Abstract
Anterior gradient protein (AGR) 3 is a highly related homologue of pro-oncogenic AGR2 and belongs to the family of protein disulfide isomerases. Although AGR3 was found in breast, ovary, prostate, and liver cancer, it remains of yet poorly defined function in tumorigenesis. This study aimed to determine AGR3 expression in a cohort of 129 primary breast carcinomas and evaluate the clinical and prognostic significance of AGR3 in these tumors. The immunohistochemical analysis revealed the presence of AGR3 staining to varying degrees in 80% of analyzed specimens. The percentage of AGR3-positive cells significantly correlated with estrogen receptor, progesterone receptor (both P<0.0001) as well as low histological grade (P=0.003), and inversely correlated with the level of Ki-67 expression (P<0.0001). In the whole cohort, AGR3 expression was associated with longer progression-free survival (PFS), whereas AGR3-positive subgroup of low-histological grade tumors showed both significantly longer PFS and overall survival. In conclusion, AGR3 is associated with the level of differentiation, slowly proliferating tumors, and more favorable prognosis of breast cancer patients.
Keywords: AGR3, patient survival, protein disulfide isomerase, ER-positive breast cancer, immuno histochemistry
Introduction
Breast cancer is the most common female malignancy and a leading cause of deaths among women worldwide. Only in 2012, in Europe, roughly 464,000 new incidences were registered, and 131,000 women died from breast cancer.1 Despite intensive research on various diagnostic and/or prognostic markers, thorough understanding of factors affecting breast cancer patients’ outcome remains of great importance. In recent years, an increasing number of reports have linked anterior gradient protein (AGR) 2 with many aspects of breast tumor biology. AGR2 is a human homologue of Xenopus laevis-secreted protein XAG-2 and belongs to an evolutionary broad family with prominent role in developmental processes and regeneration of body appendages.2,3 There are three subfamilies of AGRs: AGR1, AGR2, and AGR3, all showing the highest homology to non-secreted protein disulfide isomerase (PDI) of the TLP19 subfamily.3 PDIs are involved in proper folding and maturation of newly synthesized proteins and the regulation of endoplasmic reticulum homeostasis.4
Following the first characterization of AGR2 in the estrogen receptor (ER)-positive breast cancer cell line MCF-7,5 AGR2 has been frequently shown as an estrogen-responsive gene/protein. It was demonstrated that AGR2 is upregulated in response to estradiol treatment both in vitro5 and in vivo,6 and its high expression correlates with ER status7 and predicts poor prognosis in ER-positive breast cancers8,9 as well as resistance to tamoxifen.10 Moreover, chromatin immunoprecipitation (ChIP) confirmed direct AGR2 regulation by ER.10–12 In normal mammary gland, AGR2 induces cell proliferation and differentiation as shown in the mouse models,13 whereas in breast tumors, it promotes cell progression and survival through, among others, ER, cyclin D1, c-Myc, and survivin signaling pathways.14 Furthermore, when introduced into benign rat mammary epithelial cell line, AGR2 was found to contribute to metastasis development.15
Closely related AGR2 homologue, AGR3,7 has also been identified in breast cancer cell lines using proteomics screen as one of the membrane-associated proteins.16 Although both molecules share 71% sequence identity and lie adjacent to one another at chromosomal position 7p21,7,17 AGR2, but not AGR3, is a dominant factor identified in many OMICS screens. Thus, to date, only few reports describing AGR3 expression in various tumors were published, and there are limiting amount of data depicting AGR3 prognostic relevance in these malignancies. It has been shown that AGR3 is strongly expressed in breast carcinomas when compared to healthy tissues16 and that its expression correlates with ER status in breast tumors.7 In another study, single ER-binding site on AGR3 promoter has been found using ChIP-Seq approach.12 Our group has recently demonstrated that intrahepatic cholangiocarcinomas (ICCs) express AGR3 protein, while hepatocellular carcinomas are predominantly AGR3 negative. Furthermore, we postulated that together with acid mucopolysaccharides, AGR3 could serve as a diagnostic marker of well-differentiated ICCs.18 It has also been shown that AGR3 is overexpressed in different histological types of ovarian cancers. In non-mucinous types (including serous papillary, endometrioid, and clear cell), AGR3 expression was found to be ER independent and uncoupled with AGR2 expression, whereas in mucinous ovarian cancers, both AGR2 and AGR3 showed cognate expression patterns.19 In serous type, AGR3 staining correlated with the level of differentiation and was associated with longer patient survival.20 Additionally, AGR3 was found to be androgen-regulated gene,21,22 expression of which was highly elevated in human prostate cancer.21 The aim of this study is to examine the significance between AGR3 expression, clinicopathological characteristics, and patient outcome in primary breast carcinomas.
Materials and methods
Study group and tissue specimens
The study group consisted of 129 patients undergoing surgical procedure for primary breast cancer at the Masaryk Memorial Cancer Institute (MMCI) between 2003 and 2006. Patient age at the time of diagnosis ranged from 29 years to 84 years (median 57 years). The clinical, histological, and molecular characteristics of the analyzed set of tumors are summarized in Table 1. Histological typing of tumors was carried out according to the criteria of World Health Organization.23 Tumor stage was determined according to the guidelines of the Union for International Cancer Control (UICC).24 Tumor grade was established according to Bloom and Richardson in the modification of Elston and Ellis.25 ER, progesterone receptor (PR), human epidermal growth factor receptor 2 (Her2/neu), and Ki-67 statuses were extracted from pathological records obtained from the MMCI database. For the evaluation of AGR3 prognostic relevance without regard to ER status, additional ER-negative group of 90 breast cancer patients treated at MMCI between 1995 and 2006 were included for survival analysis. Informed consent has been obtained from all patients involved in this study. The study was approved by ethical committee of MMCI, and the data used were anonymized and were handled according to Czech Republic existing legislation.
Table 1.
Clinicopathological characteristics of primary breast carcinomas
| Variablea | Group | Nb | %c |
|---|---|---|---|
| Histology | Ductal | 95 | 73.6 |
| Lobular | 18 | 14 | |
| Other | 9 | 7 | |
| NA | 7 | 5.4 | |
| Histological grade | G1 | 27 | 20.9 |
| G2 | 43 | 33.4 | |
| G3 | 56 | 43.4 | |
| NA | 3 | 2.3 | |
| Tumor size | Pt1 | 44 | 34.1 |
| pT2 | 65 | 50.3 | |
| pT3 | 6 | 4.7 | |
| pT4 | 9 | 7 | |
| NA | 5 | 3.9 | |
| Nodal status | Negative | 45 | 34.9 |
| Positive | 73 | 56.6 | |
| NA | 11 | 8.5 | |
| ER status | Negative | 29 | 22.5 |
| Positive | 100 | 77.5 | |
| NA | 0 | 0 | |
| PR status | Negative | 34 | 26.4 |
| Positive | 94 | 72.8 | |
| NA | 1 | 0.8 | |
| Her2/neu status | Negative | 92 | 71.3 |
| Positive | 36 | 27.9 | |
| NA | 1 | 0.8 | |
| Ki-67d | <15% | 55 | 42.6 |
| ≥15% | 61 | 47.3 | |
| NA | 13 | 10.1 | |
| AGR3 expression | 1 | 25 | 19.4 |
| 2 | 25 | 19.4 | |
| 3 | 79 | 61.2 |
Notes:
Defined in the “Materials and methods” section.
Number of patients.
Percentage of total patients, out of a total of 129.
Cut-off for Ki-67 was used according to St Gallen Consensus in 2009. AGR3 expression: 1 – negative/border, 2 – weakly/moderately positive, and 3 – strongly positive.
Abbreviations: NA, not available; ER, estrogen receptor; PR, progesterone receptor; Her2/neu, human epidermal growth factor receptor 2; AGR3, anterior gradient protein 3.
Immunohistochemistry
Tumor samples were fixed in 10% neutral buffered formalin for 24 hours and then embedded in paraffin wax. Immunohistochemical analysis was performed on 4 μm thick sections cut from formalin-fixed, paraffin-embedded archival tissue blocks, mounted on slides, deparaffinized in xylene, and rehydrated in phosphate-buffered saline through a graded ethanol series. Endogenous peroxidase activity was quenched in 3% hydrogen peroxide in phosphate-buffered saline for 15 minutes. Antigen retrieval was performed in citrate buffer pH 6 at 94°C for 20 minutes. For AGR3 immunodetection, the sections were incubated overnight at 4°C with mouse monoclonal antibody to AGR3 (clone 1, in house).19 A streptavidin–biotin peroxidase detection system was used according to the manufacturer’s protocol (Vectastain Elite ABC Kit; Vector Laboratories, Burlingame, CA, USA). Signal was visualized by 3,3′-diaminobenzidine (Liquid DAB+ Substrate Chromogen System; Dako Denmark A/S, Glostrup, Denmark). Nuclear counterstaining was performed with Gill’s hematoxylin. For immunohistochemical evaluation, three conventional categories according to the number of positive cells were assessed: 1 – negative/border (0%–5% of positive cells); 2 – weakly/moderately positive (5%–50% of positive cells); 3 – strongly positive (more than 50% of positive cells).26
Reverse transcription and quantitative PCR
Under the supervision of a pathologist, corresponding samples of tumor tissue were collected and used for extraction of total cellular RNA by TRI Reagent (MRC, Cincinnati, OH, USA). cDNA synthesis was carried out using the M-MLV reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA). Triplicate samples were subjected to quantitative polymerase chain reaction (PCR) analysis using SYBR Green (Sigma-Aldrich, St Louis, MO, USA) for AGR2 and AGR3. The primer pairs used were as follows: for AGR2 – forward: 5′-GGAGCTCTATAT AAATCCAAGACAAGCA-3′ and reverse: 5′-GCCAAT TTCTGGATTTCTTTATTTTC-3′; for AGR3 – forward: 5′-GCCTAGAATCATGTTTGTAGACC-3′ and reverse: 5′-GCTTTCTTCATGTTTTCTATCAAT-3′. PCR was performed using default conditions: initial denaturation at 95°C, and then 40 cycles at 95°C for 15 seconds and at 60°C for 1 minute. To obtain absolute quantification, dilution series of plasmids pDEST12.2 with cloned respective sequences were used in range from 20 to 2 millions of copies to generate standard curves. For data normalization, 18S rRNA levels were determined using TaqMan assay for 18S rRNA (Thermo Fisher Scientific, Waltham, MA, USA).
Statistical analysis
All statistical analyses were performed using STATISTICA Version 12 (StatSoft, Inc., Tulsa, OK, USA) and IBM SPSS Statistics 20.0. Fisher’s exact test and Pearson’s chi-squared test were applied to assess the associations of immunohistochemical staining for AGR3 with clinicopathological variables. Progression-free survival (PFS) was defined as the time from the date of surgery to the date of death or relapse of disease. Overall survival (OS) was defined as the time from surgery to death or last record. Patients who had not died or who were lost to follow-up were censored when they were last known to be alive. Differences between survival curves were assessed with the Breslow test. Unadjusted hazard ratios (HRs) ±95% confidence intervals (CIs) were obtained using Cox’s multivariate analysis with backward selection. Differences at P≤0.05 were considered to be statistically significant.
Results
Association of AGR3 expression with other tumor variables
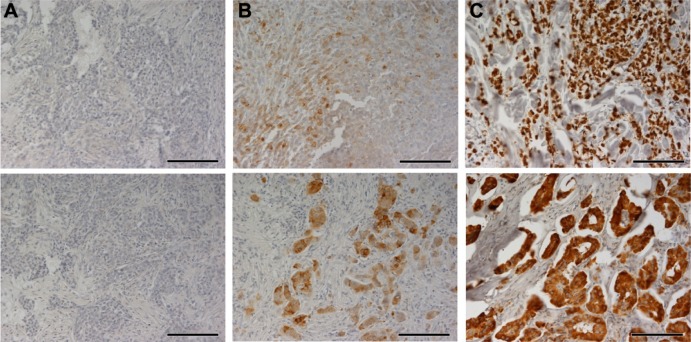
Due to the high homology between AGR2 and AGR3, protein specificity of the anti-AGR3 antibody was tested (Figure S1). The analyzed cohort composed of 95 (73.6%) tumors classified as ductal breast carcinomas, 18 (14%) as lobular type, and remaining 16 (12.4%) specimens were either of different or unknown origin. The remaining clinicopathological characteristics of the study group and their distributions are summarized in Table 1. Staining of primary breast carcinomas for AGR3 varied from tumor to tumor and was mainly cytoplasmic. Overall, of the 129 cases, 25 (19.4%) were classified as negative or borderline stained for AGR3 (<5% of positive cells), and the remaining 104 (80.6%) showed AGR3 positivity to different degrees (from weak to strong) (Figure 1). Immunohistochemical staining for AGR3 was then cross-tabulated with selected tumor features including histological type, tumor size, nodal status, histological grade, ER, PR, and Her2/neu status, and Ki-67 expression level. AGR3 positivity was significantly correlated with ductal type and slowly proliferating tumors as measured by expression level of Ki-67 marker (P<0.0001) as well as lower tumor grade (P<0.0001). Moreover, the degree of staining for AGR3 was significantly associated with that for the ER (P<0.0001) and PR (P<0.0001). There was no significant correlation between AGR3 positivity and tumor size, nodal status, or Her2/neu status (Table 2).
Figure 1.
Immunohistochemical staining for AGR3.
Notes: The level of AGR3 expression in primary breast carcinomas was determined by immunostaining in 3-point scale: (A) negative or border; (B) weak to moderate; and (C) strong. Scale bars represent a length of 100 μm.
Abbreviation: AGR3, anterior gradient protein 3.
Table 2.
Association of immunohistochemical staining for AGR3 with other tumor variables
| Variable | N (%)a
|
Statistical significance | |||
|---|---|---|---|---|---|
| Patients | AGR3 negative/border | AGR3 weak/moderate | AGR3 strong | ||
| Histological grade | |||||
| G1 | 27 | 3 (11.1) | 4 (14.8) | 20 (74.1) | <0.0001b |
| G2 | 43 | 3 (7) | 7 (16.3) | 33 (76.7) | |
| G3 | 56 | 18 (32.1) | 14 (25) | 24 (42.9) | |
| Tumor size | |||||
| pT1 | 44 | 8 (18.2) | 9 (20.4) | 27 (61.4) | 0.664b |
| pT2 | 65 | 11 (16.9) | 11 (16.9) | 43 (66.2) | |
| pT3 + pT4 | 6 | 5 (33.3) | 2 (13.3) | 8 (53.3) | |
| Nodal status | |||||
| Negative | 45 | 10 (22.2) | 10 (22.2) | 25 (55.6) | 0.332c |
| Positive | 73 | 13 (17.8) | 10 (13.7) | 50 (68.5) | |
| ER status | |||||
| Negative | 29 | 20 (69) | 8 (27.6) | 1 (3.4) | <0.0001b |
| Positive | 100 | 5 (5) | 17 (17) | 78 (78) | |
| PR status | |||||
| Negative | 34 | 19 (55.9) | 9 (26.5) | 6 (17.6) | <0.0001c |
| Positive | 94 | 6 (6.4) | 16 (17) | 72 (76.6) | |
| Her2/neu status | |||||
| Negative | 92 | 18 (19.6) | 15 (16.3) | 59 (64.1) | 0.318c |
| Positive | 36 | 7 (19.4) | 10 (27.8) | 19 (52.8) | |
| Ki-67 | |||||
| <15% | 55 | 6 (10.9) | 9 (16.4) | 40 (72.7) | <0.0001c |
| ≥15% | 61 | 14 (23.0) | 14 (23.0) | 33 (54.0) | |
Notes:
Number (percentage) of patients with tumors characterized by negative/border, weak/moderate, or strong expression of AGR3.
Probability, P, from Fisher’s exact test with the Freeman–Halton extension.
Probability, P, from Pearson’s chi-squared test.
Abbreviations: AGR3, anterior gradient protein 3; ER, estrogen receptor; PR, progesterone receptor; Her2/neu, human epidermal growth factor receptor 2.
AGR3 expression determined by immunohistochemistry was also compared with AGR3 mRNA levels and evaluated in relation to other clinicopathological variables. Interestingly, except Ki-67, whose elevated expression was associated predominantly with negative or weak AGR3 expression (Table 2), we found similar trends for AGR3 on both protein and mRNA level in relation to other clinicopathological parameters (Tables 2 and S1).
We also examined AGR2 mRNA levels under the same parameters and found almost similar association between AGR2 gene expression and clinicopathological variables as seen for AGR3 (Table S1). In line with these observations, we also confirmed a strong correlation between AGR2 and AGR3 mRNA levels (P<0.0001, R=0.6327) according to Spearman Rank Order correlation. On the other hand, we also observed several statistically significant differences in the association between AGR2 expression and clinicopathological variables with respect to AGR3 indicating that the expression of these genes is similar but not identical. The evaluation of AGR2 and AGR3 mRNA levels revealed only marginal correlation of AGR2 mRNA levels with ER (P=0.083) in comparison with AGR3 and ER (P<0.001). In accordance with immunohistochemical staining (P=0.003), determination of AGR3 transcription levels showed significant association (P=0.037) with grade as well. Conversely, determination of AGR2 mRNA levels did not show this trend (P=0.166; Table S1).
Association of AGR3 with patient survival
For the survival analysis, follow-up was determined for 10 years since surgical removal. Median PFS was 92 months (range 1–120), and median OS was 103 months (range 1–120). As there was almost no difference in survival curves between negative/border and weak/moderate subgroups (data not shown), for further statistical analyses, the above subgroups were combined (further denoted as AGR3 “low”) and were compared with patients whose tumors showed strong AGR3 positivity (more than 50% of stained cells, denoted as AGR3 “high”). While OS was not significantly affected by AGR3 expression, despite the fact that Kaplan– Meier curves indicated some trend in favor of increased AGR3 expression (P=0.111), these patients had significantly longer PFS (P=0.037) (Figure 2).
Figure 2.
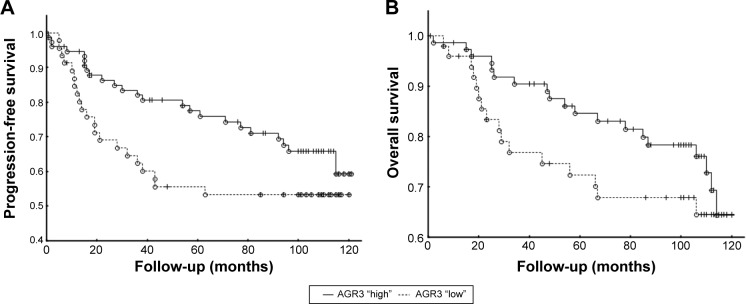
Association of immunohistochemical staining for AGR3 with patient survival.
Notes: (A) Determination of progression-free survival by Kaplan–Meier analysis in patients with “high” AGR3 expression (more than 50% of positive cells) and patients with “low” AGR3 expression (less than 50% of positive cells) using Breslow test (P=0.037). (B) Determination of overall survival by Kaplan–Meier analysis in patients with “high” AGR3 expression and patients with “low” AGR3 expression using Breslow test (P=0.111).
Abbreviation: AGR3, anterior gradient protein 3.
Association of AGR3 and other tumor variables with patient survival
As expected, patients with larger tumor size, higher histological grade, positive nodal status, and positive Her2/neu status had significantly poorer prognosis at 10 years of follow-up (Table S2). For multivariate survival analysis, the following clinicopathological parameters were included in Cox’s model with backward selection: histological type, tumor grade, tumor size, nodal status, and ER, PR, Her2/neu, and AGR3 status. As a result, tumor size and Her2/neu status were found to be independent prognostic factors for PFS, whereas tumor size and grade reached statistical significance for OS time in the studied cohort (Table 3). The remaining clinical and histological characteristics, including AGR3, failed statistical significance and were removed from the analysis during the selection process. When further pairwised with other variables (Table S3), AGR3 positivity was associated with better outcome in the subgroup of patients with tumors defined by smaller histological grade (G≤2; OS: P=0.005; PFS: P=0.024) but not by higher histological grade (G.2; OS: P=0.583; PFS: P=0.945). In Her2/neu-negative set of tumors, AGR3 expression significantly correlated with longer PFS (P=0.019) as well as OS (P=0.009). On the other hand, when ER-positive cases were considered separately, AGR3 expression did not reach statistical significance for improved survival (for PFS: P=0.228; for OS: P=0.234). Therefore, the subgroup of ER- and PR-negative patients was extended to determine the impact of AGR3 on patients’ outcome. However, within the additional ER-negative group of 90 patients, no significant association between AGR3 expression and patient outcome was observed as well with regard to both PFS and OS (P=0.282 and P=0.867, respectively; Figure S2). Statistical analysis of AGR3 IHC staining patterns with other clinicopathological parameters in cohort of ER- and PR-negative breast tumors revealed significant association between AGR3 expression and presence and Her2/neu status only (Table S4).
Table 3.
Independent prognostic factors for the analyzed set of tumors according to Cox’s multivariate survival analysis
| Variable | HR | 95% CI | Statistical significance |
|---|---|---|---|
| Progression-free survival | |||
| pT1 | 1.00 | 0.003 | |
| pT2 | 1.99 | 0.83–4.73 | 0.121 |
| pT3 | 8.25 | 2.68–25.44 | <0.0001 |
| Her2/neu status | 3.60 | 1.64–7.88 | 0.001 |
| Overall survival | |||
| pT1 | 1.00 | 0.006 | |
| pT2 | 1.38 | 0.51–3.76 | 0.531 |
| pT3 | 13.56 | 3.06–60.04 | 0.001 |
| G1 | 1.0 | 0.015 | |
| G2 | 1.87 | 0.20–17.34 | 0.582 |
| G3 | 6.38 | 0.82–49.42 | 0.076 |
Abbreviations: HR, hazard ratio; CI, confidence interval; Her2/neu, human epidermal growth factor receptor 2.
Discussion
AGR2 and AGR3 are conserved human homologues of X. laevis XAG-2 protein implicated in development and regeneration.2 AGR2 and AGR3 share high-sequence homology, localize to the same chromosomal position 7p21,7 and both respond to estrogen12 and androgen stimulation,21,22 which suggests their possible functional overlap. AGR2 is a well-studied pro-oncogene, promoting aggressive tumor phenotype and less favorable patient outcome in various malignancies.27–29 On the other hand, AGR3 function in health and disease remains ambiguous, since data published so far are relatively contradictory. AGR3 expression was demonstrated in various cancers, including breast,7 prostate,21 ovary,19,20 and liver.18 Moreover, it was shown that AGR3 binds to metastasis-associated GPI-anchored C4.4a protein and extracellular alpha-dystroglycan (DAG-1)7 and mediates resistance to cisplatin in mouse xenograft model,19 providing clear evidence for its important involvement in tumor biology. In our descriptive study, we analyzed a cohort of 129 primary breast carcinomas in order to assess clinical and prognostic relevance of AGR3 expression. We have detected AGR3 in 104 (80%) out of 129 specimens, hence confirming previously reported predominant expression of AGR3 protein in breast tumors.7,16 In the analyzed group, AGR3 was significantly associated with ER and PR positivity and tumor grades G≤2 but not with tumor size and nodal status, which is consistent with other studies.7,8,30 Moreover, we observed that increase in AGR3 positivity negatively correlated with the proliferation rate defined by the level of Ki-67 expression. Notably, similar trends in relation to other clinicopathological parameters were also found for AGR3 mRNA level. Correlation with ER and PR positivity and slowly proliferating and well-differentiated tumors suggests that AGR3 expression is associated with less aggressive tumors that are more prone to effective treatment and therefore favorable outcome. Indeed, in our work, we demonstrated for the first time that the presence of immunohistochemical staining for AGR3 is associated with improved patient PFS. Although, in the whole cohort, AGR3 expression did not predict longer OS, patients whose tumors were characterized by strong AGR3 positivity showed better response to therapy. Moreover, AGR3 predicted better outcome in the subgroup of patients with well-differentiated tumors, which is consistent with previously demonstrated significance of AGR3 expression in ovarian cancers.20 Quite the contrary, AGR2 is often described as an indicator of poor prognosis,8,9 metastasis,15,31 and resistance to commonly used treatments,10,32 indicating divergent and/or context-dependent roles of AGR proteins in breast cancer. It is of note that similar antagonistic impact of AGR proteins on patient outcome is also observed in ovarian cancers where AGR3 promotes better outcome,20 whereas AGR2 predicts shortened OS,33 possibly due to the stimulation of cell growth and migration.34 However, given that AGR3 was also shown to mediate cisplatin resistance, an explicit conclusion of AGR3-protective, antitumor role cannot be conclusively drawn. Moreover, in our recent work, we have compared AGRs distribution both in human healthy tissues and carcinomas using Genevestigator platform,35 and we found that AGR3 mirrors AGR2 expression in many cases, such as stomach, colon, pancreas, breast, female reproductive system, or respiratory system.36 In accordance, here, we have demonstrated strong correlation between AGR2 and AGR3 mRNA levels in breast carcinomas as well as similar associations of both genes with clinicopathological variables, which suggests their cognate physiological function and role in pathological conditions.
In the present work, we observed that better outcome in AGR3-positive group was independent of ER status (considered separately, neither ER-positive nor negative-subgroups had significantly longer survival time when pairwised with AGR3). These findings suggest more complex control of AGR3 expression in breast carcinomas, not solely dependent on ER, similarly to that of AGR2.8 Thus, some clues regarding AGR3 regulation could be derived from the studies focusing on AGR2 homologue. For instance, in addition to ER, AGR2 was reported to be a component of, among others, EGFR, cyclin D1, survivin, AKT, and transforming growth factor-beta signaling pathway.14,29,37,38 However, mechanisms triggering expression of AGR2 and AGR3 could be relatively unrelated as manifested by the uncoupled expression of both proteins in prostate and ovarian cancers,7,19 and thus, further in vitro and in vivo studies are warranted to understand AGR3’s function(s) in tumor biology. Relying on our in silico analyses, we have recently shown that AGR2 and AGR3 plausibly control similar aspects of tumor biology including cell cycle control, differentiation, migration, invasion, and metastasis.36 Additionally, we performed promoter analysis and demonstrated that most of the transcription factors potentially binding to AGR2 or AGR3 promoters are exclusive for each protein,36 which could partially elucidate their uncoupled expression. One possible explanation of observed AGR3 ambiguity is that dependent on the cellular context, it could support different phenotypes leading either to tumor progression or to regression.
In the light of what has been reported to date, it would be necessary not only to verify whether AGR3 plays tumor-suppressive or tumor-promoting role but also to evaluate the plausible relevance of AGR3 presence in patient’s fluids. AGR3 was firstly characterized in breast cancer cell membranes and was found to localize in secretory or endosome-like vesicles in both T47-D and MDA-MB-468 cells,16 suggesting more prominent role of secreted form of AGR3. Indeed, recent works have depicted emerging role of extracellular AGR2 in the control of tumor aggressiveness through both autocrine and paracrine effects,39,40 indicating that similar mechanism can also be valid for AGR3. Lastly, taking into account cognate expression pattern of AGR proteins in different carcinomas,7,19 it can be speculated that there is a functional cross talk between these proteins. However, whether they compete with each other, compensate for one’s lost, or support one another requires further investigation.
Supplementary materials
Determination of anti-AGR3 antibody cross-reactivity.
Notes: The specificity of our in-house anti-AGR3 antibody (AGR3.1) was confirmed by Western blot (upper panel). For comparison, we added testing of rabbit polyclonal sera raised against AGR2 protein, which recognizes AGR3 as well (bottom panel).
Abbreviation: AGR, anterior gradient protein.
Survival analysis of cohort of ER- and PR-negative breast cancer patients.
Notes: Kaplan–Meier analysis of (A) progression-free survival in relation to AGR3 expression (P=0.282, Breslow test) and (B) overall survival in relation to AGR3 expression (P=0.867, Breslow test).
Abbreviations: ER, estrogen receptor; PR, progesterone receptor; AGR3, anterior gradient protein 3.
Table S1.
Association of AGR2 and AGR3 mRNA levels with other tumor variables
| Patients (n) | AGR2 mRNA | AGR3 mRNA | |
|---|---|---|---|
| Histological grade | |||
| G1 | 26 | 0.166a | 0.037a |
| G2 | 33 | ||
| G3 | 34 | ||
| Tumor size | |||
| pT1 | 35 | 0.774a | 0.990a |
| pT2 | 50 | ||
| pT3 + pT4 | 8 | ||
| Nodal status | |||
| Negative | 37 | 0.822b | 0.541b |
| Positive | 52 | ||
| ER status | |||
| Negative | 17 | 0.081b | <0.001b |
| Positive | 76 | ||
| PR status | |||
| Negative | 19 | 0.124b | <0.001b |
| Positive | 73 | ||
| Her2/neu status | |||
| Negative | 57 | 0.364b | 0.603b |
| Positive | 36 | ||
| Ki-67 | |||
| <15% | 44 | 0.169b | 0.494b |
| ≥15% | 48 |
Notes:
Determination of P-level using Kruskal–Wallis analysis of variance.
Determination of P-level using Mann–Whitney U-test.
Abbreviations: AGR, anterior gradient protein; ER, estrogen receptor; PR, progesterone receptor; Her2/neu, human epidermal growth factor receptor 2.
Table S2.
Univariate survival analyses for the analyzed set of tumors
| Variablea | Statistical significanceb
|
|
|---|---|---|
| PFS | OS | |
| AGR3 expression | 0.037 | 0.111 |
| Histology | ||
| DU vs LO | 0.665 | 0.393 |
| DU vs OTH | 0.537 | 0.980 |
| LO vs OTH | 0.851 | 0.675 |
| Grade | ||
| G1 vs G2 | 0.159 | 0.137 |
| G1 vs G3 | 0.002 | 0.009 |
| G2 vs G3 | 0.020 | 0.055 |
| Tumor size | ||
| pT1 vs pT2 | 0.249 | 0.333 |
| pT1 vs pT3 | 0.000 | 0.000 |
| pT2 vs pT3 | 0.000 | 0.000 |
| Nodal status | 0.043 | 0.415 |
| ER status | 0.110 | 0.344 |
| PR status | 0.023 | 0.145 |
| Her2/neu status | 0.000 | 0.000 |
| Ki-67 expression | 0.004 | 0.018 |
Notes:
AGR3 expression, AGR3 “low” (less than 50% of stained cells) vs AGR3 “high” (more than 50% of stained cells); histology, ductal vs lobular vs others; nodal status, negative vs positive; estrogen receptor status, negative vs positive; progesterone receptor status, negative vs positive; Her2/neu status, negative vs positive; Ki-67 expression, <15% vs ≥15%.
Probability, P, from Breslow test.
Abbreviations: PFS, progression-free survival; OS, overall survival; AGR3, anterior gradient protein 3; DU, ductal; LO, lobular; OTH, others; ER, estrogen receptor; PR, progesterone receptor; Her2/neu, human epidermal growth factor receptor 2.
Table S3.
Survival analysis of patients with AGR3-expressing tumors
| Subgroup | Statistical significancea
|
|
|---|---|---|
| PFS | OS | |
| Histological grade | ||
| G≤2 | 0.024 | 0.005 |
| G>2 | 0.945 | 0.583 |
| Her2/neu status | ||
| Negative | 0.019 | 0.009 |
| Positive | 0.781 | 0.278 |
| PR status | ||
| Negative | 0.669 | 0.911 |
| Positive | 0.448 | 0.224 |
| ER status | ||
| Negative | 0.431 | 0.507 |
| Positive | 0.228 | 0.234 |
Note:
Probability, P, from Breslow test.
Abbreviations: AGR3, anterior gradient protein 3; PFS, progression-free survival; OS, overall survival; Her2/neu, human epidermal growth factor receptor 2; ER, estrogen receptor; PR, progesterone receptor.
Table S4.
Association of immunohistochemical staining for AGR3 with other tumor variables in a cohort of ER-negative breast cancer patients
| Variable | N (%)a
|
Statistical significanceb | |||
|---|---|---|---|---|---|
| Patients | AGR3 negative/border | AGR3 weak/moderate | AGR3 strong | ||
| Histological grade | |||||
| G1 | 1 | 1 (100.0) | 0 (0.0) | 0 (0.0) | 0.917 |
| G2 | 10 | 6 (60.0) | 3 (30.0) | 1 (10.0) | |
| G3 | 71 | 47 (66.2) | 15 (21.1) | 9 (12.7) | |
| Tumor size | |||||
| pT1 | 32 | 24 (75.0) | 5 (15.6) | 3 (9.4) | 0.729 |
| pT2 | 44 | 26 (59.1) | 11 (25.0) | 7 (15.9) | |
| pT3 + pT4 | 11 | 8 (72.8) | 2 (18.2) | 1 (9.0) | |
| Nodal status | |||||
| Negative | 34 | 25 (73.5) | 7 (20.6) | 2 (5.9) | 0.282 |
| Positive | 53 | 31 (58.5) | 13 (24.5) | 9 (17.0) | |
| Her2/neu status | |||||
| Negative | 40 | 34 (85.0) | 5 (12.5) | 1 (2.5) | 0.001 |
| Positive | 49 | 24 (49.0) | 15 (30.6) | 10 (20.4) | |
| Ki-67 | |||||
| <15% | 6 | 4 (66.7) | 2 (33.3) | 0 (0.0) | 0.626 |
| ≥15% | 42 | 30 (71.4) | 7 (16.7) | 5 (11.9) | |
Notes:
Number (percentage) of patients with tumors characterized by negative/border, weak/moderate, or strong expression of AGR3. Probability,
P, was calculated using Fisher’s exact test with the Freeman–Halton extension.
Abbreviations: AGR3, anterior gradient protein 3; ER, estrogen receptor; Her2/neu, human epidermal growth factor receptor 2.
Acknowledgments
We thank Pavlina Zatloukalova, PhD, for testing the antibody cross-reactivity. The work was supported by MH CZ – DRO (MMCI, 00209805), European Regional Development Fund and the State Budget of the Czech Republic for Regional Centre for Applied Molecular Oncology – RECAMO (CZ.1.05/2.1.00/03.0101), the projects MEYS-NPS I-LO1413 GACR 13-00956S, and IGA NT/13794-4/2012.
Footnotes
Disclosure
The authors declare that they have no competing interest.
References
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49(6):1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Sive HL, Hattori K, Weintraub H. Progressive determination during formation of the anteroposterior axis in Xenopus laevis. Cell. 1989;58(1):171–180. doi: 10.1016/0092-8674(89)90413-3. [DOI] [PubMed] [Google Scholar]
- 3.Ivanova AS, Tereshina MB, Ermakova GV, Belousov VV, Zaraisky AG. Agr genes, missing in amniotes, are involved in the body appendages regeneration in frog tadpoles. Sci Rep. 2013;3:1279. doi: 10.1038/srep01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatahet F, Ruddock LW. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11(11):2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 5.Thompson DA, Weigel RJ. hAG-2, the human homologue of the Xenopus laevis cement gland gene XAG-2, is coexpressed with estrogen receptor in breast cancer cell lines. Biochem Biophys Res Commun. 1998;251(1):111–116. doi: 10.1006/bbrc.1998.9440. [DOI] [PubMed] [Google Scholar]
- 6.Wilson CL, Sims AH, Howell A, Miller CJ, Clarke RB. Effects of oestrogen on gene expression in epithelium and stroma of normal human breast tissue. Endocr Relat Cancer. 2006;13(2):617–628. doi: 10.1677/erc.1.01165. [DOI] [PubMed] [Google Scholar]
- 7.Fletcher GC, Patel S, Tyson K, et al. hAG-2 and hAG-3, human homologues of genes involved in differentiation, are associated with oestrogen receptor-positive breast tumours and interact with metastasis gene C4.4a and dystroglycan. Br J Cancer. 2003;88(4):579–585. doi: 10.1038/sj.bjc.6600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Innes HE, Liu D, Barraclough R, et al. Significance of the metastasis-inducing protein AGR2 for outcome in hormonally treated breast cancer patients. Br J Cancer. 2006;94(7):1057–1065. doi: 10.1038/sj.bjc.6603065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barraclough DL, Platt-Higgins A, de Silva Rudland S, et al. The metastasis-associated anterior gradient 2 protein is correlated with poor survival of breast cancer patients. Am J Pathol. 2009;175(5):1848–1857. doi: 10.2353/ajpath.2009.090246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hrstka R, Nenutil R, Fourtouna A, et al. The pro-metastatic protein anterior gradient-2 predicts poor prognosis in tamoxifen-treated breast cancers. Oncogene. 2010;29(34):4838–4847. doi: 10.1038/onc.2010.228. [DOI] [PubMed] [Google Scholar]
- 11.Fullwood MJ, Liu MH, Pan YF, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462(7269):58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welboren WJ, van Driel MA, Janssen-Megens EM, et al. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28(10):1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verma S, Salmans ML, Geyfman M, et al. The estrogen-responsive Agr2 gene regulates mammary epithelial proliferation and facilitates lobuloalveolar development. Dev Biol. 2012;369(2):249–260. doi: 10.1016/j.ydbio.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanderlaag KE, Hudak S, Bald L, et al. Anterior gradient-2 plays a critical role in breast cancer cell growth and survival by modulating cyclin D1, estrogen receptor-alpha and survivin. Breast Cancer Res. 2010;12(3):R32. doi: 10.1186/bcr2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu D, Rudland PS, Sibson DR, Platt-Higgins A, Barraclough R. Human homologue of cement gland protein, a novel metastasis inducer associated with breast carcinomas. Cancer Res. 2005;65(9):3796–3805. doi: 10.1158/0008-5472.CAN-04-3823. [DOI] [PubMed] [Google Scholar]
- 16.Adam PJ, Boyd R, Tyson KL, et al. Comprehensive proteomic analysis of breast cancer cell membranes reveals unique proteins with potential roles in clinical cancer. J Biol Chem. 2003;278(8):6482–6489. doi: 10.1074/jbc.M210184200. [DOI] [PubMed] [Google Scholar]
- 17.Petek E, Windpassinger C, Egger H, Kroisel PM, Wagner K. Localization of the human anterior gradient-2 gene (AGR2) to chromosome band 7p21.3 by radiation hybrid mapping and fluorescencein situ hybridisation. Cytogenet Cell Genet. 2000;89(3–4):141–142. doi: 10.1159/000015594. [DOI] [PubMed] [Google Scholar]
- 18.Brychtova V, Zampachova V, Hrstka R, et al. Differential expression of anterior gradient protein 3 in intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Exp Mol Pathol. 2014;96(3):375–381. doi: 10.1016/j.yexmp.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Gray TA, MacLaine NJ, Michie CO, et al. Anterior gradient-3: a novel biomarker for ovarian cancer that mediates cisplatin resistance in xenograft models. J Immunol Methods. 2012;378(1–2):20–32. doi: 10.1016/j.jim.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 20.King ER, Tung CS, Tsang YT, et al. The anterior gradient homolog 3 (AGR3) gene is associated with differentiation and survival in ovarian cancer. Am J Surg Pathol. 2011;35(6):904–912. doi: 10.1097/PAS.0b013e318212ae22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaarala MH, Hirvikoski P, Kauppila S, Paavonen TK. Identification of androgen-regulated genes in human prostate. Mol Med Rep. 2012;6(3):466–472. doi: 10.3892/mmr.2012.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bu H, Schweiger MR, Manke T, et al. Anterior gradient 2 and 3 – two prototype androgen-responsive genes transcriptionally upregulated by androgens and by oestrogens in prostate cancer cells. FEBS J. 2013;280(5):1249–1266. doi: 10.1111/febs.12118. [DOI] [PubMed] [Google Scholar]
- 23.Tavassoli FA, Devilee P. In: WHO Classification of Tumours. Pathology and Genetics of Tumours of the Breast and Female Genital Organs. editors, editor. Lyon, France: IARC Press; 2003. [Google Scholar]
- 24.Sobin LH, Fleming ID. TNM classification of malignant Tumors, fifth edition (1997). Union Internationale Contre le Cancer and the American Joint Committee on Cancer. Cancer. 1997;80(9):1803–1804. doi: 10.1002/(sici)1097-0142(19971101)80:9<1803::aid-cncr16>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 25.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 26.Taiseer IR, Samar ARM, Abdelmonem HA. Immunohistochemical expression of aldehyde dehydrogenase-1 and hypoxia-inducible factor-1α in breast cancer. IJAR. 2014;2(7):822–830. [Google Scholar]
- 27.Brychtova V, Vojtesek B, Hrstka R. Anterior gradient 2: a novel player in tumor cell biology. Cancer Lett. 2011;304(1):1–7. doi: 10.1016/j.canlet.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 28.Chevet E, Fessart D, Delom F, et al. Emerging roles for the prooncogenic anterior gradient-2 in cancer development. Oncogene. 2013;32(20):2499–2509. doi: 10.1038/onc.2012.346. [DOI] [PubMed] [Google Scholar]
- 29.Salmans ML, Zhao F, Andersen B. The estrogen-regulated anterior gradient 2 (AGR2) protein in breast cancer: a potential drug target and biomarker. Breast Cancer Res. 2013;15(2):204. doi: 10.1186/bcr3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fritzsche FR, Dahl E, Pahl S, et al. Prognostic relevance of AGR2 expression in breast cancer. Clin Cancer Res. 2006;12(6):1728–1734. doi: 10.1158/1078-0432.CCR-05-2057. [DOI] [PubMed] [Google Scholar]
- 31.Smirnov DA, Zweitzig DR, Foulk BW, et al. Global gene expression profiling of circulating tumor cells. Cancer Res. 2005;65(12):4993–4997. doi: 10.1158/0008-5472.CAN-04-4330. [DOI] [PubMed] [Google Scholar]
- 32.Hrstka R, Brychtova V, Fabian P, Vojtesek B, Svoboda M. AGR2 predicts tamoxifen resistance in postmenopausal breast cancer patients. Dis Markers. 2013;35(4):207–212. doi: 10.1155/2013/761537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darb-Esfahani S, Fritzsche F, Kristiansen G, et al. Anterior gradient protein 2 (AGR2) is an independent prognostic factor in ovarian high-grade serous carcinoma. Virchows Arch. 2012;461(2):109–116. doi: 10.1007/s00428-012-1273-4. [DOI] [PubMed] [Google Scholar]
- 34.Park K, Chung YJ, So H, et al. AGR2, a mucinous ovarian cancer marker, promotes cell proliferation and migration. Exp Mol Med. 2011;43(2):91–100. doi: 10.3858/emm.2011.43.2.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hruz T, Laule O, Szabo G, et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Obacz J, Takacova M, Brychtova V, et al. The role of AGR2 and AGR3 in cancer: similar but not identical. Eur J Cell Biol. 2015;94(3–4):139–147. doi: 10.1016/j.ejcb.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Norris AM, Gore A, Balboni A, Young A, Longnecker DS, Korc M. AGR2 is a SMAD4-suppressible gene that modulates MUC1 levels and promotes the initiation and progression of pancreatic intraepithelial neoplasia. Oncogene. 2013;32(33):3867–3876. doi: 10.1038/onc.2012.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hrstka R, Murray E, Brychtova V, Fabian P, Hupp TR, Vojtesek B. Identification of an AKT-dependent signalling pathway that mediates tamoxifen-dependent induction of the pro-metastatic protein anterior gradient-2. Cancer Lett. 2013;333(2):187–193. doi: 10.1016/j.canlet.2013.01.034. [DOI] [PubMed] [Google Scholar]
- 39.Tsuji T, Satoyoshi R, Aiba N, et al. Agr2 mediates paracrine effects on stromal fibroblasts that promote invasion by gastric signet-ring carcinoma cells. Cancer Res. 2015;75(2):356–366. doi: 10.1158/0008-5472.CAN-14-1693. [DOI] [PubMed] [Google Scholar]
- 40.Arumugam T, Deng D, Bover L, Wang H, Logsdon CD, Ramachandran V. New blocking antibodies against novel AGR2-C4.4A pathway reduce growth and metastasis of pancreatic tumors and increase survival in mice. Mol Cancer Ther. 2015;14(4):941–951. doi: 10.1158/1535-7163.MCT-14-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Determination of anti-AGR3 antibody cross-reactivity.
Notes: The specificity of our in-house anti-AGR3 antibody (AGR3.1) was confirmed by Western blot (upper panel). For comparison, we added testing of rabbit polyclonal sera raised against AGR2 protein, which recognizes AGR3 as well (bottom panel).
Abbreviation: AGR, anterior gradient protein.
Survival analysis of cohort of ER- and PR-negative breast cancer patients.
Notes: Kaplan–Meier analysis of (A) progression-free survival in relation to AGR3 expression (P=0.282, Breslow test) and (B) overall survival in relation to AGR3 expression (P=0.867, Breslow test).
Abbreviations: ER, estrogen receptor; PR, progesterone receptor; AGR3, anterior gradient protein 3.
Table S1.
Association of AGR2 and AGR3 mRNA levels with other tumor variables
| Patients (n) | AGR2 mRNA | AGR3 mRNA | |
|---|---|---|---|
| Histological grade | |||
| G1 | 26 | 0.166a | 0.037a |
| G2 | 33 | ||
| G3 | 34 | ||
| Tumor size | |||
| pT1 | 35 | 0.774a | 0.990a |
| pT2 | 50 | ||
| pT3 + pT4 | 8 | ||
| Nodal status | |||
| Negative | 37 | 0.822b | 0.541b |
| Positive | 52 | ||
| ER status | |||
| Negative | 17 | 0.081b | <0.001b |
| Positive | 76 | ||
| PR status | |||
| Negative | 19 | 0.124b | <0.001b |
| Positive | 73 | ||
| Her2/neu status | |||
| Negative | 57 | 0.364b | 0.603b |
| Positive | 36 | ||
| Ki-67 | |||
| <15% | 44 | 0.169b | 0.494b |
| ≥15% | 48 |
Notes:
Determination of P-level using Kruskal–Wallis analysis of variance.
Determination of P-level using Mann–Whitney U-test.
Abbreviations: AGR, anterior gradient protein; ER, estrogen receptor; PR, progesterone receptor; Her2/neu, human epidermal growth factor receptor 2.
Table S2.
Univariate survival analyses for the analyzed set of tumors
| Variablea | Statistical significanceb
|
|
|---|---|---|
| PFS | OS | |
| AGR3 expression | 0.037 | 0.111 |
| Histology | ||
| DU vs LO | 0.665 | 0.393 |
| DU vs OTH | 0.537 | 0.980 |
| LO vs OTH | 0.851 | 0.675 |
| Grade | ||
| G1 vs G2 | 0.159 | 0.137 |
| G1 vs G3 | 0.002 | 0.009 |
| G2 vs G3 | 0.020 | 0.055 |
| Tumor size | ||
| pT1 vs pT2 | 0.249 | 0.333 |
| pT1 vs pT3 | 0.000 | 0.000 |
| pT2 vs pT3 | 0.000 | 0.000 |
| Nodal status | 0.043 | 0.415 |
| ER status | 0.110 | 0.344 |
| PR status | 0.023 | 0.145 |
| Her2/neu status | 0.000 | 0.000 |
| Ki-67 expression | 0.004 | 0.018 |
Notes:
AGR3 expression, AGR3 “low” (less than 50% of stained cells) vs AGR3 “high” (more than 50% of stained cells); histology, ductal vs lobular vs others; nodal status, negative vs positive; estrogen receptor status, negative vs positive; progesterone receptor status, negative vs positive; Her2/neu status, negative vs positive; Ki-67 expression, <15% vs ≥15%.
Probability, P, from Breslow test.
Abbreviations: PFS, progression-free survival; OS, overall survival; AGR3, anterior gradient protein 3; DU, ductal; LO, lobular; OTH, others; ER, estrogen receptor; PR, progesterone receptor; Her2/neu, human epidermal growth factor receptor 2.
Table S3.
Survival analysis of patients with AGR3-expressing tumors
| Subgroup | Statistical significancea
|
|
|---|---|---|
| PFS | OS | |
| Histological grade | ||
| G≤2 | 0.024 | 0.005 |
| G>2 | 0.945 | 0.583 |
| Her2/neu status | ||
| Negative | 0.019 | 0.009 |
| Positive | 0.781 | 0.278 |
| PR status | ||
| Negative | 0.669 | 0.911 |
| Positive | 0.448 | 0.224 |
| ER status | ||
| Negative | 0.431 | 0.507 |
| Positive | 0.228 | 0.234 |
Note:
Probability, P, from Breslow test.
Abbreviations: AGR3, anterior gradient protein 3; PFS, progression-free survival; OS, overall survival; Her2/neu, human epidermal growth factor receptor 2; ER, estrogen receptor; PR, progesterone receptor.
Table S4.
Association of immunohistochemical staining for AGR3 with other tumor variables in a cohort of ER-negative breast cancer patients
| Variable | N (%)a
|
Statistical significanceb | |||
|---|---|---|---|---|---|
| Patients | AGR3 negative/border | AGR3 weak/moderate | AGR3 strong | ||
| Histological grade | |||||
| G1 | 1 | 1 (100.0) | 0 (0.0) | 0 (0.0) | 0.917 |
| G2 | 10 | 6 (60.0) | 3 (30.0) | 1 (10.0) | |
| G3 | 71 | 47 (66.2) | 15 (21.1) | 9 (12.7) | |
| Tumor size | |||||
| pT1 | 32 | 24 (75.0) | 5 (15.6) | 3 (9.4) | 0.729 |
| pT2 | 44 | 26 (59.1) | 11 (25.0) | 7 (15.9) | |
| pT3 + pT4 | 11 | 8 (72.8) | 2 (18.2) | 1 (9.0) | |
| Nodal status | |||||
| Negative | 34 | 25 (73.5) | 7 (20.6) | 2 (5.9) | 0.282 |
| Positive | 53 | 31 (58.5) | 13 (24.5) | 9 (17.0) | |
| Her2/neu status | |||||
| Negative | 40 | 34 (85.0) | 5 (12.5) | 1 (2.5) | 0.001 |
| Positive | 49 | 24 (49.0) | 15 (30.6) | 10 (20.4) | |
| Ki-67 | |||||
| <15% | 6 | 4 (66.7) | 2 (33.3) | 0 (0.0) | 0.626 |
| ≥15% | 42 | 30 (71.4) | 7 (16.7) | 5 (11.9) | |
Notes:
Number (percentage) of patients with tumors characterized by negative/border, weak/moderate, or strong expression of AGR3. Probability,
P, was calculated using Fisher’s exact test with the Freeman–Halton extension.
Abbreviations: AGR3, anterior gradient protein 3; ER, estrogen receptor; Her2/neu, human epidermal growth factor receptor 2.