Abstract
Aberrant microRNAs (miRNAs) are reported to contribute to the pathogenesis of most human malignancies. The miRNA, miR-134, has been found to be downregulated in renal cell carcinoma (RCC), but its function in the disease is unknown. The aims of this study were to detect the expression of miR-134 in human RCC samples and explore its function in RCC cell lines. Real-time qualitative polymerase chain reaction (qPCR) was used to quantify miR-134 in human RCC samples. Assays for cell cycle, viability, migration, and invasion were performed to assess the phenotypic changes in RCC cells. A luciferase reporter assay was carried out to confirm whether KRAS (Kirsten rat sarcoma viral oncogene homolog) is a direct target of miR-134. Western blot was used to identify the potential signaling pathways that had an impact on RCC cell growth and alterations of markers for epithelial–mesenchymal transition (EMT), which affected metastasis by miR-134. miR-134 was found to be downregulated in RCC samples (p<0.05), while overexpression of miR-134 suppressed proliferation (p<0.05) by triggering G1/G0 cell cycle arrest (p<0.05). Forced expression of miR-134 could also inhibit migration (p<0.05) and invasion (p<0.05) by blocking EMT in RCC cell lines. KRAS was identified as a target of miR-134, and miR-134 may act as a tumor suppressor through the KRAS-related MAPK/ERK pathway other than PI3K/AKT signaling. Thus, miR-134 may function as a tumor suppressor in cell proliferation and EMT by targeting KRAS in RCC cells.
Introduction
Renal cell carcinoma (RCC) is a common malignant cancer in China, reported to have approximately 54.3 new diagnoses per 10,000 Chinese in 2005 (Yang et al., 2005). It is the third most common genitourinary cancer, with 54,390 predominantly male new cases and 13,010 deaths expected in the United States in 2008 (Garcia et al., 2009). Up to 40% of these patients with primary disease will eventually develop metastases and those with metastases have a poor median survival of 6–12 months, only 9% with a 5-year survival (van et al., 2005). There is a lack of effective therapeutics in RCC as the disease is highly resistant to chemotherapy and radiotherapy (van et al., 2005). Thus, early detection and prognostic markers as well as novel therapeutic modalities are required.
microRNAs (miRNAs) are small noncoding RNAs with a length of ∼22 nucleotides and can bind to 3′-UTR of the targeted message RNA, thus inhibiting translation or promoting RNA degradation (Wu et al., 2007; Ozen et al., 2008). miRNAs are implicated in physiological and pathological processes, especially in tumor pathogenesis, and can act as tumor suppressors or oncogenes (Croce, 2009; Zimmerman and Wu, 2011). Aberrant levels of the miRNA, miR-134, have been detected in a variety of malignancies and could regulate tumor development, differentiation, proliferation, invasion, and metastasis (Li et al., 2012; Liu et al., 2013). In human nonsmall cell lung cancer cells, low expression of miR-134 was correlated with invasive potential, and altered miR-134 expression was associated with the inhibition of epithelial–mesenchymal transition (EMT), by directly binding to the 3′-UTR of the forkhead box M1 (FOXM1), which was involved in TGF-β1-induced EMT in A549 cells (Li et al., 2012). However, in head and neck carcinoma cells, miR-134 was found to be upregulated and induced oncogenicity and metastasis by targeting the 3′-UTR of the WW domain containing oxidoreductase (WWOX) (Liu et al., 2013). Thus, a given miRNA may have many mRNA targets, and the biological effects of changes in miRNA expression are likely to depend on the cellular context.
Kirsten rat sarcoma viral oncogene homolog (KRAS) oncogene, one of the RAS gene family members, encodes a small guanosine triphosphatase, which has crucial functions in various biological processes, including cell proliferation and EMT (Johnson et al., 2005; Chen et al., 2009; Yu et al., 2010; Wang et al., 2012). The downregulation of KRAS reduced tumor growth in pancreatic cancer, glioma, and oral cancer, among others (Yu et al., 2010; Shin et al., 2011; Wang et al., 2012; Ma et al., 2013; Tanaka et al., 2013). Recently, KRAS was found to inhibit EMT (Buonato et al., 2014). EMT has been reported to be a key process contributing to cancer metastasis and is characterized by the loss of the epithelial marker, E-cadherin, and an increase in the mesenchymal markers, vimentin and N-cadherin, and an increase in the migratory and invasive behavior (Kraljevic et al., 2011). Therefore, miR-134 and KRAS may be viable biomarkers or potential therapeutic targets for RCC.
In this study, we used qualitative polymerase chain reaction (qPCR) to investigate the miR-134 levels in RCC samples and RCC cells. Then, to confirm that miR-134 directly targets KRAS and the possible antiproliferative and antimetastatic properties of miR-134, a series of in vitro assays had been conducted. To further understand how miR-134 controls cell phenotypes by targeting KRAS, we then detected a number of markers indicating the alterations of EMT and proteins in signaling pathways downstream of KRAS.
All these results may give us insight into how miR-134 acts as a tumor suppressor in RCC cells and suggest a novel biomarker or therapeutic strategy for treatment of RCC.
Materials and Methods
Study design
To investigate the miR-134 levels in RCC samples and RCC cells, total RNA was isolated from tissues and cultured cells. Then, qPCR was conducted to measure the relative expression of miR-134 in RCC samples versus paired adjacent nontumor tissues or in RCC cells versus normal renal cells.
Cells of the cell lines, 786-O and caki-1, transfected with nothing, negative control (NC), or miR-134 mimics were defined as mock, NC, or miR-134 mimic groups, respectively. After transfection, cell proliferation assay and cell cycle assay were performed on cells to study the impact of miR-134 on cell proliferation. Cell migration and invasion assays were carried out to investigate the antimetastatic properties of miR-134. In addition, dual-luciferase assays were also conducted to confirm that miR-134 directly targets KRAS.
Proteins were isolated from transfected cells and western blot was used to detect the KRAS levels in different groups, alterations of markers, taking E-cadherin and N-cadherin, for example, in EMT, and the changes of proteins in signaling pathways downstream of KRAS so that we can further understand how miR-134 controls cell phenotypes by targeting KRAS.
Cell culture and tissue samples
The human RCC cell lines (786-O, caki-1, 769-P, and ACHN), normal renal proximal tubular cells (HK-2), and human embryonic kidney cells (HEK-293T) were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells of the RCC cell lines, 786-O and 769-P, were maintained in RPMI-1640 (Gibco), and HK-2, HEK-293T, and ACHN cells were maintained in Dulbecco's modified Eagle's medium (Gibco), and caki-1 was maintained in McCoy's 5A (Gibco), all supplemented with 10% fetal bovine serum (FBS; Gibco) within a humidified atmosphere containing 5% CO2 at 37°C.
In accordance with the local Ethics Committees of the First Affiliated Hospital with Nanjing Medical University, China, 24 paired tumor specimens and tissue samples used to detect miR-134 expression were obtained with informed consent from RCC patients (Table 1). These patients underwent radical nephrectomy or partial nephrectomy. All samples were obtained during surgery, immediately frozen in liquid nitrogen, and stored at −80°C for further analysis. Diagnoses were determined by histopathological examination of all the tumor specimens and nontumor tissue samples.
Table 1.
Characteristics of Renal Cell Carcinoma Patients
| Gender | ||
| Male | 15 | |
| Female | 9 | |
| Age, median (range) | 58.5 (29–80) y | |
| T stage | ||
| T1 | 19 | |
| T2 | 1 | |
| T3 | 4 | |
| T4 | 0 | |
| Pathology | Clear cell RCC | 24 |
RCC, renal cell carcinoma.
Cell transfection
Cells of the cell lines, 786-O and caki-1, were seeded in six-well plates at 70% confluence on the day before transfection. Cell transfection was performed with Lipofectamine2000 (Invitrogen) in accordance with the manufacturer's instructions. Five hours post-transfection, the culture medium was replaced with RPMI-1640 or McCoy's 5A containing FBS as described before. The sequences of the miR-134 mimics were sense, 5′-UGUGACUGGUUGACCAGAGGGG-3′; and antisense, 5′-CCUCUGGUCAACCAGUCACAUU-3′. RNA with no sequence homology to any human genomic sequence was used as the NC: sense, 5′-UCCUCCGAACGUGUCACGUTT-3′; and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. The sequence of the miR-134 inhibitor (Inhibitor) was 5′-CCCCUCUGGUCAACCAGUCACA-3′. The sequence of the negative control inhibitor (Inhibitor NC) was 5′-CAGUACUUUUGUGUAGUACAA-3′. miR-134 mimics, NC, Inhibitor, or Inhibitor NC used for each transfection were designed and synthesized by Invitrogen. For functional assays, cells grown in six-well plates were transfected with 100 pM of synthetic miR-134 mimics or NC. For the dose-dependent experiment, cells were transfected with 50pM of synthetic miR-134 mimics or NC to eliminate the dose effect of high dose of miR-134 transfection. Total RNA was prepared 24 h after transfection and used for qPCR analysis. Total protein was prepared 60 h after transfection and used for western blot analysis.
RNA isolation and qPCR
Total RNA was isolated from tissues and cultured cells using Trizol (Invitrogen) in accordance with the manufacturer's instructions for miRNA and mRNA analyses. RNA concentration was measured using NanoDrop (Thermo Scientific). Analysis of mature miR-134 expression was performed using a TaqMan miRNA assay in accordance with the manufacturer's instructions (Applied Biosystems). Levels of miR-134 relative to U6 were calculated using the 2−ΔΔCt method (Yuan et al., 2006). The reaction for miRNA detection was performed under the following conditions: 95°C for 10 min, 40 cycles at 95°C for 15 s, and 60°C for 1 min. The reactions were performed and analyzed using an Applied Biosystems StepOne Plus Real-Time PCR System (Applied Biosystems). All reactions were run in triplicate. Differences between the tumor and paired adjacent nontumor samples were analyzed using Student's paired t-test using SPSS software (Version 13.0 SPSS); meanwhile, the differences between the tumor cells and normal cells were analyzed using Student's t-test. A value of p<0.05 was considered as statistically significant.
Cell proliferation assay
Cells of the cell lines, 786-O and caki-1, were transfected with miR-134 mimics or NC to investigate the impact of miR-134 on cell proliferation of RCC cells. Forty-eight hours after transfection, cells were seeded into 96-well plates at a density of 2×103 cells/well and cultured for 24, 48, 72, or 96 h. Cell proliferation was determined using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies) in accordance with the manufacturer's protocol. Absorbance was detected at the wavelength of 450 nm. Three wells were measured for cell viability in each treatment group.
Cell cycle assay
Flow cytometry was used to analyze the distribution of the cell cycle stages in transfected cells so that we could further understand the possible mechanism of miR-134 on cell growth. Forty-eight hours post-transfection, cells were harvested, washed twice with ice-cold phosphate-buffered saline, and fixed with 70% ethanol at −20°C overnight. Subsequently, cells were incubated in 50 mg/mL propidium iodide and 1 mg/mL RNase for 30 min at room temperature. The treated cells were analyzed by flow cytometry (Becton Dickinson). At least 100,000 cells were acquired for each sample. The experiments were performed in triplicate.
Cell migration and invasion assays
To investigate the possible effect on metastasis on RCC cells, migration and invasion assays were used. For the migration assays, 2×104 cells in 200 mL of serum-free medium were placed in the upper chamber of the transwell (pore size, 8 mm; BD Bioscience). For the invasion assays, 5×104 cells in 200 mL of serum-free medium were placed in the upper chamber that was coated with Matrigel (BD Bioscience) in accordance with the manufacturer's protocol. Media containing 20% FBS were added to the lower chamber. After the cells had incubated for 24 h at 37°C, the cells remaining in the upper membrane were removed and those on the lower surface of the membrane were fixed in 95% ethanol and stained with crystal violet. Five random fields were counted. All of the experiments were performed in triplicate.
Plasmid construction and dual-luciferase assay
To fully understand the mechanisms by which miRNAs execute their function, we adopted three bioinformatic algorithms (TargetScan, PicTar, and miRanda) to identify a large number of potential target genes of miR-134. Considering the reported influence of miR-134 on cell proliferation and metastasis, as well as the fact that KRAS was involved in the signaling pathways, which have an impact on cell proliferation and metastasis, KRAS was selected for further analysis. We hypothesized that KRAS might be a target of miR-134; therefore, a fragment of the KRAS 3′-UTR (KRAS-WT) and a mutated 3′-UTR of KRAS (KRAS-MUT) that contained the putative miR-134-binding sites were cloned downstream of the luciferase gene in a pGL3-control vector (Invitrogen). For reporter assays, HEK-293T cells were cotransfected with KRAS-WT plasmid and miR-134 mimics, KRAS-WT plasmid and NC, KRAS-MUT plasmid and miR-134 mimics, KRAS-MUT plasmid and NC, KRAS-WT plasmid and miR-134 inhibitor, or KRAS-WT plasmid and inhibitor NC. Luciferase activities were measured by dual-luciferase assays (Promega) 48 h after cotransfection and normalized against the activity of the Renilla luciferase gene.
Protein isolation and western blot
To further study the effects of miR-134 on protein changes in signaling pathways referring to cell proliferation and EMT in RCC cells, proteins were isolated from transfected cells. Cells were washed twice in ice-cold phosphate-buffered saline and lysed using radioimmunoprecipitation assay buffer (KeyGene Biotech) supplemented with protease inhibitors at 4°C for 30 min. Equal amounts of proteins were analyzed by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to a polyvinylidene fluoride membrane (Millipore), blocked for 1 h with 5% nonfat milk at room temperature, and incubated with primary antibodies at 4°C overnight. The membrane was incubated with a horseradish peroxidase-conjugated secondary antibody for 1 h after three washes with Tris-buffered saline and 0.1% Tween. Antibodies against KRAS, glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Bioworld Technology), p-ERK, p-AKT (Abcam), AKT, ERK, E-cadherin, N-cadherin, and vimentin (Cell Signaling Technology) were used in western blot analysis in accordance with the manufacturer's instructions. Signals were detected using enhanced chemiluminescence detection reagent (Thermo Scientific). Protein levels were determined by normalization to GAPDH.
Statistical analyses
Results are expressed as mean±standard deviation (SD). Differences between groups were subjected to Student's t-test. p<0.05 was considered statistically significant. All of the statistical calculations were performed using SPSS software (Version 13.0 SPSS).
Results
miR-134 is low in RCC samples and cells
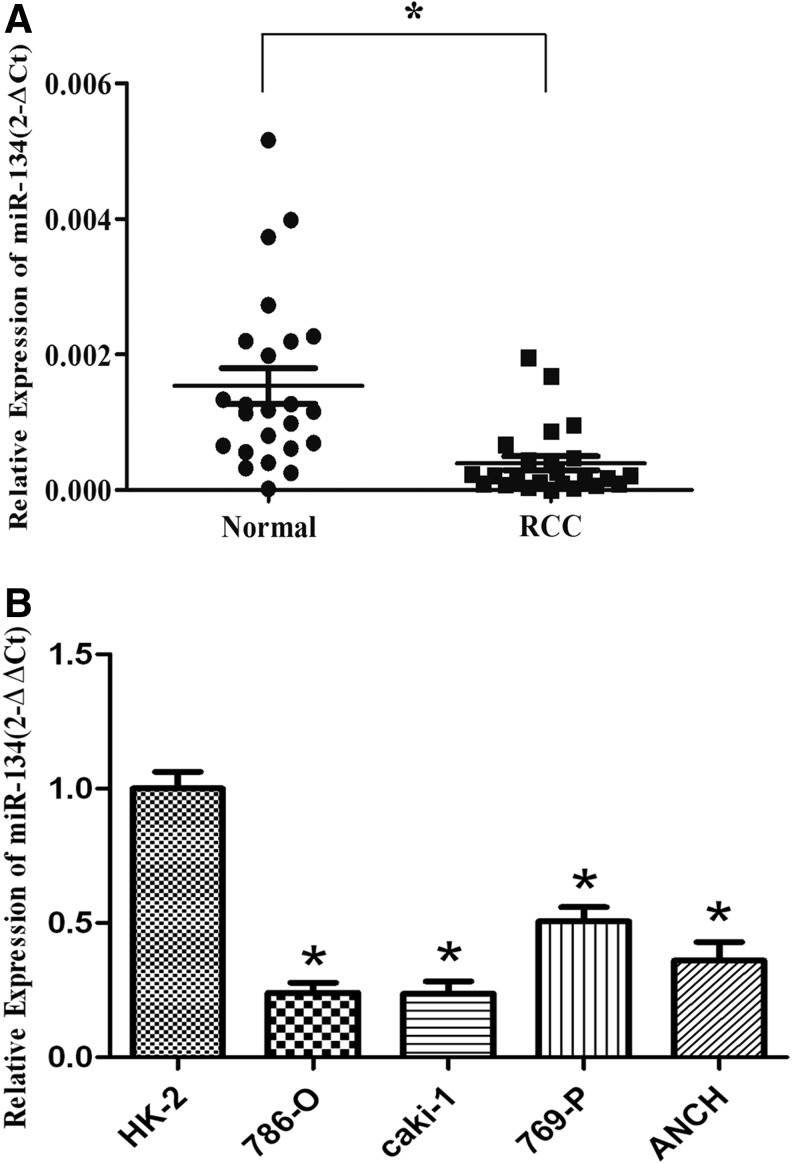
miR-134 expression was analyzed in 24 paired RCC tissues and their paired adjacent nontumor tissues samples using qPCR. There was less miR-134 in RCC tissues compared with that in adjacent nontumor tissues (p<0.05; Fig. 1A). Furthermore, analysis of miR-134 expression in the normal renal cell (HK-2) and four RCC cell lines (786-O, caki-1, ACHN, and 769-P) reveals that miR-134 is decreased in tumor cell lines as well (p<0.05; Fig. 1B).
FIG. 1.
miR-134 is downregulated in renal cell carcinoma (RCC). (A) miR-134 level in RCC samples was significantly downregulated compared with the paired adjacent nontumor tissues. The median in each triplicate was used to calculate the relative miR-134 concentration using the comparative 2−ΔCt method (2-ΔCt was used to detect the expression of miR-134 in tissue samples). (B) The expression level of miR-134 in RCC cell lines was relatively low compared with the normal renal cell line. The median in each triplicate was used to calculate the relative miR-134 concentration using the comparative 2−ΔΔCt method (2-ΔΔCt and normalized data were used to identify the comparative expression among RCC cell lines). *p<0.05 compared with the nontumor tissues or normal cell line. Black circle, normal tissue of RCC patients; black square, tumor tissue of RCC patients; each circle/square means the relative expression of miR-134 of normal/tumor tissue of one patient.
miR-134 inhibits cell proliferation of RCC cells
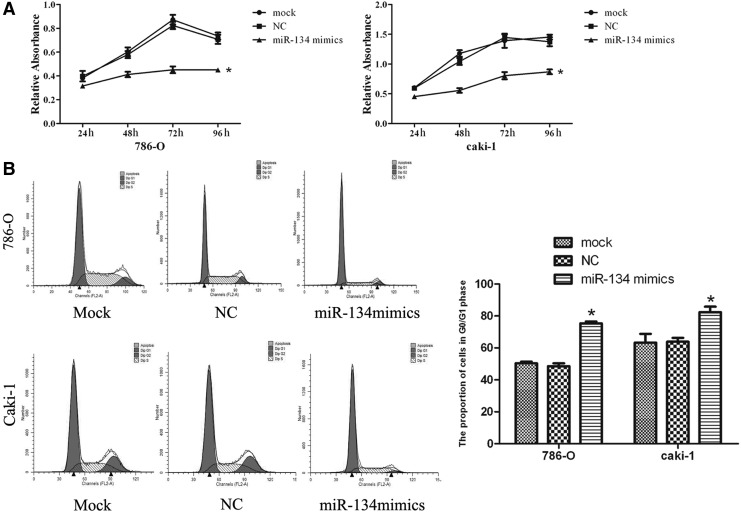
In the CCK-8 assay, the forced expression of miR-134 significantly inhibited the growth of RCC cells starting at 24 h (p<0.05; Fig. 2A and Supplementary Fig. S1A; Supplementary Data are available online at www.liebertpub.com/dna). The percentages of 786-O and caki-1 cells transfected with miR-134 mimics in the G0/G1 phase were higher compared with the NC and mock, providing evidence that high miR-134 expression may induce G0/G1 cell cycle arrest in RCC cells (p<0.05; Fig. 2B and Supplementary Fig. S1B).
FIG. 2.
miR-134 inhibits cell proliferation and induces cell cycle arrest in 786-O and caki-1 cell lines. (A) Assessment of cell proliferation by CCK-8 assay. The proliferation of 786-O and caki-1 cells was significantly inhibited by overexpression of miR-134. (B) Flow cytometry analysis showed that the cell cycle of 786-O and caki-1 cells was arrested at G1 after upregulation of miR-134. The histogram indicates the percentage of cells in G1. *p<0.05 compared with the mock or negative control group. NC, negative control.
miR-134 inhibits EMT in RCC cells
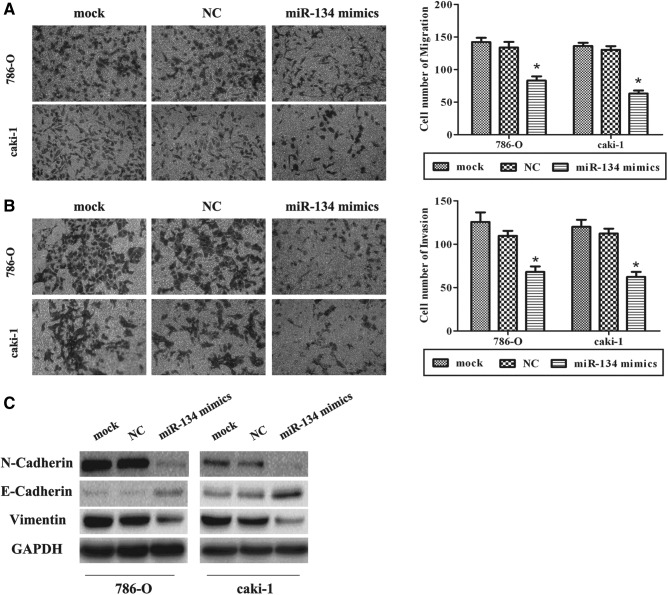
As previously described, EMT is a key process in cancer metastasis. Migration and invasion assays reveal lower migratory and invasive abilities in the cell in which miR-134 expression was forced than the NC and mock (p<0.05; Fig. 3A, B and Supplementary Fig. S2A, B). Overexpression of miR-134 triggered changes in EMT markers' expression with a gain in E-cadherin expression and a loss of N-cadherin and vimentin (Fig. 3C and Supplementary Fig. S2C), which indicate that miR-134 could inhibit EMT.
FIG. 3.
miR-134 inhibits epithelial–mesenchymal transition (EMT) in RCC cells. (A, B) Overexpression of miR-134 inhibited migration and invasion in RCC cell lines. (C) Western blot analysis was used to detect the changes in EMT markers in 786-O and caki-1 cells treated with miR-134 mimics. A gain in E-cadherin expression and a loss of N-cadherin and vimentin were observed in treated cells. Data are mean±SD of at least three independent experiments. *p<0.05 compared with the mock or negative control group. Original magnification 200×. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; SD, standard deviation.
miR-134 suppresses KRAS levels and can specifically bind to KRAS 3′-UTR
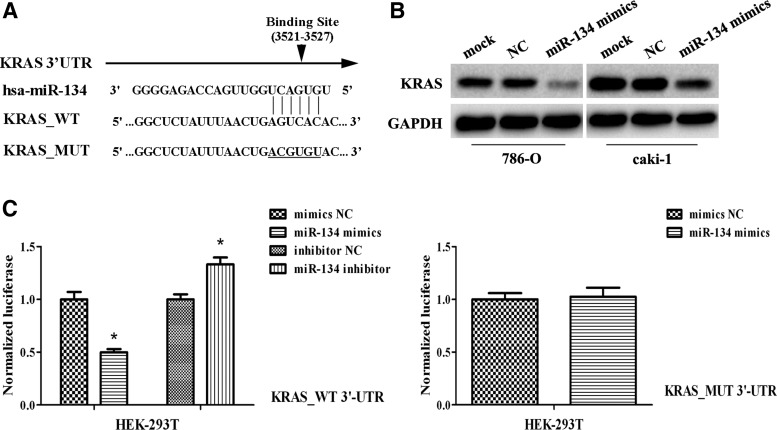
A binding site of miR-134 was observed in the 3′-UTR of KRAS (Fig. 4A). Western blot analysis indicated an inverse correlation between miR-134 and KRAS expression; KRAS levels were significantly decreased in cells treated with miR-134 mimics (p<0.05; Fig. 4B). Dual-luciferase assays revealed that miR-134-transfected cells showed a marked reduction (∼53%) of luciferase activity (Fig. 4C) and the inhibition of luciferase activity by miR-134 mimics was almost abolished in the KRAS-MUT (Fig. 4C), suggesting that the conserved region was fully responsible for miR-134 function.
FIG. 4.
KRAS is a direct target of miR-134. (A) Sequence of the miR-134-binding sites within the human KRAS 3′-UTR and a schematic diagram of the reporter constructs showing the entire KRAS 3′-UTR sequence (KRAS_WT) and the mutated KRAS 3′-UTR sequence (KRAS_MUT: the mutant nucleotides of the miR-134-binding site are underlined). (B) Western blot analysis of KRAS in 786-O and caki-1 cells transfected with miR-134 mimics or negative control. KRAS was significantly decreased with increasing miR-134. GAPDH was used as a housekeeping control. (C) Dual-luciferase assay results for HEK-293T cells suggest that KRAS is a target gene of miR-134. *p<0.05 compared with the mock or negative control group. All data are shown as mean±SD.
miR-134 suppresses the RAS/MAPK/ERK pathway
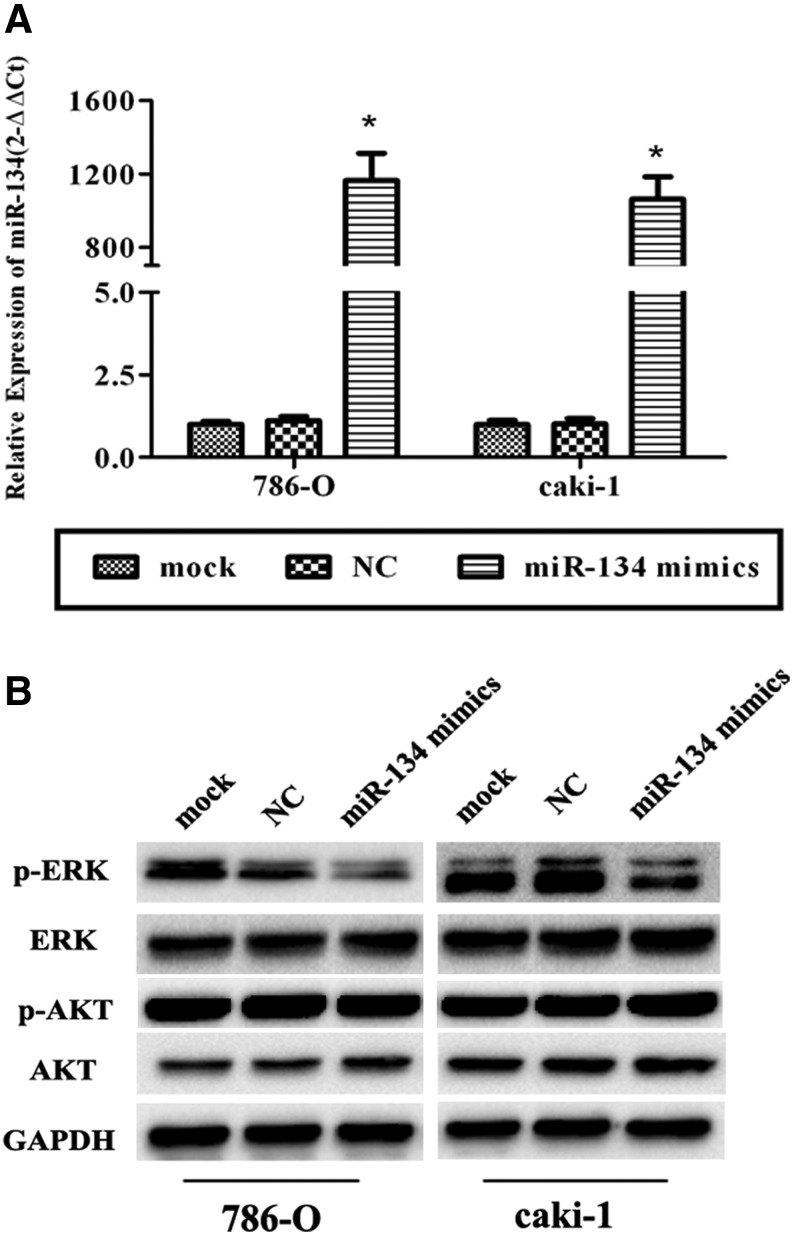
After transfection, the expression level in transfected cells, which were treated with miR-134 mimics, was over 1000-fold that of cells transfected with NC (p<0.05; Fig. 5A), while the expression level in transfected cells treated with a small dose of miR-134 mimics was about 200-fold that of cells transfected with NC (p<0.05; Supplementary Fig. S3A). Since our results suggest that the miR-134 might suppress cell proliferation and EMT, to assess the mechanism by which KRAS contributed to these progressions, we examined the effect of transient miR-134 upregulation on RAS/MAPK/ERK and PI3K/Akt signaling. KRAS was effectively reduced in miR-134 mimic-transfected RCC cells (p<0.05; Fig. 4B, Supplementary Fig. S3B), and p-ERK was significantly decreased (p<0.05; Fig. 5B, Supplementary Fig. S3B) with a gain of miR-134 and a loss of KRAS. However, no differences were found in p-Akt expression in the treated cells (p>0.05; Fig. 5B, Supplementary Fig. S3B). All these suggested that miR-134 might suppress cell proliferation and metastasis through the RAS/MAPK/ERK pathway other than PI3K/AKT signaling.
FIG. 5.
miR-134 suppresses the RAS/MAPK/ERK pathway. (A) Relative expression of miR-134 in transfected cells. The expression of miR-134 was extremely high in miR-134 mimic-transfected 786-O and caki-1 cells. (B) Western blot analysis of the RAS/MAPK/ERK pathway and AKT pathway. p-ERK was found to be decreased with a high expression of miR-134 and a low expression of KRAS, but no changes were found in the expression of p-Akt. *p<0.05 compared with the mock or negative control group. All data are shown as mean±SD.
Discussion
Recently, large numbers of aberrant miRNAs have been revealed by microarray in urologic cancers, including RCC. According to White et al.'s (2011) and Yi et al.'s (2010) microarray profiling, miR-134 was downregulated in RCC. Therefore, we investigated the expression of miR-134 in RCC samples in an eastern Chinese population. We found that miR-134 was significantly downregulated in RCC specimens relative to adjacent nontumor tissues. Meanwhile, we tested the miR-134 levels in RCC cell lines and found that there was less miR-134 in RCC cells than that in normal renal cells.
Although our study reported the deregulation of miR-134 in RCC, the function of miR-134 in RCC remains unknown. A considerable amount of evidence indicates that aberrant miRNAs can either suppress or promote the differentiation, proliferation, apoptosis, and metastasis of RCC (Catto et al., 2011). Because of the apparent loss of miR-134 in RCC patients, we forced the expression of miR-134 through transfected miR-134 mimics into RCC cells. We found that upregulating miR-134 suppressed cell proliferation due to an arrest in the G1 phase of the cell cycle. The gain in miR-134 also contributed to the inhibition of migration and invasion in the cancer cells by blocking EMT. Therefore, we conclude that miR-134 would be a novel tumor suppressor in RCC due to its negative effect on tumor growth and metastasis.
With the results of the present study showing a possible role of miR-134 in RCC pathogenesis as well as knowing that miRNAs could regulate the expression of target genes by interacting with the 3′-UTR of their target genes' mRNAs (Chou et al., 2013), we continued investigations to understand the mechanism of miR-134 in tumor pathogenesis. Three bioinformatic algorithms (TargetScan, PicTar, and miRanda) were selected to help find the target gene. The KRAS oncogene, a member of the well-known RAS family and the most likely to take part in the functional changes, was chosen as the candidate target gene. A recent research reported that miR-134 was a novel RTK (receptor tyrosine kinase)-related tumor suppressor and could mediate RTK and RTK inhibitor effects on glioblastoma by targeting KRAS and STAT5B (Zhang et al., 2014). As a result of the upregulation of miR-134, KRAS decreased significantly. This implies an inverse correlation between miR-134 and KRAS expression. Results of the luciferase reporter assay suggest that miR-134 may bind with the 3′-UTR of KRAS. Thus, we show that KRAS is a direct target gene of miR-134.
KRAS is reported to have a key role in cellular signal transduction and its mutation appears to be crucial in colorectal, pancreatic, and other cancers (Chen et al., 2009; Yu et al., 2010; Magudia et al., 2012). Activation of KRAS signaling could stimulate multiple downstream pathways, including the RAS/MAPK/ERK and PI3K/Akt, thus promoting tumorigenesis (Schubbert et al., 2007; Bodemann and White, 2008). A recent research revealed that an ERK1/2 blockade prevented EMT in lung cancer cells. This suggested a possible role of KRAS in tumor metastasis [17]. Our results in the present study show that a low expression of KRAS induced a decrease of p-ERK, indicating the inactivation of the ERK pathway. However, when we analyzed the activation state of the PI3K/Akt pathway, there were no changes found in the expression of p-Akt. All these results imply that miR-134 could directly regulate KRAS; the loss of KRAS inhibits the ERK pathway rather than the PI3K/Akt pathway and finally causes the suppression of cell proliferation and EMT in RCC. However, there are other targets of miR-134, which may contribute to the inhibition of cell proliferation and EMT in RCC; further studies need to determine if other pathways are cooperatively inhibited as well.
In summary, our results show that miR-134 is frequently downregulated in RCC and may function as a potential tumor suppressor in RCC by targeting KRAS. Consequently, miR-134 may have application in miRNA-based therapy. However, our data were derived from cell lines and as such cannot be considered accurate representations of a clinical RCC study. Further studies are still required to assess the roles of miR-134 in vivo and in a clinical context.
Supplementary Material
Acknowledgments
This work was supported by the project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), by the Jiangsu Provincial Special Program of Medical Science (BL2012027), by the Program for Development of Innovative Research Team in the First Affiliated Hospital of Nanjing Medical University, and by the National Natural Science Foundation of China [grant numbers 81171963, 81201998, 81201571]. The authors would like to thank Drs. Jie Zhao, Zhenzhen Wu, and Feifei Chen for providing antibodies for the research and also thank all the colleagues who helped in designing and performing the experiment.
Disclosure Statement
No competing financial interests exist.
References
- Bodemann B.O., and White M.A. (2008). Ral gtpases and cancer: Linchpin support of the tumorigenic platform. Nat Rev Cancer 8, 133–140 [DOI] [PubMed] [Google Scholar]
- Buonato J.M., and Lazzara M.J. (2014). Erk1/2 blockade prevents epithelial-mesenchymal transition in lung cancer cells and promotes their sensitivity to egfr inhibition. Cancer Res 74, 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catto J.W., Alcaraz A., Bjartell A.S., De Vere White R., Evans C.P., Fussel S., Hamdy F.C., Kallioniemi O., Mengual L., Schlomm T., and Visakorpi T. (2011). Microrna in prostate, bladder, and kidney cancer: a systematic review. Eur Urol 59, 671–681 [DOI] [PubMed] [Google Scholar]
- Chen X., Guo X., Zhang H., Xiang Y., Chen J., Yin Y., Cai X., Wang K., Wang G., Ba Y., Zhu L., Wang J., Yang R., Zhang Y., Ren Z., Zen K., Zhang J., and Zhang C.Y. (2009). Role of mir-143 targeting kras in colorectal tumorigenesis. Oncogene 28, 1385–1392 [DOI] [PubMed] [Google Scholar]
- Chou J., Shahi P., and Werb Z. (2013). Microrna-mediated regulation of the tumor microenvironment. Cell Cycle 12, 3262–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce C.M. (2009). Causes and consequences of microrna dysregulation in cancer. Nat Rev Genet 10, 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J.A., Cowey C.L., and Godley P.A. (2009). Renal cell carcinoma. Curr Opin Oncol 21, 266–271 [DOI] [PubMed] [Google Scholar]
- Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., and Slack F.J. (2005). Ras is regulated by the let-7 microrna family. Cell 120, 635–647 [DOI] [PubMed] [Google Scholar]
- Kraljevic Pavelic S., Sedic M., Bosnjak H., Spaventi S., and Pavelic K. Metastasis. (2011). New perspectives on an old problem. Mol Cancer 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang Y., Luo J., Fu Z., Ying J., Yu Y., and Yu W. (2012). Mir-134 inhibits epithelial to mesenchymal transition by targeting foxm1 in non-small cell lung cancer cells. FEBS Lett 586, 3761–3765 [DOI] [PubMed] [Google Scholar]
- Liu C.J., Shen W.G., Peng S.Y., Cheng H.W., Kao S.Y., Lin S.C., and Chang K.W. (2013). Mir-134 induces oncogenicity and metastasis in head and neck carcinoma through targeting wwox gene. Int J Cancer 134, 811–821 [DOI] [PubMed] [Google Scholar]
- Ma Y., Gu Y., Zhang Q., Han Y., Yu S., Lu Z., and Chen J. (2013). Targeted degradation of kras by an engineered ubiquitin ligase suppresses pancreatic cancer cell growth in vitro and in vivo. Mol Cancer Ther 12, 286–294 [DOI] [PubMed] [Google Scholar]
- Magudia K., Lahoz A., and Hall A. (2012). K-ras and b-raf oncogenes inhibit colon epithelial polarity establishment through up-regulation of c-myc. J Cell Biol 198, 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozen M., Creighton C.J., Ozdemir M., and Ittmann M. (2008). Widespread deregulation of microrna expression in human prostate cancer. Oncogene 27, 1788–1793 [DOI] [PubMed] [Google Scholar]
- Schubbert S., Shannon K., and Bollag G. (2007). Hyperactive ras in developmental disorders and cancer. Nat Rev Cancer 7, 295–308 [DOI] [PubMed] [Google Scholar]
- Shin K.H., Bae S.D., Hong H.S., Kim R.H., Kang M.K., and Park N.H. (2011). Mir-181a shows tumor suppressive effect against oral squamous cell carcinoma cells by downregulating k-ras. Biochem Biophys Res Commun 404, 896–902 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Suzuki H.I., Shibahara J., Kunita A., Isagawa T., Yoshimi A., Kurokawa M., Miyazono K., Aburatani H., Ishikawa S., and Fukayama M. (2013). Evi1 oncogene promotes kras pathway through suppression of microrna-96 in pancreatic carcinogenesis. Oncogene 33, 2454–2463 [DOI] [PubMed] [Google Scholar]
- van Spronsen D.J., de Weijer K.J., Mulders P.F., and De Mulder P.H. (2005). Novel treatment strategies in clear-cell metastatic renal cell carcinoma. Anticancer Drugs 16, 709–717 [DOI] [PubMed] [Google Scholar]
- Wang X.F., Shi Z.M., Wang X.R., Cao L., Wang Y.Y., Zhang J.X., Yin Y., Luo H., Kang C.S., Liu N., Jiang T., and You Y.P. (2012). Mir-181d acts as a tumor suppressor in glioma by targeting k-ras and bcl-2. J Cancer Res Clin Oncol 138, 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N.M., Bao T.T., Grigull J., Youssef Y.M., Girgis A., Diamandis M., Fatoohi E., Metias M., Honey R.J., Stewart R., Pace K.T., Bjarnason G.A., and Yousef G.M. (2011). Mirna profiling for clear cell renal cell carcinoma: Biomarker discovery and identification of potential controls and consequences of mirna dysregulation. J Urol 186, 1077–1083 [DOI] [PubMed] [Google Scholar]
- Wu W., Sun M., Zou G.M., and Chen J. (2007). Microrna and cancer: current status and prospective. Int J Cancer 120, 953–960 [DOI] [PubMed] [Google Scholar]
- Yang L., Parkin D.M., Ferlay J., Li L., and Chen Y. (2005). Estimates of cancer incidence in china for 2000 and projections for 2005. Cancer Epidemiol Biomarkers Prev 14, 243–250 [PubMed] [Google Scholar]
- Yi Z., Fu Y., Zhao S., Zhang X., and Ma C. (2010). Differential expression of mirna patterns in renal cell carcinoma and nontumorous tissues. J Cancer Res Clin Oncol 136, 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Lu Z., Liu C., Meng Y., Ma Y., Zhao W., Liu J., Yu J., and Chen J. (2010). Mirna-96 suppresses kras and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res 70, 6015–6025 [DOI] [PubMed] [Google Scholar]
- Yuan J.S., Reed A., Chen F., and Stewart C.N., Jr. (2006). Statistical analysis of real-time pcr data. BMC Bioinform 7, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Kim J., Mueller A.C., Dey B., Yang Y., Lee D.H., Hachmann J., Finderle S., Park D.M., Christensen J., Schiff D., Purow B., Dutta A., and Abounader R. (2014). Multiple receptor tyrosine kinases converge on microrna-134 to control kras, stat5b, and glioblastoma. Cell Death Differ 21, 720–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman A.L., and Wu S. (2011). Micrornas, cancer and cancer stem cells. Cancer Lett 300, 10–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.