enrichment and diversity.
Traditional analyses methods – flow cytometry facilitated by specific V-β antibodies, or, PCR-based CDR3 spectratyping – only reveal a limited scope of the intratumoral T-cell repertoire.24 More recently, large-scale TCR repertoire analysis by deep sequencing methods has provided a systematic and comprehensive snapshot of various adaptive immune responses.25 These studies demonstrated the great potential of this technology in immune monitoring, which could facilitate the advancement of personalized medicine and propose immunotherapeutic strategies for clinical intervention. In this study, we assembled a cohort of patients to investigate the association between compartmented T-cell repertoires and clinical outcomes in gastric cancer. We developed a TCRB sequencing strategy to balance the demands of high-throughput and cost, which makes large-scale deep sequencing of clinical samples feasible. By mining our TCR repertoire data, we attempted to depict: (i) the expansion and diversity of tumor infiltrating T-cell repertoire; (ii) the origin and destination of expanded T-cell clonotypes in tumor tissue; and, (iii) correlations between clinical outcome and T-cell repertoire diversity.
Result
Deep sequencing of cryopreserved tissue samples enables detailed repertoire characterization
To characterize the landscape associated with T-cell-mediated antitumor immunity, we developed a high-throughput platform to analyze TCR repertoire. From various quantities of tissue samples, in the absence of additional cell separation procedures, mRNAs coded for TCRB chains were captured and barcoded by multiplex PCR, and then subjected to Ion Torrent PGM-based deep sequencing. Our sequencing strategy was designed to survey the TCR repertoire within a small quantity of clinical tissue, while balancing high-throughput with cost efficiency. To assess the coverage and reproducibility of this sequencing strategy, we collected 10 milliliters of blood twice from the same healthy donor 2 mo apart. In addition, we varied the coverage of sequencing chip for these two samples: we allocated 1/16 of a chip to cover the library generated from time point 1, and, 1/32 of the same type of chip for the library from the same healthy donor at time point 2 (Fig. S1A). About 137,000 and 34,000 valid TCR reads were generated for time points 1 and 2, respectively (Fig. S1C). Based on unique nucleotide (NT) sequences of TCRB transcripts, only 20% of T-cell clones were shared between these two sequenced PBMC samples. However, focusing only on highly expanded clones (HECs, defined as clones occupying more than 0.5% of total reads26), we found that every clone identified in the first sampling can be identified in blood collected 2 mo later and vice versa (Fig. S1D). Accordingly, a comparison of the entire pool of unique amino acid (AA) sequences at the CDR3 region – the most critical region for antigen recognition18 – demonstrated that these two samples are strongly correlated (R = 0.92 after the Pearson assay) at the global level (Fig. S1F). This indicated that our sequencing strategy has sufficient depth to cover high frequency clones, and, in the absence of immune challenge, HECs are largely stable.
To determine the minimum quantity of tissue needed to cover the HECs, we collected 10 milliliters, 2 milliliters, and 250 microliters of blood from the same healthy donor in one sitting and sequenced these three samples independently (Fig. S1B). Differences in sampling space generated very significant variations between each sample: only 15% unique TCRB NT sequences can be reliably detected in all three samples, with only 20% of unique CDR3 AA clones in common (Fig. S1G). However, despite the different sampling volumes, HECs were again repeatedly captured in all three samples and the sequencing results for CDR3 regions were strongly correlated (R = 0.9, Fig. S1H and I). Taken together, these quality control experiments indicated that although the sampling size and sequencing depth are too limited to capture every individual T-cell, our sequencing strategy can adequately and reproducibly detect HECs of a TCR repertoire with as little as 250 uL blood.
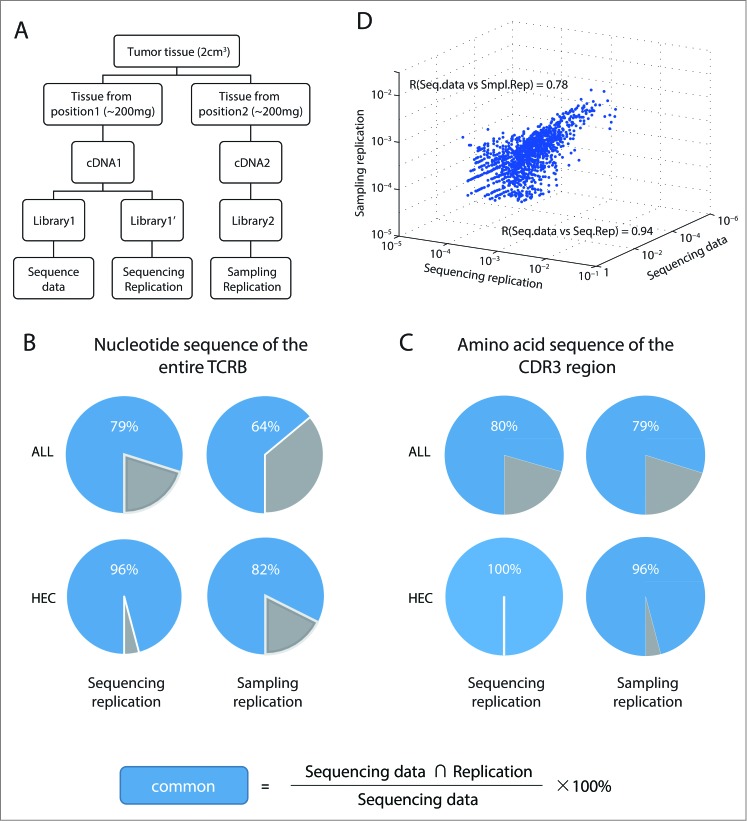
We further validated the sequencing reproducibility with cryopreserved tissue samples (Fig. 1A) by repeatedly sequencing a cDNA library obtained from 200 mg tumor tissue. Again, the sequencing depth sufficiently captures almost all (96%) HECs. In contrast to the healthy PBMC sample that was largely heterogeneous, 79% of the sequencing reads from tumor tissue are identical in NT sequence (Fig. 1B and C, left, experimental group vs. sequencing replicate). Recent deep sequencing data established the genetic and functional heterogeneity within a tumor,27,28 which suggest a heterogeneous expression of TAAs at different tumor sites. In addition, Gerlinger and colleagues revealed differences in the T-cell repertoires among different regions of a single renal cell carcinoma lesion, demonstrating intratumoral heterogeneity of the T-cell response.29 To validate the representative value of the repertoire data collected from a single site of a tumor, we randomly picked a tumor sample and collected two pieces of 200 mg tissue samples from the opposite end of the tumor (Fig. 1A). For all sequenced TCR clonotypes, 64% of TCRB NT sequence and 79% CDR3 AA sequences were in common between sampling replicates (Fig. 1B and C, upper right panel). Focusing on HECs, 82% NT reads were shared and 96% CDR3 sequences were identical between sampling replicates (Fig. 1B and C, lower right panel). When these clonotype profiles were globally analyzed for all sequenced clones, the Pearson's correlation coefficient (R) was 0.94 and 0.78 for the sequencing replicate test and sampling replicate test, respectively (Fig. 1D). This suggested that repertoire profiles for HECs from a single sampling site remain highly reproducible and representative of the whole tumor sample.
Figure 1.
Highly expanded clonotypes shape the major characteristics of TCRB repertoire and can be reliably detected with cryopreserved tissue samples.(A) Experimental approach. Gastric tumor tissue from patient sy014 was collected during surgical resection. Tumor tissue samples from two separate locations were homogenized, and total RNA extracted. Sequence data, sequencing replication, and sampling replication libraries were prepared and sequenced as shown. (B, C) Pie charts illustrating overlapping filtered T-cell receptor sequences between the experimental data set and two replicates for all clonotypes, or only for highly expanded clonotypes (HECs; defined as a proportion > 0.5% in the repertoires of the respective tissue compartments) for both (B) TCRB NT sequence (C) CDR3 AA sequence. (D) Reproducibility evaluation. The frequency of TCRB NT sequence shared among the sequence data, sequencing, and sampling replication as shown in a three-dimensional scatterplot.
Inter-individual sample variation diminishes potential CDR3 over-representation among TILs at the group level
We established a clinical cohort in 2009 to study tumor infiltrating T-cells from patients with diagnosed gastric cancer (described in Table S1). From each patient, we collected peripheral blood samples right before surgical resection, as well as tumor tissues (collected from invasive margins) and normal mucosal tissues (>5 cm distance from the edge of tumor, Fig. S2). Within this cohort, 28 patients were followed up prospectively for more than 48 mo and we subject paired samples – 600 uL blood, ∼200 mg of tumor tissue, and ∼200 mg of mucosal tissue – to repertoire analysis (summarized in Table S2).
Previous studies of TIL clones with low-resolution sequencing or in small cohort studies suggested that the usage of certain TCRB V regions would gain dominance within the tumor tissue, which indicated TAA-specific T-cell responses.30 We first evaluated whether there was a general bias of certain V or CDR3 usage by TILs. We assessed the CDR3 length distribution of the TCR repertoire collected from different tissues. When all clonotypes were combined, regardless of whether total (Fig. 2A) or only unique (Fig. 2B) CDR3 AA sequences were taken into account, there were no significant differences in CDR3 length distribution in tumor tissue, normal mucosa, or peripheral blood of gastric cancer patients. In addition, we recruited 28 age-matched healthy subjects and sequenced their TCR repertoires to establish the baseline distribution. Comparing the CDR3 length distribution of PBMC samples between tumor patients and healthy donors, we found no significant differences in either the total (Fig. 2C) or unique (Fig. 2D) CDR3 AA clonotypes. Consistent with this, a detailed analysis of V and J usage of each sample group again identified no specific usage bias between healthy donor PBMC, and tumor patient PBMC, TIL, and mucosal T-cells (Fig. 2E).
Figure 2.
Repertoires derived from patients and healthy donors exhibit similar CDR3 length distributions and V β and J β segment usage preferences at the group level.(A, B) The distributions of CDR3 lengths for (A) total (accounting for all CDR3 AA clonotypes, including the size of each individual clonotype) and (B) unique CDR3 AA clonotypes (disregarding the size of each clonotype) in the tumor site, adjacent normal mucosa, and peripheral blood from all patients. (C, D) CDR3 length distributions in the peripheral blood of 28 patients and 28 age-matched healthy donors. The proportion of TCRB sequences with CDR3 regions of each length are presented in a histogram for each tissue type. (E) VJ usage frequencies in the respective tissues. (F) Overall comparative analyses were performed for all 112 sequenced T-cell repertoires to determine their preferences for V/J-β segment usage, as represented by a color-coded heatmap from p = 1 (black; indistinguishable usage difference) to p = 0.01 (blue; significantly distinct usage frequencies). This difference matrix revealed a high inter-individual variation in V-β (upper triangle) and J-β (lower triangle) segment usage frequencies within the TCRB chain. p-values were calculated based on a two-tailed Kolmogorov-Smirnov test. CDR3 length was defined as the number of residues between the conserved cysteine in the V β segment and the phenylalanine in the J β segment.
To understand why the biased segment usage was not detected among different sample groups, we generated a heatmap to characterize the V- and J-β segment usage frequencies of each individual sample (Fig. 2F). This analysis demonstrated a high inter-individual variation within each of these four groups: even when T-cells were collected from the same tissue origin of healthy subject, V and J usages were distinct between individual subjects. Therefore, this intra-group variation diminished any potential significant differences between tissue compartments (Fig. S3C–F). Similarly, when an unsupervised clustering algorithm was applied to organize these samples based on their V or J usage, no clusters were distinguishable by tissue origin (Fig. S3G). Therefore, on an average, no V or J biases could be detected among TILs, in comparison to T-cells from mucosa or PBMC. Taken together, these analyses indicated that in a large cohort analysis, unbiased V/J usages by T-cells within tumor tissue does not necessarily reflect equal contribution of each V/J segment in tumor antigen recognition in each individual patient. More likely, different subjects may utilize different CDR3 to respond to their tumor antigens. Therefore, in a larger cohort analysis, it is difficult to predict the magnitude of an individual's immune response by searching for a specific V/J segment bias.
TIL repertoires are characterized by a remarkably reduced diversity
We reasoned that the individual TILs repertoire from gastric cancer patients consists of clonotypes that have been preferentially primed and expanded by TAAs.8 This clonal expansion would change the clonotype diversity (species and abundance), and we hypothesize that this change of diversity index would distinguish repertoires of the TIL group from others. To test this hypothesis, we plotted the cumulative distribution curve for each tissue type from patients to evaluate any potential skewing of the repertoire composition. Indeed, the most dominant 10% of TIL CDR3b clonotypes occupied 73.7% of the total TCR repertoire within patient tumors; in contrast, the most dominant 10% of mucosal and peripheral blood CDR3b clonotypes contributed to only 57.3% and 69.4% of the total TCR repertoires, respectively (Fig. 3A and Fig. S4). Specifically, the size distribution of TIL clonotypes was highly skewed: the proportion of HECs at tumor sites is 32.0%; it includes several highly dominant clonotypes whose individual frequency exceeds 10% of the entire TCR pool. By contrast, only 12.4% of mucosal and 14.5% of peripheral blood T-cell repertoires were composed of HECs; and, individual clonotypes with frequencies above 10% were not detected in mucosal compartment (Fig. 3B). Across all 28 patients, at the group level, the proportion of HECs is significantly different (with two-tailed Wilcoxon matched pair test) within different tissue compartments: based on their TCRB NT sequences, HECs are 19.3% ± 12.5% in tumors; 5.1 ± 5.6% in mucosa, and 7.4 ± 6.5% in peripheral blood (Fig. 3C); based on their CDR3 AA sequences, HECs are 34.0% ± 19.0% in tumors; 10.8% ± 9.1% in mucosa, and 14.0% ± 12.0% in peripheral blood (Fig. 3D).
Figure 3.
Clonal size analysis revealed clonal expansion and distinct diversity in different tissue types.(A) Distribution of clonotype size in each tissue from all patients represented as the cumulative percentage of the total CDR3 AA repertoire versus the cumulative percentage of unique CDR3 AA clones, in order of increasing clonal size. In this representation, the extent of deviation from the diagonal line indicates the unevenness of the distribution and the extent of expansion. (B) Expanded CDR3 AA clones that make up >10%, >5%, >2%, >1%, >0.5%, and<0.5% (rare) are shown as color gradient segments. Segments shaded in darker colors correspond to CDR3 AA clones found above the cutoff percentage and those shaded in lighter colors correspond to CDR3 AA clones that are rarer in the repertoire. (C, D) The proportion of rare (clonal frequency < 0.5%) and high frequency (clonal frequency ≥ 0.5%) clonotypes, as determined from (C) TCRB NT and (D) CDR3 AA clones for each patients. The abundance of high-frequency clonotypes were compared between tumor and other tissue repertoires based on the Wilcoxon matched pairs test. (E, F). The NSDE was calculated for each patient tissue type for both the TCRB NT (E) and CDR3 AA (F) clones. Statistics are based on the Wilcoxon matched paired test.
Given our findings that HECs accumulate in the tumor microenvironment, we speculated that TILs would show a relatively narrow and restricted repertoire. To quantitatively evaluate the expansion and diversity of the T-cell repertoire among samples, we measured the relative degree of diversity by calculating the normalized Shannon diversity entropy (NSDE) for TCR repertoires from each tissue origin. NSDE is based on the weighted geometric mean of the proportional abundances of the clonotypes, while factoring in the influence of repertoire size. This measure reflects the comprehensive oligoclonal vs. polyclonal nature of each T-cell repertoire (Fig. 3E and F). Based on this measure, the TIL repertoire tended to show markedly lower NSDE compared to adjacent mucosal tissue and peripheral blood in terms of both TCRB NT and CDR3 AA sequences (median entropy of entire TCRB NT sequence pool: tumor, 0.84; mucosa, 0.92; peripheral blood, 0.90, and mean entropy of CDR3 AA sequences: tumor, 0.77; mucosa, 0.88; peripheral blood, 0.86; two-tailed Wilcoxon matched pair test). These observations are consistent with results of a previous study by Sherwood and colleagues, which showed that colorectal cancer patients had substantially lower diversity within the TIL repertoire in both gastrointestinal tract tumors and tumors of epithelial origin.31 We postulate that the more restricted T-cell repertoire observed in tumor samples may be indicative of a specific and oligoclonal T-cell response to TAAs that are present in and restricted to the tumor microenvironment during tumorigenesis.
TAA-driven tumorous accumulation of high-frequency and coding-degenerate clonotypes
Besides the speculated TAA-driven expansion, the diminished TIL repertoire diversity described above may also arise from a limitation of T-cell sampling (with 200 mg tumor tissues), or from antigen-nonspecific expansion32 driven by the inflammatory tumor microenvironment.33 To investigate whether the expansion of these HECs was antigen-dependent, we included another perimeter, the level of coding degeneracy, into our analysis. The coding degeneracy level measures how many unique TCRB NT sequences present in the sample encode a single CDR3 AA clonotype. Since antigen specificity of a TCR is determined by the AA sequence of the CDR3 region, antigen-induced T-cell proliferation should expand every NT clonotype that codes for the same antigen-specific CDR3. Therefore, we expect to observe a correlated increase between the clonotype size and the level of coding degeneracy of a TAA-specific CDR3. On the contrary, this correlation cannot be expected if the diminished repertoire diversity is a sampling artifact or the consequence of antigen-independent expansion.
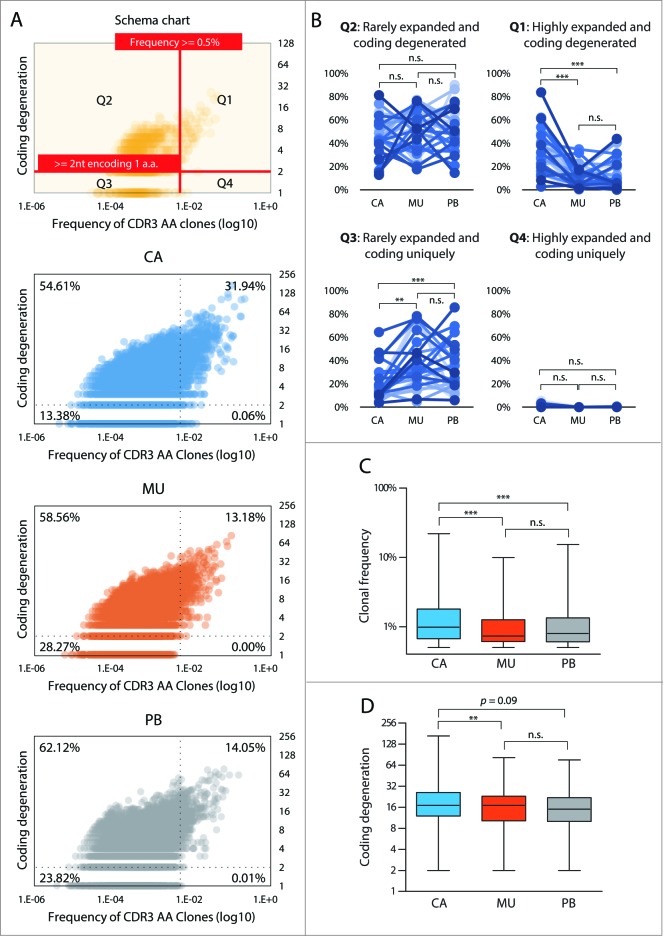
Combining all sequencing data from the 28 patients, we plotted the level of coding degeneracy against the frequency of that CDR3 AA clonotype among the T-cell repertoire in each respective tissue compartment. As shown in Fig. 4A, TCR repertoires were divided into four quadrants corresponding to high frequency-coding degenerate (Q1; with a cutoff as > 0.5% of the whole repertoire and employing more than two coding strategies), rare frequency-coding degenerate (Q2), rare frequency-coding non-degenerate, and high frequency-coding non-degenerate (Q4) clonotypes. Analysis of all three tissue compartments revealed a general correlative trend within the high frequency-coding degenerate quadrant: higher frequency clones were associated with higher levels of coding degeneracy, which indicates that antigen-driven T-cell expansion occurred in all compartments. Comparing repertoires from different tissue origins, 31.94% of clonotypes found in the TIL pool fell into this quadrant, reaching a maximum abundance to occupy 21.96% of repertoire space and a degeneracy level of 168. In the other two tissue compartments, T-cell clonotypes in this quadrant were markedly reduced to around 13.18%, with a maximum abundance of 9.95% and degeneracy level of 82 in mucosa and a maximum abundance of 14.05% and degeneracy level of 76 in peripheral blood (Fig. 4A).
Figure 4.
Expanded and coding degenerate clonotypes are significantly enriched at tumor sites.(A) Coding degeneracy level (number of unique TCRB NT sequences encoding a common CDR3 AA sequence) of CDR3 AA clones are plotted against their frequency in each tissue compartment. As shown in the schema chart, the scatter is divided into four quadrants, corresponding to highly expanded and coding degenerate (Q1), rarely expanded and coding degenerate (Q2), rarely expanded and coded uniquely (Q3) and highly expanded and coded uniquely (Q4), based on specific cutoff values. The percentages shown in each quadrant indicate the total proportion from all patients. (B) Proportion of clonotypes from the respective tissue compartments in each quadrant. Each dot represents the total frequency of the clonotypes that fall in each quadrant from one sample. Statistics are based on the Wilcoxon matched paired test. (C) Comparison of the frequency of clonotypes within the highly expanded and coding degenerate quadrant Q1 among tumor (CA), mucosa (MU), and peripheral blood (PB) T-cells from all patients. (D) Comparison of the coding degeneracy level of clonotypes in Q1 among the three tissue compartments from all patients. Data are displayed as box-and-whisker plots, with whiskers extending to the minimum and maximum values. Statistics are based on the Mann-Whitney U test.
We further performed the same analysis for each of the 28 patients’ individual repertoires (Fig. 4B and Fig. S5). Using the two-tailed Wilcoxon matched pair test, clonotypes in two reciprocal quadrants, Q1 and Q3, showed significant differences in different compartments. Compared to T-cells from mucosal tissue and PBMC, highly expanded and coding degenerate clonotypes (Q1) are enriched in TILs by 3.12- and 2.41-folds, respectively; Reciprocally, in the TIL group, the frequency of rarely expanded clonotypes with unique coding sequences (Q3) was half of that found in mucosa or PBMC (Fig. 4B). Strikingly, we found that for most patients (23/28), the Q1 population constituted a higher proportion in the TIL repertoire than in the paired mucosa and blood counterparts (Fig. 4B). Even after pooling and averaging all 28 patient samples, the Q1 clonotypes from the tumor compartment still contained a significantly higher occupancy of repertoire (Fig. 4C, median frequency for TILs, 0.98%; T-cells in mucosa, 0.73%; and T-cells in peripheral blood, 0.80%; two-tailed Mann–Whitney U test) and a higher level of coding degeneracy (Fig. 4D, median degeneracy level of 17 for TILS, 14 for mucosal T-cells, and 15 for T-cells in peripheral blood; two-tailed Mann–Whitney U test).
The correlation between the clonal size and the level of coding degeneracy can also be affected by the CDR3 AA composition, since different AA sequence have different levels of codon degeneracy. As a control, we also analyzed quadrant Q2 that contains rarely expanded and coding degenerate clonotypes. The general trend between higher frequency and higher codon degeneracy exists across all three tissue compartments (Fig. 4A), which indicates that the intrinsic properties of CDR3 AA composition may play a role. However, clonal sizes in quadrant Q2 were not distinguishable among these three compartments (Fig. 4A, B). In addition, a small proportion of clonotypes (0.06%) occurring at high frequency and with a unique NT coding sequence was observed in the tumor pool, which was not detected in the pooled non-tumor samples (0.00% for adjacent mucosa and 0.00% for peripheral blood). Taken together, these data showed that CDR3 clonotypes with degenerate CDR3 AA coding are the major contributors that reshape the repertoire at tumor sites during tumorigenesis, and, the multiplicity of NT clonotype for each individual HEC CDR3 is the likely consequence of TAA-driven selection and expansion.
Coding degenerate tumor infiltrating HECs contract in the mucosal compartment
Following these observations, we focused on these coding-degenerate and highly expanded tumor infiltrating clonotypes ‒ potential TAA-specific T-cell clones. To trace their origin and destination, we intersected these TIL clones with paired TCR repertoires from mucosa or peripheral blood (Fig. 5A). Combining data from all 28 patients, 376 clonotypes can be simultaneously detected in paired TIL and mucosal repertoires; 291 clonotypes are shared between paired TIL and circulating T-cells. To analyze whether these intersecting clonotypes were responsible for reshaping the repertoire, we inspected the frequencies of shared clonotypes in the tumor and mucosa (Fig. 5B)/peripheral blood (Fig. 5C) repertoires. In addition, we designated the area of each bubble to represent the degeneracy level of each clonotype. The majority of intersected CDR3 clonotypes (352/376 overlap between TIL and mucosal repertoires and 274/291 overlap between TIL and circulating TCR repertoires) exhibited a tendency to expand in tumor tissues (dots on the right of the gray diagonal line in the chart, Fig. 5B and C). This strongly suggested that, in comparison to normal mucosal and peripheral blood compartments, these potential TAA-specific T-cell clones expanded at the tumor site (Fig. 5D and E).
Figure 5.
(see previous page) Highly expanded TILs contract in mucosal and circulating compartments.(A) Workflow for tracking shared TIL clonotypes in other tissue compartments. To retrospectively determine the shared clonotypes between different tissue compartments, the highly expanded and coding degenerate clonotypes (with a frequency >0.5% and ≥ 2 coding strategies) in tumor sites were intersected in paired mucosal and PBMC repertoires for analysis. (B, C) The bubble chart shows the size and coding degeneracy of intersected clonotypes between the tumor and (B) mucosal/(C) circulating compartments. Each dot represents a CDR3 AA clones detected in both tissue types; the area of each dot corresponds to the coding degeneracy level, and the horizontal and vertical axes indicate the clonotype frequency in CA and MU/PB samples, respectively. The gray lines represent the diagonal of the coordinate. (D, E) Frequencies of shared clonotypes in the mucosa (D) and peripheral blood (E) from all patients. Two-tailed paired t-tests were performed to compare the frequency of each shared clonotype within the tumor or other tissue repertoires. (F) The Bhattacharyya similarity index was calculated for each individual patient. The median similarity is 0.2088 between tumor and mucosal repertoires, vs. 0.1468 between tumor and PBMC repertoires. Statistics are based on the Wilcoxon matched pairs test. (G, H) FACS analysis of resident T-cells in tumor-adjacent mucosal tissues freshly isolated from gastric cancer patient.
We then compared the intersection of these coding-degenerate clonotypes between normal mucosal tissue and peripheral blood. 335/376 shared CDR3 clonotypes remained coding degenerate in the mucosa, which is significantly higher than that of circulating T-cells (231/291, p = 0.0007 by Fisher's-exact test). This indicated that these potential TAA-specific clones in the mucosa compartment contracted to a lesser extent. To quantify the relative contribution of these two repertoires to the final clonotypes present in tumors, we also calculated the Bhattacharyya similarity index to measure the overlap between the tumor repertoire and paired normal mucosa/peripheral blood repertoires for each individual. Despite falling short of statistical significance (p = 0.086, two-tailed Wilcoxon matched pair test, Fig. 5F), the overall trend suggests that mucosal repertoires are more similar to TIL repertoires than circulating T-cells.
To assess compositions of mucosal T-cells, we digested mucosal tissue adjacent to gastric tumor from fresh surgery samples and performed FACS analysis. Our data showed that within the CD3+ T-cell population, the ratio between the CD8+ and CD8- T-cell (majority of them are CD4+ T-cells) is very close to 1:1. For those CD8+ mucosal T-cells, more than 2/3 are CD103hiCD45RO+CD69hiCD62L- (Fig. 5G), perfectly resembling characteristics of the resident TMs.16 Taken together, we propose that coding-degenerate HECs in tumor tissues of gastric cancer patients are derived from circulating T-cells, and, a portion of these expanded clonotypes egressed from the tumor, underwent clonal contraction and became mucosal-resident T lymphocytes.
High diversity of mucosal-resident TCR repertoire is associated with increased survival of gastric cancer patients
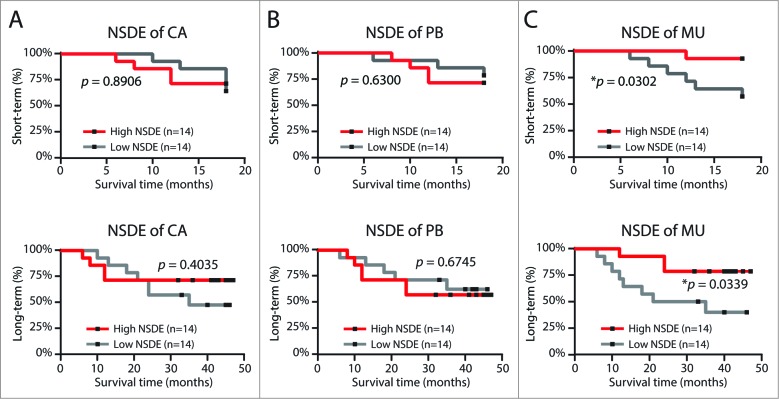
Due to the complexity of immune responses against tumors, it was inconceivable to define a single parameter for immune monitoring. Based on studies from various cancer types, e.g., melanoma,34 ovarian,35 and colorectal cancers,36 the established consensus is that tumor-infiltrating T lymphocytes, especially CTLs,34-36 appear to prevent tumor development and are associated with improved clinical prognosis.6 The antigen-driven activation and expansion of tumor specific T-cells will reshape the local repertoire and change the value of the NSDE: the larger expansion and longer persistence of particular T-cell clonotype(s) will narrow the relative broadness of the repertoire and consequently dampen the value of NSDE. Therefore, we hypothesized that a lower NSDE in TIL repertoire will predict at least a better short-term survival benefit in gastric cancer patients.
To test this hypothesis, we grouped the 28 patients based on their follow-up period and TIL repertoire diversities. Using the median value of NSDE as the cutoff, we divided these 28 patients into high and low groups. Surprisingly, we found that NSDE of TIL repertoire does not impact either short-term (with 18 mo follow-up) or overall (with 48 mo follow-up) survival rate (Fig. 6A). The TIL repertoire NSDE was similar between patients who did and did not experience a cancer relapse (p = 0.085, Fig. S6A); and, TIL repertoire diversity was very similar between the Stage I–II and Stage III–IV patient groups (Fig. S6B), across all T classifications (Fig. S6C), and N classifications (Fig. S6D).
Figure 6.
Diversity of the mucosal T-cell repertoire correlates with cancer outcome.Kaplan-Meier plot for short-term (upper panel)/overall (lower panel) patient survival time as a function of the median NSDE value for tumor (A), circulating (B), and tumor-adjacent normal mucosal (C) compartments.
Using the same strategy, we grouped these patients based on diversity of their adjacent mucosal and peripheral blood repertoires. The NSDE of circulating T lymphocytes also do not predict patient survival (Fig. 6B), TNM stage, T classification, or N classification (Fig. S7). However, although the mucosal TCR repertoire was also not impacted by the stage of tumor development (Fig. S8), the diversity index of mucosal repertoire can clearly predict both short-term and long-term survival of cancer patients: more restricted repertoires with low diversity indices were correlated with a poor clinical prognosis in patients (Fig. 6C). Furthermore, in this moderate size cohort, we found that NSDE of mucosal TCR repertoire is the most significant marker to predict prognosis in COX univariate analysis; and, COX multivariate analysis indicated that NSDE of mucosal TCR repertoire predicts the survival of patients independently, not relying on any other clinical features recorded for this cohort (Table 1, Fig. S3, S4, and S9).
Table 1.
Univariate and Multivariate Analysis of Survival Associated Factors
| Variables | Multivariate | |||
|---|---|---|---|---|
| Univariate p value | Hazard ratio | 95% confidence interval | pvalue | |
| Age (>= 60 vs. <60 years) | 0.795 | |||
| Gender (male vs. female) | 0.704 | |||
| Tobacco (smoking vs. non-smoking) | 0.603 | |||
| Alcoholism (drinking vs. non-drinking) | 0.564 | |||
| H.Pylori antibody (positive vs. negative) | 0.251 | |||
| Carcinoembryonic antigen (>=5 vs. <5 U/L) | 0.415 | |||
| Tumor location (Upper vs. Middle vs. Lower) | 0.736 | |||
| Surgical method (Laparoscopy vs. Laparotomy) | 0.559 | |||
| Resection Type (Distal vs. Proximal vs. Total) | 0.377 | |||
| Tumor invasion (T1+T2 vs. T3+T4) | 0.153 | |||
| Lymphoid node status (N0+N1 vs. N2+N3) | 0.108 | |||
| Distant metastasis (M0 vs. M1) | 0.635 | |||
| TNM stage (stage I+II vs. stage III+IV) | 0.060 | 0.212 | 0.019–2.310 | 0.203 |
| Number of metastatic LNs (>=5 vs. <5 LNs) | 0.060 | 1.018 | 0.096–10.766 | 0.988 |
| Tumor size (>=5 vs. <5 cm) | 0.388 | |||
| Differentiation (poorly vs. differentiated) | 0.856 | |||
| NSDE of CA (High vs. Low diversity) | 0.413 | |||
| NSDE of MU (High vs. Low diversity) | 0.050 | 4.806 | 1.145–20.170 | 0.032 |
| NSDE of PB (High vs. Low diversity) | 0.678 | |||
1. Laboratory testing was performed at least 1 d before surgical sampling.
2. Tumor size was determined according to the maximal diameter of tumor mass.
3. For diversity test, patients were separated by the median value of NSDE for CA/MU/PB.
4. Associated factors with univariate p < 0.1 were included into COX multivariate analysis.
Discussion
The interplays between mutated tumor cells and the immune system play a profound role throughout stages of tumorigenesis.37 Specifically, an immune-stimulatory milieu, especially efficacious IFNγ-secreting type I T-cell responses within tumors, is closely associated with favorable prognosis in patients.6 Furthermore, recent clinical trials demonstrated that, for many patients, the blockade of co-inhibitory signaling in T-cells generates robust antitumor responses and significant clinical benefits.38 These progresses indicated that parameters of antitumor immune responses could serve as biomarkers to predict cancer prognosis, or, to measure clinical response toward therapies. There is, therefore, an urgent need to develop such biomarkers to guide personalized care for cancer patients.
Facilitated by the analytical power of next-generation sequencing, we developed a molecular biology method and computational platform to profile the TCR repertoire within various human tissues. Our strategy is streamlined and cost-effective, as it requires minimal sample quantity, minimal sample processing, and, only a moderate sequencing depth. These features enable it to become a high throughput platform for multi-sample screenings, which makes it feasible to characterize the major repertoire features of multiple clinical samples with high reproducibility. Taking advantage of this platform, we sequenced 112 samples including PBMCs from healthy donors and various tissues from gastric cancer patients; and, we established that the repertoire diversity index within the tumor-adjacent mucosal tissue is a biomarker associated with gastric cancer prognosis. To our knowledge, this is the first identification of a single biomarker for immune repertoire monitoring that can independently predict patients’ survival rate. The TCR repertoire diversity in adjacent normal mucosa may provide an additional dimension to the proposed Immunoscore39 This immune monitoring index may provide clinical benefit in terms of disease prognosis, guidance for immunotherapy, and categorization of high-risk patients. Furthermore, enhancement of the density and diversity of mucosal-resident T-cells may be a potential immunotherapeutic strategy for improving the efficacy of tumor intervention.
Consistent with our hypothesis that TILs undergo a selective expansion upon TAA encounter, we observed a significantly restricted TCRB repertoire within tumor sites, which was characterized by a lower diversity index, accumulation of high-frequency clonotypes, and greater multiplicity of CDR3 codon degeneracy compared to repertoires in adjacent healthy mucosa and peripheral blood (Figs. 3, 4). However, it was a rather surprising observation that the TIL repertoire does not correlate at all with the prognosis with gastric cancer patients. It has been shown in colorectal tumors that, despite immunoediting, the strength and quality of in situ TH1 responses associate with patients’ long-term clinical outcome.4,10 The assumption was that these in situ T-cell responses mirrored the strength and quality of systemic antitumor immune activation in patients. Since surgery remains the primary form of therapy for solid tumors and intratumoral immunity was assessed with surgically removed samples, it may be difficult to draw a relationship between the qualities of removed T-cells – which can no longer participate in tumor rejection – with the long-term outcome of cancer patients. We speculated that in our study, surgical removal was the reason why TIL repertoire diversity alone failed to correlate with prognosis. Alternatively, the intratumoral recruitment or differentiation of regulatory T-cells (Treg) fosters tolerance by suppressing tumor-specific immunity.40 Our current repertoire analysis with tissue-sampling strategy cannot distinguish this pro-tumorigenesis population. Therefore, without exclusion of enriched Treg cells, the TIL repertoire diversity may never predict tumor outcomes. Interestingly, in this gastric cancer cohort, although their diversity index values are close to those from mucosal compartment, repertoire of patients’ PBMCs right before surgery also failed to predict gastric cancer prognosis, suggesting that levels of systemic T-cell responses do not accurately reflect local T-cell responses in the tumor microenvironment. This was further supported by our sequencing data, which clearly revealed that the diversity of TIL repertoire was distinct from the repertoire of circulating T-cells, specifically for those expanded clones potentially driven by TAAs (coding-degenerate HECs, Figs. 3 and 4). These led us to conclude that in gastric cancer patients, the TIL repertoire and the PBMC repertoire both do not represent the overall quality of antitumor responses and therefore do not predict the disease outcome.
It has been shown that the abundance of TMs markers in tumors is associated with a low incidence of colorectal tumor relapse. This directly led to the hypothesis that TAA-specific TMs play a central role in post-surgery surveillance against tumor recurrence.4,10 Despite further study on larger cohort is needed to determine the precise magnitude of this favorable effect, our tissue-specific repertoire analyses indicated that the prognosis of gastric cancer is strongly associated with the diversity of T-cells residing within tumor-adjacent normal mucosa (Fig. 6). In this compartment, we identified a portion of T-cells that share the same CDR3 clonotypes with coding-degenerated HECs in tumors, which are very likely expanded by TAAs (Fig. 5). Furthermore, the size of these shared clonotypes in the mucosa are significantly contracted compared to TILs, but are less contracted than in the peripheral blood (Fig. 5F). We hypothesized that these overlapping clonotypes identified in tumor-adjacent mucosa represents a population of TRM cells that egressed from the tumor.
The gastrointestinal tract mucosa is a well-studied niche that harbors viral antigen-specific TRMs.16 When forced into circulation, TRMs isolated from gut mucosa were severely impaired in survival and proliferation during antigen re-challenge,41 indicating that the mucosal microenvironment provides critical instructions to reprogram effector T-cells locally. In addition, in various types of cancer, such as colon carcinoma, glioma, and ovarian carcinoma, tumorigenesis also induces the local accumulation of CD103+CD8+ TRM cells.42,43 And, at least for high-grade serous ovarian cancer, increased patient survival positively associates with TRM accumulation in the intraepithelial compartment of the tumor.44 We speculated that mucosal tissues adjacent to gastric tumors provide a perfect microenvironment for TRM cell differentiation and maintenance. Firstly, TGF-b signaling comprehensively regulates the formation of the gut-resident TRM population,42,45 and, TGF-b is highly enriched in the gastric tumor microenvironment and adjacent mucosal tissues46; Secondly, both the inflammatory environment14 and locally presented antigens47 surrounding tumors could serve as differentiation and retention signals for the formation of a persistent TRM population. Thirdly, despite lack of functional evidence, immunological profiling showed that the majority of mucosal resident T cells are CD103hiCD45RO+CD69hiCD62L-, which fully matches the characteristic surface marker expression of the resident memory T cell population. Collectively, we propose that the gastric mucosa is a specific niche to harbor TRM cells, which in turn enforce local surveillance against tumor recurrence. Besides directly executing cytolysis, recognition of specific antigens also activates local TRMs to function as producers of “danger signals,” which initiate chemokine storms to recruit other circulating effector or TMs .48 Hence, the diversity of mucosal repertoire, comprised by, or even determined by, these identified TRM cells, represents the spectrum of immunosurveillance in restraining a wide range of escaped primary tumor cells and mutant clones resulting from TIL immunoediting.8 Although the detailed mechanism remains elusive, the clinical relevance of the properties of the mucosal repertoire to predisposed outcomes support the notion that mucosa-resident T-cells might serve as a sentinel population for maintaining immunosurveillance of residual tumor cells in postoperative patients.
Materials and Methods
Study design
Gastric tumor tissue, normal mucosa (at least 5 cm distant from the edge of tumor), and peripheral blood samples were collected from 28 patients who were recruited into a clinical trial at the Third Military Medicine University (TMMU IRB identifier: KY200905) during debulking surgery in collaboration with the Southwest Hospital (Chongqing, China). Individuals with autoimmune diseases, infectious diseases, and other primary cancers were excluded, and none of the included patients had received chemotherapy or radiotherapy before sampling. Peripheral blood from 28 age-matched healthy donors was used for control experiments. Clinical staging of tumors was determined according to the TNM classification system of the International Union Against Cancer (7th edition). All samples were processed and frozen within 45 min of excision (cold ischemia), placed into a cryogenic vial labeled with a unique patient identifier, immersed in liquid nitrogen, and stored at −80°C until analysis. The study protocol was approved by institutional review board of TMMU in accordance with the Helsinki Declaration, and written informed consent was obtained from each subject. Clinical characteristics of 28 patients were described in Table S1.
High-throughput sequencing of TCRB
Analysis of the TCRB CDR3 regions of gastric cancer patients was performed on cryopreserved samples. Briefly, total RNA was extracted from 600 μL peripheral blood, 200 mg frozen tumor, or 200 mg healthy mucosal tissue samples using the RNeasy Mini Kit (QIAGEN) and converted to cDNA (RevertAid First Strand cDNA Synthesis Kit; Fermentas) with a constant region-specific primer (RT primer: 5′-ATCTCTGCTTCTGATGGCTCA-3′). A multiplex PCR system was introduced to amplify the CDR3 region of rearranged TCRB loci. A set of forward primers, each specific to one or a set of functional TCR V β segments, and a reverse primer specific to the constant region of TCRB, were used to generate amplicons that cover the entire CDR3 region. PCR products were loaded on 3% agarose gels (Sigma–Aldrich), and bands centered at ∼220–240 bp were excised and purified using the QIAquick Gel Extraction kit (QIAGEN). Purified products were sequenced using the Ion Torrent PGM platform (Life Technologies).
Processing of raw reads
Ion Torrent Suite software filters were used for data pre-processing to exclude low quality reads and erroneous sequences derived from unrecognized multiplex barcodes. Raw sequence data were converted to FASTQ format using an Ion Torrent PGM built-in plugin. The resulting FASTQ files were imported to the MATLAB software. The TCRB CDR3 region was identified according to the International ImMunoGeneTics (IMGT) collaboration, beginning with the second conserved cysteine encoded by the 3′ portion of the V β gene segment and ending with the conserved phenylalanine encoded by the 5′ portion of the J β gene segment. The number of NT s between these codons determines the length of the CDR3 region. A manual algorithm was used to identify which V and J segments contributed to each TCRB CDR3 sequence. Sequences with lengths shorter than 110 bp, an average Phred quality score < 25, minimum Phred score < 20, or those with no exact match to the TRBC constant region primer were discarded. In addition, sequences with out-of-frame rearrangements, ambiguous V- and J-β segment alignment, V-β segment pseudogenes, or a CDR3 AA junction lacking a 5′ cysteine or 3′ phenylalanine were discarded. Resulting sequences were further analyzed using MATLAB 2013b (MathWorks) via manual scripts, and graphed using Excel (Office 2013, Microsoft) and Prism 5 software (GraphPad).
Statistical analysis
Repertoire diversity based on RNA-template was evaluated as previously described.49 NSDE was calculated as:
where n is the number of unique sampling clones and p(i) is the abundance of sampling clones in its repertoire.50 Formula was factored by log10(n) to eliminate the influence of sampling size. Diversity index is a statistics variable derived from ecologic studies to estimate the biodiversity of a given environment based on limited sampling size (counting the absolute number of organism inside is somehow impossible). Similarly, since the measuring for every T-cell clones in the whole healthy mucosa/tumor mass is impracticable, we introduced this formulation to evaluate the diversity of interest repertoire. More uneven clonal frequencies results in a smaller corresponding NSDE. If all clonotypes in the repertoire of interest have equal distribution, the index takes the unity value. To quantify the similarity between each TCR repertoire, we adopted the Bhattacharyya similarity index based on the percentage as well as homogeneity of abundance of shared sequences within two populations51 as follows:
where n is the number overlap clones and f(i,1/2) is the abundance of overlapped clones in two repertoires. A value ranging from zero (no overlap) to one (identical repertoire) was calculated for each pair of clonotypes in the repertoire.
Differences between two groups were determined by the Mann–Whitney U test. Correlations between parameters were assessed using Pearson correlation analysis. Kolmogorov–Smirnov tests were used to assess overall difference in V and J segment usage frequencies between distinct repertoire datasets. Overall survival was defined as the interval between surgery and death or between surgery and the last observation for surviving patients. Any known tumor-unrelated deaths were excluded from the death record for this study. Cumulative survival time was calculated using the Kaplan–Meier method, and survival was measured in months; the log rank test was applied to compare survival between two groups. MATLAB software was used for all statistical analysis. All data were analyzed using two-tailed tests, and p < 0.05 was considered statistically significant unless otherwise specified.
Acknowledgments
We thank Drs. Bo Zhu and Michael Krangel for critical suggestions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Q.-J.L. is a Whitehead Family Foundation Scholar and is supported by grants from the American Cancer Society (RSG-10-157-01-LIB). N.J. is a Cancer Prevention and Research Institute of Texas Scholar and a recipient of Damon Runyon-Rachleff Innovation Award. This work was partially supported by the National Program on Key Basic Research Project in China (973 Program, 2010CB911800), National Natural Science Foundation of China (Grant No. 31000401) and the specific fund of Third Military Medical University (Grant No. 2012XJQ13)
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Data Access
Sequencing data generated for this study have been submitted to the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) under study accession number ERP008698.
References
- 1. Japanese Gastric Cancer A Japanese gastric cancer treatment guidelines 2010 (ver. 3). Gastric Cancer 2011; 14:113-23; PMID:21573742; http:dx/doi.org/10.1007/s10120-011-0042-4 [DOI] [PubMed] [Google Scholar]
- 2. Galon J, Mlecnik B, Bindea G, Angell HK, Berger A, Lagorce C, Lugli A, Zlobec I, Hartmann A, Bifulco C, et al. Towards the introduction of the 'Immunoscore' in the classification of malignant tumours. J Pathol 2014; 232:199-209; PMID:; http://dx.doi.org/ 10.1002/path.4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Halpern AC, Schuchter LM. Prognostic models in melanoma. Semin Oncol 1997; 24:S2-7; PMID: [PubMed] [Google Scholar]
- 4. Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006; 313:1960-4; PMID:; http://dx.doi.org/ 10.1126/science.1129139 [DOI] [PubMed] [Google Scholar]
- 5. Whiteside TL. Immune responses to cancer: are they potential biomarkers of prognosis? Front Oncol 2013; 3:107; PMID:; http://dx.doi.org/ 10.3389/fonc.2013.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012; 12:298-306; PMID:; http://dx.doi.org/ 10.1038/nrc3245 [DOI] [PubMed] [Google Scholar]
- 7. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:; http://dx.doi.org/ 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 8. Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 2011; 331:1565-70; PMID:; http://dx.doi.org/ 10.1126/science.1203486 [DOI] [PubMed] [Google Scholar]
- 9. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science (New York, NY) 2013; 339:1546-58; PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, Mlecnik B, Kirilovsky A, Nilsson M, Damotte D, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Eng J Med 2005; 353:2654-66; PMID:; http://dx.doi.org/ 10.1056/NEJMoa051424 [DOI] [PubMed] [Google Scholar]
- 11. Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science 2001; 291:2413-7; PMID:; http://dx.doi.org/ 10.1126/science.1058867 [DOI] [PubMed] [Google Scholar]
- 12. Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nature immunology 2003; 4:225-34; PMID:; http://dx.doi.org/ 10.1038/ni889 [DOI] [PubMed] [Google Scholar]
- 13. Jiang X, Clark RA, Liu L, Wagers AJ, Fuhlbrigge RC, Kupper TS. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature 2012; 483:227-31; PMID:; http://dx.doi.org/ 10.1038/nature10851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mackay LK, Stock AT, Ma JZ, Jones CM, Kent SJ, Mueller SN, Heath WR, Carbone FR, Gebhardt T. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc Natl Acad Sci U S A 2012; 109:7037-42; PMID:; http://dx.doi.org/ 10.1073/pnas.1202288109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, Vega-Ramos J, Lauzurica P, Mueller SN, Stefanovic T, et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol 2013; 14:1294-301; PMID:; http://dx.doi.org/ 10.1038/ni.2744 [DOI] [PubMed] [Google Scholar]
- 16. Cauley LS, Lefrancois L. Guarding the perimeter: protection of the mucosa by tissue-resident memory T cells. MucosalIimmunol 2013; 6:14-23; PMID:; http://dx.doi.org/ 10.1038/mi.2012.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davis MM, Krogsgaard M, Huse M, Huppa J, Lillemeier BF, Li QJ. T cells as a self-referential, sensory organ. Annu RevImmunol 2007; 25:681-95; PMID:; http://dx.doi.org/ 10.1146/annurev.immunol.24.021605.090600 [DOI] [PubMed] [Google Scholar]
- 18. Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Ligand recognition by alpha beta T cell receptors. Ann Rev Immunol 1998; 16:523-44; PMID:; http://dx.doi.org/ 10.1146/annurev.immunol.16.1.523 [DOI] [PubMed] [Google Scholar]
- 19. Ebert PJ, Li QJ, Huppa JB, Davis MM. Functional development of the T cell receptor for antigen. Prog Mol Biol Transl Sci 2010; 92:65-100; PMID:; http://dx.doi.org/ 10.1016/S1877-1173(10)92004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chien YH, Gascoigne NR, Kavaler J, Lee NE, Davis MM. Somatic recombination in a murine T-cell receptor gene. Nature 1984; 309:322-6; PMID:; http://dx.doi.org/ 10.1038/309322a0 [DOI] [PubMed] [Google Scholar]
- 21. Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol 2009; 21:133-9; PMID:; http://dx.doi.org/10.1016/j.coi.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol 2011; 11:251-63; PMID:; http://dx.doi.org/ 10.1038/nri2941 [DOI] [PubMed] [Google Scholar]
- 23. Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science 1996; 274:209-19; PMID:; http://dx.doi.org/ 10.1126/science.274.5285.209 [DOI] [PubMed] [Google Scholar]
- 24. Thor Straten P, Schrama D, Andersen MH, Becker JC. T-cell clonotypes in cancer. J Transl Med 2004; 2:11; PMID:; http://dx.doi.org/ 10.1186/1479-5876-2-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Emerson RO, Sherwood AM, Rieder MJ, Guenthoer J, Williamson DW, Carlson CS, Drescher CW, Tewari M, Bielas JH, Robins HS. High-throughput sequencing of T-cell receptors reveals a homogeneous repertoire of tumour-infiltrating lymphocytes in ovarian cancer. J Pathology 2013; 231:433-40; PMID:; http://dx.doi.org/ 10.1002/path.4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klarenbeek PL, de Hair MJ, Doorenspleet ME, van Schaik BD, Esveldt RE, van de Sande MG, Cantaert T, Gerlag DM, Baeten D, van Kampen AH, et al. Inflamed target tissue provides a specific niche for highly expanded T-cell clones in early human autoimmune disease. Ann Rheum Dis 2012; 71:1088-93; PMID:; http://dx.doi.org/ 10.1136/annrheumdis-2011-200612 [DOI] [PubMed] [Google Scholar]
- 27. Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012; 366:883-92; PMID:; http://dx.doi.org/ 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martinez P, Birkbak NJ, Gerlinger M, McGranahan N, Burrell RA, Rowan AJ, Joshi T, Fisher R, Larkin J, Szallasi Z, et al. Parallel evolution of tumour subclones mimics diversity between tumours. J Pathol 2013; 230:356-64; PMID:; http://dx.doi.org/ 10.1002/path.4214 [DOI] [PubMed] [Google Scholar]
- 29. Gerlinger M, Quezada SA, Peggs KS, Furness AJ, Fisher R, Marafioti T, Shende VH, McGranahan N, Rowan AJ, Hazell S, et al. Ultra-deep T-cell receptor sequencing reveals the complexity and intratumour heterogeneity of T-cell clones in renal cell carcinomas. J Pathol 2013; 231:424-32, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Serana F, Sottini A, Caimi L, Palermo B, Natali PG, Nistico P, Imberti L. Identification of a public CDR3 motif and a biased utilization of T-cell receptor V beta and J beta chains in HLA-A2/Melan-A-specific T-cell clonotypes of melanoma patients. J Transl Med 2009; 7:21; PMID:; http://dx.doi.org/ 10.1186/1479-5876-7-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sherwood AM, Emerson RO, Scherer D, Habermann N, Buck K, Staffa J, Desmarais C, Halama N, Jaeger D, Schirmacher P, et al. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer Immunol, immunother 2013; 62:1453-61; PMID:; http://dx.doi.org/ 10.1007/s00262-013-1446-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Unutmaz D, Pileri P, Abrignani S. Antigen-independent activation of naive and memory resting T cells by a cytokine combination. J Ex Med 1994; 180:1159-64; PMID:; http://dx.doi.org/ 10.1084/jem.180.3.1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Olurinde MO, Shen CH, Drake A, Bai A, Chen J. Persistence of tumor-infiltrating CD8 T cells is tumor-dependent but antigen-independent. Cell Mol Immunol 2011; 8:415-23; PMID:; http://dx.doi.org/ 10.1038/cmi.2011.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grotz TE, Vaince F, Hieken TJ. Tumor-infiltrating lymphocyte response in cutaneous melanoma in the elderly predicts clinical outcomes. Melanoma Res 2013; 23:132-7; PMID:; http://dx.doi.org/ 10.1097/CMR.0b013e32835e5880 [DOI] [PubMed] [Google Scholar]
- 35. Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, Jungbluth AA, Frosina D, Gnjatic S, Ambrosone C, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci U S A 2005; 102:18538-43. Epub 2005 Dec 12; PMID:; http://dx.doi.org/ 10.1073/pnas.0509182102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Di Caro G, Bergomas F, Grizzi F, Doni A, Bianchi P, Malesci A, Laghi L, Allavena P, Mantovani A, Marchesi F. Occurrence of tertiary lymphoid tissue is associated to T cell infiltration and predicts better prognosis in early stage colorectal cancers. Clin Cancer Res 2014; 12:12; PMID:24523438; http://dx.doi.org/10.1158/1078-0432.CCR-13-2590 [DOI] [PubMed] [Google Scholar]
- 37. Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol 2004; 22:329-60; PMID:; http://dx.doi.org/ 10.1146/annurev.immunol.22.012703.104803 [DOI] [PubMed] [Google Scholar]
- 38. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl Journal Med 2012; 366:2443-54; PMID:; http://dx.doi.org/ 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Galon J, Pages F, Marincola FM, Thurin M, Trinchieri G, Fox BA, Gajewski TF, Ascierto PA. The immune score as a new possible approach for the classification of cancer. J Transl Med 2012; 10:1; PMID:; http://dx.doi.org/ 10.1186/1479-5876-10-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 2004; 10:942-9. Epub 2004 Aug 22; PMID:; http://dx.doi.org/ 10.1038/nm1093 [DOI] [PubMed] [Google Scholar]
- 41. Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R. Cutting edge: gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J Immunol (Baltimore, Md : 1950) 2006; 176:2079-83. [DOI] [PubMed] [Google Scholar]
- 42. Ling KL, Dulphy N, Bahl P, Salio M, Maskell K, Piris J, Warren BF, George BD, Mortensen NJ, Cerundolo V. Modulation of CD103 expression on human colon carcinoma-specific CTL. J Immunol (Baltimore, Md : 1950) 2007; 178:2908-15. [DOI] [PubMed] [Google Scholar]
- 43. Kilinc MO, Gu T, Harden JL, Virtuoso LP, Egilmez NK. Central role of tumor-associated CD8+ T effector/memory cells in restoring systemic antitumor immunity. J Immunol (Baltimore, Md : 1950) 2009; 182:4217-25; PMID:19299720; http://dx.doi.org/10.4049/jimmunol.0802793 [DOI] [PubMed] [Google Scholar]
- 44. Webb JR, Milne K, Watson P, Deleeuw RJ, Nelson BH. Tumor-infiltrating lymphocytes expressing the tissue resident memory marker CD103 are associated with increased survival in high-grade serous ovarian cancer. Clin Cancer Res 2014; 20:434-44; PMID:; http://dx.doi.org/ 10.1158/1078-0432.CCR-13-1877 [DOI] [PubMed] [Google Scholar]
- 45. Zhang N, Bevan MJ. Transforming growth factor-beta signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity 2013; 39:687-96; PMID:; http://dx.doi.org/ 10.1016/j.immuni.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ebert MP, Yu J, Miehlke S, Fei G, Lendeckel U, Ridwelski K, Stolte M, Bayerdorffer E, Malfertheiner P. Expression of transforming growth factor beta-1 in gastric cancer and in the gastric mucosa of first-degree relatives of patients with gastric cancer. Br JCancer 2000; 82:1795-800; PMID:; http://dx.doi.org/ 10.1054/bjoc.1999.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wakim LM, Woodward-Davis A, Bevan MJ. Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc Natl Acad Sci U S A 2010; 107:17872-9; PMID:20923878; http://dx.doi.org/10.1073/pnas.1010201107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schenkel JM, Fraser KA, Vezys V, Masopust D. Sensing and alarm function of resident memory CD8(+) T cells. Nature Immunol 2013; 14:509-13; PMID:; http://dx.doi.org/ 10.1038/ni.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature 2013; 497:258-62; Epub 2013 Apr 28; PMID:; http://dx.doi.org/ 10.1038/nature12079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stewart JJ, Lee CY, Ibrahim S, Watts P, Shlomchik M, Weigert M, Litwin S. A Shannon entropy analysis of immunoglobulin and T cell receptor. Mol Immunol 1997; 34:1067-82; PMID:; http://dx.doi.org/ 10.1016/S0161-5890(97)00130-2 [DOI] [PubMed] [Google Scholar]
- 51. Rempala GA, Seweryn M. Methods for diversity and overlap analysis in T-cell receptor populations. J of Math Biol 2013; 67:1339-68; PMID:; http://dx.doi.org/ 10.1007/s00285-012-0589-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.