Abstract
Mutation of the adenomatous polyposis coli (APC gene), an early event in the adenoma-carcinoma sequence, is present in 70-80% of sporadic human colorectal adenomas and carcinomas. To test the hypothesis that mutation of the APC gene alters microbial interactions with host intestinal mucosa prior to the development of polyposis, culture-independent methods (targeted qPCR assays and Illumina sequencing of the 16S rRNA gene V1V2 hypervariable region) were used to compare the intestinal microbial composition of 30 six-week old C57BL/6 APCMin/+ and 30 congenic wild type (WT) mice. The results demonstrate that similar to 12-14 week old APCMin/+ mice with intestinal neoplasia, 6 week old APCMin/+ mice with no detectable neoplasia, exhibit an increased relative abundance of Bacteroidetes spp in the colon. Parallel mouse RNA sequence analysis, conducted on a subset of proximal colonic RNA samples (6 APCMin/+, 6 WT) revealed 130 differentially expressed genes (DEGs, fold change ≥ 2, FDR <0.05). Hierarchical clustering of the DEGs was carried out by using 1-r dissimilarity measurement, where r stands for the Pearson correlation, and Ward minimum variance linkage, in order to reduce the number of input variables. When the cluster centroids (medians) were included along with APC genotype as input variables in a negative binomial (NB) regression model, four of seven mouse gene clusters, in addition to APC genotype, were significantly associated with the increased relative abundance of Bacteroidetes spp. Three of the four clusters include several downregulated genes encoding immunoglobulin variable regions and non-protein coding RNAs. These results support the concept that mutation of the APC gene alters colonic-microbial interactions prior to polyposis. It remains to be determined whether interventions directed at ameliorating dysbiosis in APCMin/+mice, such as through probiotics, prebiotics or antibiotics, could reduce tumor formation.
Introduction
Alterations in the gut microbiome (dysbiosis) have been reported in human colonic neoplasia [1–6]. However it remains unclear as to whether dysbiosis represents a response to tumorigenesis or whether it precedes tumor formation. One of the most prominent genetic mutations associated with the pathogenesis of sporadic and hereditary colorectal cancers (CRC) lies in the tumor suppressing adenomatous polyposis coli (APC) gene [7–13]. A germ-line mutation of the APC gene causes familial adenomatous polyposis (FAP), which results in the development of multiple colorectal adenomas at an early age that unequivocally lead to CRC if no surgical interventions are taken. APC mutations also represent an early event in the adenoma-carcinoma sequence and are present in about 70–80% of sporadic human colorectal adenomas and carcinomas.
The multiple intestinal neoplasia (Min) mouse model of FAP carries a truncation mutation at codon 850 of the Apc gene [14]. Studies comparing the number of intestinal polyps in germ-free and conventionally raised C57Bl/6 APCMin/+ mice suggest that the gut microbiome may promote development of intestinal neoplasia [15, 16]. One study reported decreased incidence of polyps in only the mid small intestinal segment, however a subsequent study reported a significant reduction of intestinal adenomas in both the small and large intestine of germ-free mice compared with conventionally raised mice. Antibiotic treatment of C57BL/6 APCMin/+MSH2-/-mice, which carry both the APC mutation and an HNPCC DNA mismatch repair mutation, reduced the number of polyps in both the small and large intestine [17].
We hypothesize that mutation of the APC gene results in alterations in host-microbiota interactions prior to tumor formation. To test this hypothesis, gut microbial composition was compared between 6 week-old C57Bl/6 APCMin/+, prior to the development of detectable neoplasia [18], and congenic WT mice.
Materials and Methods
Animal Type and Housing
All of the mice were acclimated for two weeks in order to reduce stress from traveling. Carbon dioxide was used during euthanasia of the mice. This study was approved by the Institutional Animal Care and Use Committee (#202449) and Division of Laboratory Animal Resources at Stony Brook University. Three shipments of 10 four-week-old female C57BL/6J APCMin/+ and 10 four-week-old female C57BL/6J WT mice were received from The Jackson Laboratory (Bar Harbor, ME) between June 2012 and May 2013. APCMin/+ mice and WT mice were housed separately in groups of three to four in specific pathogen free (SPF) cages for two weeks prior to euthanization. All the experiments strictly followed guidelines from the Institutional Animal Care and Use Committee and Division of Laboratory Animal Resources at Stony Brook University.
Tissue and Luminal Content Sample Collection
All of the mice were euthanized at 6 weeks of age using carbon dioxide. Immediately after sacrifice, the gastrointestinal tract was divided along its cephalocaudal axis as previously described [19]. The segments analyzed included the ileum, cecum, proximal colon, and distal colon. Each small intestinal segment was washed in sterile phosphate buffered saline to remove the luminal content. A 1.0–1.5-cm section was obtained from the proximal ends of duodenum, jejunum, distal ends of ileum, proximal colon, and distal colon, and placed into RNAlater solution (Life Technologies, Grand Island, NY, USA) for RNA/DNA studies. The cecum was placed in its entirety in RNAlater. Three pellets of distal colonic luminal content (formed stool) were collected from the distal colon and stored in RNAlater. All the samples were kept at room temperature overnight and then stored in -80°C. In the first cohort, the remainder of each intestinal segment was processed into “swiss rolls” and fixed in 10% buffered formalin for histological analyses. In the second and third cohort, the remainder of each intestinal segment was stained with 0.25% methylene blue and inspected under a dissecting microscope for adenomas and aberrant crypt foci [20]. Stools were collected from nine 12–14 week APCMin\+ female mice with intestinal neoplasia [18] and six WT female mice, and placed into RNAlater.
DNA and RNA Extraction of Intestinal Tissue and Luminal Content Samples
Total RNA and DNA, (host and associated bacterial mixed community) were extracted from the duodenum, jejunum, ileum, cecal pouch, proximal colon and distal colon tissues using TRI Reagent (Sigma, St. Louis, MO) according to the manufacturer’s recommendations. For distal colonic luminal content samples, DNA was extracted using the UltraClean Fecal Kit (Mo BIO Laboratories, Inc., Carlsbad, CA).
Quantitative PCR (qPCR) for targeted bacterial clades
QPCR assays were performed using established primers for Bacteroides–Prevotella–Porphyromonas [21], Lachnospiraceae [21], and total bacteria [22] on all tissue and distal colonic luminal content samples as previously described [23, 24]. The relative abundance of taxa within the Bacteroidetes phylum measured by ΔCt = Ct (threshold cycle)total bacteria - CtBacteroides–Prevotella–Porphyromonas. The relative abundance of taxa within the Lachnospiraceae clade (i.e., Clostridia Group Xi’an clade) was measured by ΔCt = Ct (threshold cycle)total bacteria - CtLachnospiraceae as previously described [23, 24]. All assays were carried out in triplicate. Plasmid quantification standards were prepared from representative clones of the target organisms. The Mann-Whitney test using GraphPad Prism 5 (La Jolla, California) was performed to compare APCMin/+ and WT ΔCt values. The Bonferroni correction was made to correct for multiple comparison, thus significance required p< 0.025.
16S rRNA Amplicon Library Construction and Illumina V1V2 Sequencing Analysis
Bacterial profiles were determined by broad-range amplification and sequence analysis of 16S rRNA genes following our previously described methods [25, 26]. In brief, amplicons were generated using primers that target approximately 300 bp. of the V1V2 variable region of the 16S rRNA gene (primers 8F and 338R, modified by addition of Illumina adapter and dual index sequences). PCR products were normalized using a SequalPrep kit (Invitrogen, Carlsbad, CA), pooled, lyophilized, purified and concentrated using a DNA Clean and Concentrator Kit (Zymo, Irvine, CA). Pooled amplicons was quantified using Qubit Fluorometer 2.0 (Invitrogen, Carlsbad, CA). The pool was diluted to 4nM and denatured with 0.2 N NaOH at room temperature. The denatured DNA was diluted to 15pM and spiked with 25% of the Illumina PhiX control DNA prior to loading the sequencer. Illumina paired-end sequencing was performed on the Miseq platform with version v2.3.0.8 of the Miseq Control Software and version v2.3.32 of MiSeq Reporter, using a 600 cycle version 3 reagent kit.
Paired-end sequences were sorted by sample via barcodes in the paired reads with a python script [25]. Sorted paired end sequence data were deposited in the NCBI Short Read Archive under BioProject Accession Number: PRJNA270112 (www.ncbi.nlm.nih.gov/bioproject/PRJNA270112)PRJNA270112. The sorted paired reads were assembled using phrap [27, 28] and paired reads that did not assemble were discarded. Assembled sequence ends were trimmed over a moving window of 5 nucleotides until average quality met or exceeded 20. Trimmed contigs with more than 1 ambiguity or shorter than 200 nt were discarded. Potential chimeras identified with Uchime (usearch6.0.203_i86linux32) [29] using the Schloss [30] Silva reference sequences were removed from subsequent analyses. Assembled sequences were aligned and classified with SINA (1.2.11) using the 418,497 bacterial sequences in Silva 115NR99 as reference configured to yield the Silva taxonomy [31, 32]. Operational taxonomic units (OTUs) were produced by clustering sequences with identical taxonomic assignments. OTU counts were normalized between samples by dividing sequence counts by the total number of sequences generated per sample. Phylum-level and family-level OTU tables were generated by collapsing lower level OTUs into higher-level categories. OTUs with a relative abundance <0.0001 and with a prevalence <0.01 were culled and relative abundances then transformed using the square root function [33]. The software package Explicet (v2.9.4, www.explicet.org) was used to display OTU data and estimate alpha diversity indices (i.e., SChao1, Shannon complexity [H], and Shannon Evenness [H/Hmax]) through 1000 replicate samplings of rarefied datasets [34].
Comparisons of individual phyla and families passing the initial filtering step, were conducted as follows. Because of the commonly observed over-dispersion in microbiome count data [35], the effects of APCMin/+ genotype on individual OTU abundances were examined using the negative binomial (NB) regression model as follows:
Yijk denotes the OTU k’s observed count for mouse j in shipment i, μ ijk is the mean parameter for OTU k’s count distribution of mouse j in shipment I and ϕ k is the dispersion parameter. Shipment refers to three different deliveries of mice (mice 1–20, mice 21–40 and mice 41–60), which has a zero-mean random coefficient. Coefficients b0 and β 1 are fixed constant representing grand mean and APC genotype respectively. The log total sequence count for each mouse is considered as an offset. In addition, the skewness of the distribution of low abundance OTUs causes a large proportion of zero counts. Therefore, a zero-inflated version of the negative binomial (NB) model is also fitted to OTUs with zero counts in additional to the NB model:
The NB and zero-inflated NB models are chosen based on AIC criterion [36]. The p values for the genotype effects on each OTU were then adjusted by the Benjamini-Hochberg [37] procedure to calculate the FDR. Significance was set as FDR<0.05. In addition, a 10-fold cross-validation was performed to validate the significant OTUs that were identified. The square root transformation was then applied to the relative abundances to correct for the skewness and to reduce the coefficient of variation. Comparisons of overall microbial composition between APCMin/+mice versus the wild type mice were subsequently conducted using the permutation Hotelling T2 test with 10,000 permutations using the R package ‘Hotelling’ [38]. Principle coordinate analysis (PCoA) was conducted at the lowest taxonomic level (genus) using the wcmdscale function implemented by the vegan R package [39] and using Morisita-Horn dissimilarity scores.
Histological Analysis of the Intestinal Sections
Histological analysis was carried out in a subset of 10 APCMin/+ mice and 10 wild type mice (1st cohort) by constructing “Swiss rolls” of intestinal segments. These segments were stained with hematoxylin and eosin and scored for adenomas and inflammation by a pathologist who was blinded with respect to the genotype of the mice (N.O.), as previously described [40]. Inspection of intestinal segments stained with 0.25%methylene blue was carried out on the second and third cohorts of mice with the aid of a Zeiss dissecting scope for detection of adenomas and aberrant crypt foci [20].
Comparison of Mouse Proximal Colonic mRNA Expression in APCMin/+ and Wild type Mice
IL-1β mRNA expression relative to actin mRNA, was measured in cecal, proximal colonic, and distal colonic intestinal tissue RNA samples in all 30 APCMin/+ and 30 WT mice as previously described [8]. RNA extracted from the proximal colon of 9 week-old WT mice treated with 3% DSS in water for 7 days was used as a positive control for the assay. The IL-1β ΔCt values (ΔCt = Ctactin-CtIL1β) were compared between APCMin/+ and WT groups using the Mann-Whitney test. Significance was set at a threshold of p <0.05.
Aliquots (1 μg) of proximal colon RNA samples from 6 APC were subjected to paired-ends 100 bp Illumina sequencing. The RNA-Seq libraries were prepared and sequenced at the New York Genome Center. Between 81 and 314 million reads were generated for each of the RNA samples. The RNA-Seq data were deposited in NCBI's Gene Expression Omnibus database with accession number GSE67634. The short reads were aligned to the GRCm38 genome (http://useast.ensembl.org/Mus_musculus/Info/Annotation) using STAR (Spliced Transcripts Alignment to a Reference) [41], and then converted to raw gene counts using featureCounts [42]. The edgeR package [43] was used to identify differentially expressed (FDR<0.05) genes (DEGs) between the APCMin/+ and wild type mice, using additional cutoff of 2-fold differential expression between groups. Hierarchical clustering based on the reads per kilobase of exon per million mapped reads (RPKM) value of the 130 DEGs was carried out by using 1-r dissimilarity measurement and Ward linkage, and the cluster number (n = 7) was chosen based on inspection of the coefficient of determination (R2) plot as previously described [44]. Second, a negative binomial (NB) regression model was fit with gene clusters as following:
xijh is gene cluster h’s expressions centroids (medians) of mouse j in shipment i. μijk is the mean parameter for phylum k’s count distribution of mouse j in shipment i and ϕk is the dispersion parameter. “Shipment” is as defined above. b0 and β1 are fixed coefficients representing grand mean and APC genotype respectively. The log total count of each mice is considered as an offset. Best subset model selection was conducted to choose the model with lowest AIC. All models were fitted with R package: glmmADMB [45].
Results
The relative abundance of Bacteroidetes spp is increased in APCMin/+mice colonic mucosa and luminal content prior to the development of intestinal neoplasias.
In preliminary targeted qPCR studies, we observed that the relative abundance of Bacteroidetes spp. in fecal DNA was higher in 12–14 week-old APCMin/+ female mice compared to age-matched WT female mice (ΔCt = -2.4 vs. ΔCt = -5.1, p = 0.0004). To test the hypothesis that the increase in the relative abundance of Bacteroidetes spp. preceded polyposis, we compared the relative abundance of this clade in the ileal, cecal, proximal colonic, distal colonic mucosa and the distal colonic luminal content in 6 week-old mice. Because gender effects have been previously reported on the number and location of polyps, we restricted our analysis to female mice [46]. Previous studies conducted on 6 week APCMin/+ female mice had established the absence of detectable neoplasias at that age [18]. The absence of intestinal adenomas and aberrant crypt foci was confirmed in the mice included in this study by microscopic inspection of the intestinal segments. The mean histological scores for inflammation were 0.1 and 0 (p = 0.37) for APCMin/+ and WT colons, respectively (n = 10 in each category). We observed a significant increase in the relative abundance of Bacteroidetes spp. in proximal colonic, distal colonic and distal luminal contents between the APCMin/+ and WT-control mice, but no significant difference in the ileal or cecal mucosal samples (Table 1). In contrast we observed no significant difference is Lachnospiriceae spp., a prominent group of Firmicutes, except in the luminal content of the distal colon, which exhibited significantly higher loads in WT mice (p = 0.003).
Table 1. QPCR comparison of the relative abundances of the Bacteroidetes phylum and the Lachnospiriceae clade within the Firmicutes phylum in 6 week old APCMin/+ and WT mice.
| Bacteroidetes | APC Min/+ ΔCt Median (range) | Wild type ΔCt Median (range) | P-value |
| Ileal mucosa | -2.5 (-6.4, 0.5) | -3.0 (-7.5, 0.4) | 0.65 |
| Cecal mucosa | -4.0 (-8.0, -1.0) | -4.3 (-6.2, -3.1) | 0.10 |
| Proximal colonic mucosa | -3.4 (-6.9, -1.6) | -5.0 (-6.4, -2.2) | 0.0008 |
| Distal colonic mucosa | -3.1 (-7.1, -1.6) | -3.9 (-7.9, -1.7) | 0.005 |
| Distal colonic luminal content | -1.4 (-2.6, -0.1) | -2.3 (-5.3, -1.0) | <0.0001 |
| Lachnospiriciae | APC ΔCt Median (range) | Wild type ΔCt Median (range) | P-value |
| Ileal mucosa | -4.2 (-12.1, -0.9) | -3.9 (-11.6, -2.4) | 0.89 |
| Cecal mucosa | -2.9 (-8.6, -1.3) | -2.4 (-6.1, -0.7) | 0.06 |
| Proximal colonic mucosa | -2.2 (-9.2, -0.3) | -1.9 (-11.4, -0.3) | 0.15 |
| Distal colonic mucosa | -3.6 (-8.3, -1.5) | -2.8 (-7.4, -0.5) | 0.06 |
| Distal colonic luminal content | -4.8 (-9.0, -3.4) | -3.9 (-10.4, -1.6) | 0.003 |
The qPCR assays were conducted using established primers as described in Methods. The median and range of ΔCt values (~Log2 relative abundance of targeted taxa) are listed for.the ileal, cecal, proximal colonic, distal colonic mucosal samples and the distal colonic luminal samples collected from 30 6 week old APCMin/+ and 30 wild type mice. The p-values were carried out using the Mann-Whitney U test. The Bonferroni correction was made to the p-value, so that significance was set at p<0.025.
16S rRNA sequence analysis of proximal colonic mucosal samples from APCMin/+ and WT mice
Illumina sequencing of the 16S rRNA gene V1V2 hypervariable region was carried out for the proximal colonic DNA samples. A total of 13,248,412 high-quality sequences were generated (average sequence length: 317 nt; average sample size: 220,807 sequences/sample; minimum: 9,381 sequences; maximum: 411,636 sequences, exclusive of negative controls which were near zero). The median Good’s coverage score was ≥ 99.9987% at the rarefaction point of 9,381 sequences, indicating deep sequence coverage of the intestinal microbiome.
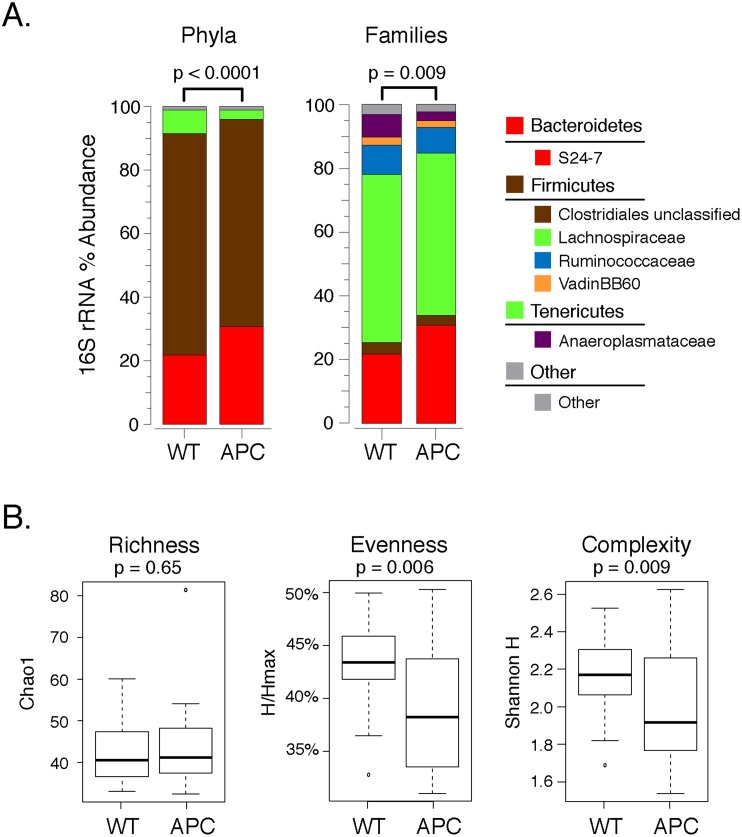
The 16S rRNA sequencing results confirmed the targeted qPCR results in demonstrating a significant increase in the relative abundance of taxa within the Bacteroidetes phylum (FDR = 0.0009) in the APCMin/+ mice (Fig 1A and Table 2). Within the Bacteroidetes phylum, the predominant family was S24-7, whose relative abundance was also significantly increased in APCMin/+ mice (Fig 1A and Table 3, FDR = 0.0015). 16S rRNA sequencing detected a significant reduction in the relative abundance of taxa within the Tenericutes phylum (FDR< 0.0001), as well as a significant reduction in the relative abundance of the Anaeroplasmataceae family (FDR < 0.0001), which was the most prevalent family in the Tenericutes phylum. The results were also confirmed in all 10 cross-validations. The relative abundance of the Cyanobacteria phylum and the Chloroplast family, which is the most prevalent family in this phylum), was decreased in APCMin/+ mice (FDR = 0.047). However this observation was confirmed in only 4 out of 10 cross-validations.
Fig 1. Comparison of phyla and families between wildtype (WT) and APCMin/+ (APC) mice.
A. The mean relative abundances of phyla (left panel) and families (right panel) as inferred from the 16S rRNA sequence analysis. Only phyla and families with relative abundances >0.5% are shown. The Hotelling T2 test was used to compare the overall microbial composition, with p-values noted above each barchart.
Table 2. 16S rRNA Sequence comparison of the relative abundances of phyla in the proximal colonic mucosa of 6 week old APCMin/+ and WT mice.
| Phyla | Mean Relative Abundance | Test type, P-value and FDR | |||
|---|---|---|---|---|---|
| APC | WT | Test Type | P-value | FDR | |
| Firmicutes | 0.65247 | 0.69756 | NB regression | 0.10125 | 0.19302 |
| Bacteroidetes | 0.30827 | 0.21810 | NB regression | 0.00022 | 0.00089 |
| Tenericutes | 0.029 | 0.07425 | NB regression | 0.00001 | 0.00008 |
| Bacteria | 0.00535 | 0.00672 | NB regression | 0.16889 | 0.19302 |
| Proteobacteria | 0.00432 | 0.00276 | NB regression | 0.15898 | 0.19302 |
| Actinobacteria | 0.00055 | 0.00047 | NB regression | 0.31061 | 0.31061 |
| Cyanobacteria | 0.00004 | 0.00012 | NB regression | 0.01753 | 0.04675 |
| Verrucomicrobia | 0 | 0.00002 | NB regression | 0.16798 | 0.19302 |
Seven phyla remained after preprocessing filtering (maximum relative abundance < 0.0001, prevalence <0.01). Significant differences were detected in the bolded phyla, with the threshold set as FDR <0.05.
Table 3. 16S rRNA Sequence comparison of the relative abundances of families in the proximal colonic mucosa of 6 week old APCMin/+ and WT mice.
| Families | Mean Relative Abundance | Test type, P-value and FDR | |||
|---|---|---|---|---|---|
| APC | WT | Test Type | P value | FDR | |
| Firmicutes phylum | |||||
| Lachnospiriceae | 0.50691 | 0.52577 | NB regression | 0.4327 | 0.5024 |
| Ruminococcaceae | 0.08136 | 0.09148 | NB regression | 0.1694 | 0.2072 |
| Unassigned Clostridiales | 0.03145 | 0.03665 | NB regression | 0.0589 | 0.0727 |
| vadinBB60 | 0.02152 | 0.02471 | NB regression | 0.2689 | 0.3203 |
| Erysipelotrichaceae | 0.00369 | 0.00622 | NB regression | 0.0020 | 0.0027 |
| Lactobacillaceae | 0.00288 | 0.00402 | NB regression | 0.0399 | 0.0497 |
| Peptococcaceae | 0.00237 | 0.00650 | NB regression | 0.0000 | 0.0000 |
| Unassigned Firmicutes | 0.00118 | 0.00121 | NB regression | 0.9504 | 0.9717 |
| Family XIII Incerta Sedis | 0.00081 | 0.00076 | NB regression | 0.6219 | 0.6762 |
| Peptostreptococcaceae | 0.00010 | 0.00012 | NB regression | 0.0001 | 0.0001 |
| Clostridiaceae | 0.00006 | 0.00007 | NB regression | 0.9443 | 0.9717 |
| Paenibacillaceae | 0.00005 | 0.00000 | NB regression | 0.0000 | 0.0001 |
| Bacillaceae | 0.00004 | 0.00003 | NB regression | 0.7316 | 0.7831 |
| Staphylococcaceae | 0.00003 | 0.00001 | NB regression | 0.0157 | 0.0203 |
| Thermoactinomycetaceae | 0.00000 | 0.00001 | zero-inflated NB regression | 0.9933 | 0.9975 |
| Unassigned Bacilli | 0.00001 | 0.00000 | zero-inflated NB regression | 0.0000 | 0.0000 |
| Bacteroidetes phylum | |||||
| S24-7 | 0.30782 | 0.21769 | NB regression | 0.0002 | 0.0015 |
| Unassigned Bacteroidales | 0.00016 | 0.00007 | NB regression | 0.0000 | 0.0000 |
| Bacteriodaceae | 0.00014 | 0.00018 | NB regression | 0.5226 | 0.5818 |
| Rickenellaceae | 0.00009 | 0.00008 | NB regression | 0.5008 | 0.5670 |
| Prevotellaceae | 0.00003 | 0.00003 | NB regression | 0.7769 | 0.8251 |
| Porphyromonadaceae | 0.00002 | 0.00004 | NB regression | 0.4825 | 0.5508 |
| Unassigned Bacteroidetes | 0.00001 | 0.00000 | NB regression | 0.6390 | 0.6893 |
| Ternicutes phylum | |||||
| Anaeroplasmataceae | 0.02664 | 0.07133 | NB regression | 0.0000 | 0.0000 |
| RF9 | 0.00236 | 0.00293 | NB regression | 0.2202 | 0.2647 |
| Proteobacteria phylum | |||||
| Enterobacteriaceae | 0.00319 | 0.00039 | NB regression | 0.0002 | 0.0003 |
| Phyllobacteriaceae | 0.00046 | 0.00144 | NB regression | 0.9975 | 0.9975 |
| Bradyrhizobiaceae | 0.00038 | 0.00065 | NB regression | 0.8941 | 0.9280 |
| Sphingomonadaceae | 0.00008 | 0.00011 | NB regression | 0.3601 | 0.4253 |
| Methylobacteriaceae | 0.00010 | 0.00007 | NB regression | 0.8034 | 0.8467 |
| Ricketsiella/mitochondria | 0.00001 | 0.00004 | NB regression | 0.0338 | 0.0425 |
| Burkholderiaceae | 0.00001 | 0.00001 | NB regression | 0.5053 | 0.5674 |
| Alcaligenaceae | 0.00001 | 0.00001 | NB regression | 0.5501 | 0.6029 |
| Moraxellaceae | 0.00001 | 0.00001 | NB regression | 0.0336 | 0.0425 |
| Pseudomonadaceae | 0.00001 | 0.00001 | NB regression | 0.9914 | 0.9975 |
| Desulfovibrionaceae | 0.00001 | 0.00000 | NB regression | 0.0120 | 0.0156 |
| Actinobacteria phylum | |||||
| Coriobacteriaceae | 0.00050 | 0.00042 | NB regression | 0.3842 | 0.4499 |
| Proprionibacteriaceae | 0.00003 | 0.00004 | NB regression | 0.4533 | 0.5218 |
| Corynebacteriaceae | 0.00000 | 0.00001 | NB regression | 0.5266 | 0.5818 |
| Cyanobacteria phylum | |||||
| Chloroplast | 0.00004 | 0.00012 | NB regression | 0.0213 | 0.0273 |
| Verrucomicrobia phylum | |||||
| Verrucomicrobiaceae | 0.00000 | 0.00002 | zero-inflated NB regression | 0.8836 | 0.9240 |
| Other Phyla | |||||
| Other Bacteria | 0.00535 | 0.00672 | NB regression | 0.2041 | 0.2475 |
Forty-two families remained after preprocessing filtering (maximum relative abundance < 0.0001, prevalence <0.01). The 42 families are listed within their respective phyla in order of their relative abundance, with the remaining families combined in Other categories. The phyla are underlined in the table. Significant differences were detected in the bolded families, with the threshold set as FDR <0.05.
Although 16S rRNA sequence analysis did not detect a significant difference in the overall relative abundance of the Firmicutes phylum, it also confirmed the lack of any change in the relative abundance of taxa within Lachnospiriciae clade, thus confirming the qPCR data (see Table 1). Furthermore decreases as well as increased were observed in some of the individual families within the Firmicutes phylum (Table 3). Similarly, although 16S rRNA sequence analysis did not detect a significant difference in the overall relative abundance of the Proteobacteria phylum, increases as well as decreases were observed in some of the individual families within the Proteobacteria phylum.
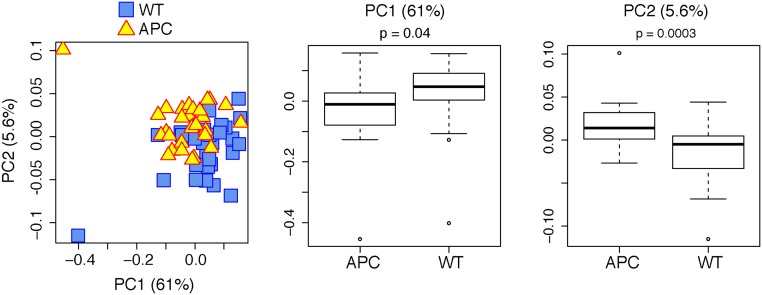
Overall, the proximal colonic microbiomes differed between APCMin/+ and WT mice at both the phylum (p<0.0001) and family (p <0.0001) levels, as assessed by a 10000-permutation Hotelling T2 test of the 7 phyla and 42 families that passed the initial filtering step (Fig 1A). The proximal colonic microbiomes of APCMin/+ mice were also characterized by significantly lower OTU complexity (Shannon H; p = 0.009) and evenness (Shannon H/Hmax; p = 0.006) compared with WT mice (Fig 1B); OTU richness (SChao1) was comparable in the two groups. Finally, principle coordinates analysis (PCoA) demonstrated significant associations of principle component axes 1 and 2 with APC genotype (see Fig 2).
Fig 2. Principle Coordinate Analysis.
PCoA was conducted at the family taxonomic level using a pairwise dissimilarity matrix calculated using the Morisita-Horn beta-diversity index. Each triangle is representative of a single APCMin/+ (APC) mouse and each square is representative of a single WT mouse, plotted along the first two principal component axes (left panel). PC1 and PC2 accounted for 61% and 5.6%, respectively, of total variance. The middle and right panels display the PC scores along axes 1 and 2, respectively. Differences in scores between genotypes were assessed by student t-test.
Differentially expressed genes in 6 week-old APCMin/+mice
We reasoned that differences in colonic microbial composition associated with the APCMin/+ genotype must be linked to alterations in mouse colon gene expression, despite the lack of histological evidence of neoplasias. To examine how the APC mutation could alter the host colon gene expression, we conducted parallel RNA-sequence analysis on 6 APCMin/+ mice and 6 WT mice, (sampling all three cohorts). A total of 130 host genes (fold change > 2 fold, FDR <0.05) were selected using edgeR [43]. The DEGs were grouped into seven clusters as described in Methods (see Table 4), with 106 upregulated genes distributed among three clusters and the 24 downregulated genes distributed among four clusters.
Table 4. Hierarchical clustering of DEGs in APCMin/+ proximal colonic RNA transcripts vs. control RNA samples.
| Gene name | Log2 FC | FDR | Gene name | Log2 FC | FDR |
|---|---|---|---|---|---|
| Cluster 1 | Up ↑ | Cluster 4 | Down↓ | ||
| Hoxd13 | 7.95 | 5.00E-04 | Rmrp | -3.9 | 4.00E-04 |
| Tgm3 | 9.07 | 6.00E-04 | Gm22513 | -3.97 | 8.00E-04 |
| Atp12a | 6.4 | 8.00E-04 | Gm26035 | -5.49 | 8.00E-04 |
| Ly6g | 9.39 | 8.00E-04 | Igkv5-48 | -3.23 | 0.0022 |
| Sval1 | 7.44 | 8.00E-04 | Gm24146 | -4.69 | 0.0023 |
| Fut9 | 6.44 | 0.001 | Igkv12-98 | -5.86 | 0.0023 |
| Gm8540 | 8.63 | 0.0018 | Metazoa_SRP | -3.23 | 0.0034 |
| AI854703 | 4.65 | 0.0022 | Rn7sk | -3.04 | 0.0036 |
| B3gnt7 | 5.27 | 0.0023 | Igkv3-5 | -5.88 | 0.0044 |
| Cpn2 | 4.92 | 0.0023 | Rpph1 | -2.62 | 0.0073 |
| Evx1 | 8.64 | 0.0023 | Igkv3-1 | -5.52 | 0.0224 |
| Gm15053 | 8.81 | 0.0023 | Vaultrc5 | -2.57 | 0.0267 |
| Itih2 | 5.16 | 0.0023 | Ighv1-7 | -2.54 | 0.0352 |
| Mptx1 | 5.36 | 0.0023 | Klk1b22 | -2.09 | 0.0352 |
| Myh2 | 3.28 | 0.0023 | Olfr424 | -4.87 | 0.0352 |
| Thbs4 | 4.23 | 0.0023 | Gm22179 | -3.19 | 0.0353 |
| Casp14 | 10.53 | 0.0029 | Igkv9-124 | -3.76 | 0.0445 |
| Gm16341 | 5.86 | 0.0032 | Cluster 5 | Down↓ | |
| Best4-ps | 6.75 | 0.0034 | Ighv5-15 | -6.07 | 5.00E-04 |
| Ctse | 4.5 | 0.0034 | Igkv2-109 | -5.08 | 0.0066 |
| Fxyd4 | 9.83 | 0.0034 | Cluster 6 | Down↓ | |
| Gm2539 | 10.01 | 0.0034 | Ighv9-4 | -3.72 | 0.0176 |
| Sval3 | 7.01 | 0.0034 | Igkv9-120 | -3.07 | 0.0432 |
| Klk15 | 9.86 | 0.0035 | Ighv1-80 | -3.7 | 0.0456 |
| Tmprss13 | 7.92 | 0.0035 | Cluster 7 | Down↓ | |
| Hoxb13 | 10.06 | 0.0039 | Igkv14-100 | -3.87 | 0.0388 |
| Hoxa13 | 3.92 | 0.0044 | Sel1l2 | -7.57 | 0.0444 |
| Mptx2 | 5.49 | 0.0044 | |||
| Insl5 | 4.45 | 0.0045 | |||
| Pla2g4f | 3.83 | 0.0045 | |||
| Slc28a3 | 4.12 | 0.0046 | |||
| Vsig1 | 3.39 | 0.0046 | |||
| Rims4 | 3.76 | 0.0047 | |||
| Vtcn1 | 5.41 | 0.0049 | |||
| Anxa8 | 3.61 | 0.0053 | |||
| Pdzd7 | 3.17 | 0.0053 | |||
| 4930552P12Rik | 3.66 | 0.0059 | |||
| HOXA11-AS1_5 | 4.62 | 0.0059 | |||
| Gpr83 | 4.27 | 0.006 | |||
| B3gnt5 | 2.98 | 0.0064 | |||
| Gjb5 | 3.34 | 0.0066 | |||
| Tnip3 | 2.94 | 0.0066 | |||
| Eno3 | 2.53 | 0.007 | |||
| Gjb4 | 3.84 | 0.0074 | |||
| Nxpe4 | 3.51 | 0.0074 | |||
| Spink3 | 3.63 | 0.0083 | |||
| Trpv3 | 3.11 | 0.0091 | |||
| Hoxa11os | 3.58 | 0.0098 | |||
| 2310079G19Rik | 8.03 | 0.01 | |||
| Gm11535 | 7.68 | 0.01 | |||
| Cyp2f2 | 3.9 | 0.0112 | |||
| HOXA11-AS1_4 | 3.93 | 0.0112 | |||
| Muc1 | 3.11 | 0.0112 | |||
| St8sia5 | 7.08 | 0.0126 | |||
| HOXB13-AS1_2 | 7.9 | 0.015 | |||
| Cyp2a12 | 6.98 | 0.0156 | |||
| Ttr | 2.01 | 0.0172 | |||
| Nccrp1 | 3.85 | 0.0173 | |||
| Evx2 | 7.43 | 0.0187 | |||
| Slc46a1 | 2.38 | 0.0187 | |||
| Csta | 4.42 | 0.0188 | |||
| Gm16556 | 3.57 | 0.0191 | |||
| Sycn | 2.76 | 0.0191 | |||
| Slc15a1 | 3.24 | 0.0204 | |||
| Grin2b | 3.81 | 0.022 | |||
| Cyp2d12 | 3.51 | 0.0223 | |||
| Gm11830 | 3.09 | 0.0236 | |||
| HOTTIP_2 | 7.46 | 0.0236 | |||
| HOXB13-AS1_1 | 6.11 | 0.0236 | |||
| Psg17 | 3.46 | 0.0236 | |||
| Cela1 | 2.1 | 0.0258 | |||
| Iqch | 3.58 | 0.0258 | |||
| Foxq1 | 2.17 | 0.0259 | |||
| Ms4a10 | 1.48 | 0.0259 | |||
| Nt5c1a | 3.4 | 0.026 | |||
| Il18 | 1.6 | 0.0264 | |||
| Myo16 | 2.71 | 0.0264 | |||
| Hoxd12 | 5.36 | 0.0271 | |||
| Brinp2 | 2.7 | 0.0291 | |||
| Ankdd1b | 2.84 | 0.0292 | |||
| Defb45 | 4.97 | 0.0292 | |||
| Gp6 | 3.93 | 0.0328 | |||
| Ggh | 2.36 | 0.0336 | |||
| Gm15401 | 1.69 | 0.0336 | |||
| A930011G23Rik | 1.78 | 0.035 | |||
| Rdh16 | 1.75 | 0.035 | |||
| Hrg | 4.05 | 0.0369 | |||
| 1700042G15Rik | 5.12 | 0.0411 | |||
| Wnt8b | 3.96 | 0.0413 | |||
| Ctgf | 1.44 | 0.0432 | |||
| Slc36a1 | 2.23 | 0.0432 | |||
| Nt5e | 1.75 | 0.0434 | |||
| Sh3d21 | 1.87 | 0.0461 | |||
| Slc16a12 | 1.93 | 0.0466 | |||
| Gm16557 | 5.58 | 0.0469 | |||
| Ano4 | 2.8 | 0.0477 | |||
| Pla2g5 | 1.87 | 0.0477 | |||
| Gpr137b | 2.36 | 0.0483 | |||
| Cd207 | 6.32 | 0.0493 | |||
| Gm17384 | 6.35 | 0.0494 | |||
| Cluster 2 | Up ↑ | ||||
| Gm10800 | 10 | 0.0023 | |||
| Gm10801 | 9.72 | 0.0034 | |||
| Gm21738 | 9.54 | 0.0035 | |||
| Gm26870 | 7.7 | 0.0191 | |||
| Gm10718 | 7.06 | 0.0236 | |||
| Cluster 3 | Up↑ | ||||
| Apon | 2.18 | 0.0419 |
One hundred thirty DEGs were selected by edgeR analysis of RNA sequence data (see Methods) and grouped into seven clusters by hierarchical clustering. Shown on the left are the three upregulated clusters (1–3), and shown on the right are the four dowregulated clusters (4–7).
NB regression (see Methods) selected four (1, 4, 6, 7) out of seven gene clusters (see Table 5) significantly (p-value < 0.05) in addition to APC genotype, which were positively associated with the relative abundance of Bacteroidetes. Among those four clusters, cluster 1 (coefficient 0.051) is composed of 100 out of 106 upregulated genes. The downregulated gene clusters 4,6,7 covers 22 out of 24 downregulated genes. While APC genotype had a dominant effect on the relative abundance of Bacteroidetes, detection of additional associations with mouse colonic gene expression, suggest that alterations in host colonic gene expression play a role in influencing mucosal associated microbial composition.
Table 5. Association between gene cluster expression (centroid medians), APC genotype and the relative abundance of Bacteroidetes in the proximal colonic mucosa of 6 week old APCMin/+ and WT mice.
| Bacteroidetes | Coefficient | p-value |
|---|---|---|
| APC genotype | 0.915 | 8.36E-21 |
| cluster1 | 0.051 | 0.000632 |
| cluster4 | 0.031 | 7.17E-10 |
| cluster6 | 0.006 | 0.000556 |
| cluster7 | 0.029 | 0.000158 |
The cluster medians of seven geneclusters along with APC genotype were used in the following model as described in Methods. The significant effects are bolded, with the threshold set as p-value <0.05. Regression coefficients are also reported as index of effect size.
Because elevated IL-1β levels have previously been reported in 18–25 week old APCMin/+ compared with WT mice, RT-PCR assays were conducted on proximal colonic RNA samples in all of the mice as previously described [15]. No significant difference was observed in the ΔCtIL1β-actin values between APCMin/+ and WT mice (-13.5 vs. -16.2, p = 0.485). These values were both very low compared to that measured in DSS treated mice (-4.9), indicating that IL-1β was not highly expressed in the colons of either mouse group in our study.
Discussion
Alterations in the gut microbiome (dysbiosis) have been reported in human colonic neoplasia and in mouse models [1–6, 47]. This study demonstrates that alterations in the gut microbiome, characterized by an increased relative abundance of Bacteroidetes spp. observed in association with intestinal neoplasias, actually precedes the development of microscopically detectable intestinal neoplasias in 6 week old APCMin/+ mice. Increased loads of Bacteroidetes spp. have been reported in another colitis-associated mouse model of colon cancer [47], and in some but not all studies of human colorectal neoplasia [1–6]. 16S rRNA sequence analysis revealed that the increased relative abundance of Bacteroidetes spp. corresponded primarily to an increased relative abundance of taxa within the uncultured family S24-7. Similar increases in S24-7 have also been reported in conventionally raised C57BL6 mice that were fed a high fat diet [48]. This association with intestinal dysbiosis is of interest, because increased dietary fat has previously been associated with increased number and /or size in both WT and APCMin/+ mice [49,50].
The relative abundance of the phylum Tenericutes observed in this study is higher than reported by some studies of C57Bl/6 mice [51], but similar to another study using C57Bl/6 mice purchased from the same vendor [52]. In this study, the APCMin/+ and WT mice were housed in separate cages, which could influence the reported microbial compositions [53, 54], possibly related to coprophagic behavior.
Alterations in gene expression have been previously reported in normal appearing mucosa of APC mutant mice after the development of intestinal polyposis [55]. We report differential expression of genes (DEG) in 6 week APCMin/+ mice prior to the detection of intestinal polyposis. In order to integrate host colonic gene expression with the microbial taxonomic data, we reduced the gene expression input variables by first selecting DEGs, reasoning that these genes would be most likely to be involved in disrupted colonic microbial interactions in the mutant mice. Variable dimensionality was further reduced by clustering the 130 DEGs into seven groups. The detection of significant associations between host colonic gene expression and the relative abundance of microbial taxa, after taking into consideration APC genotype, support the concept that host colonic microbial cross talk influences mucosal associated microbial composition. Cluster 4, which included downregulated genes encoding immunoglobulins and non-coding functional RNAs, demonstrated a significant linear relationship with the relative abundance of Bacteroidetes after controlling for APC genotype. The inverse correlation between immunoglobulin gene expression and the relative abundance of Bacteroidetes spp. is intriguing in light of previous reports linking an increased relative abundance of Bacteroidetes spp. with a reduction of immunoglobulin coated bacteria in humans [56, 57]. The observation that some of the non-coding RNAs in this cluster may be located in the mitochondria, is intriguing in light of the observation that mutated APC proteins in contrast to WT APC proteins are detected in mitochondria [58]
In summary, our results support the concept that APC haplo-insufficiency of the host colonic epithelial cell alters colonic microbial interactions prior to polyposis. It is thus conceivable that such microbiome changes contribute to the pathogenesis of colon cancer. An important corollary to such a notion would be that the colonic microbiome represents an important (and druggable) target for the prevention of colon cancer. Indeed, interventions directed at the microbiome (germ free and antibiotic treatment) have been reported to modulate tumor formation in mouse models of colon cancer [17]. However, it remains to be determined whether interventions directed at ameliorating dysbiosis in APCMin/+ mice, such as through probiotic, prebiotic or antibiotic interventions, could reduce tumor formation.
Acknowledgments
We thank Aaron Rosenfeld for assistance with conducting the RT-PCR assays for mouse IL-1β. We thank the New York Genome Center for providing RNA-Sequence Analysis.
Data Availability
All relevant data are within the paper. Raw 16S rRNA gene sequence data are deposited in NCBI Short Read Archive under BioProject Assession Number: PRJNA270112 (www.ncbi.nlm.nih.gov/bioproject/PRJNA270112). The RNA-Sequence data are deposited in NCBI's Gene Expression Omnibus database with accession number GSE67634.
Funding Statement
This work was supported by 269010 Simons Foundation (www.simonsfoundation.org). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, et al. (2010) Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes 1: 138–147. 10.4161/gmic.1.3.12360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, et al. (2011) Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One 6: e16393 10.1371/journal.pone.0016393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen W, Liu F, Ling Z, Tong X, Xiang C (2012) Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 7: e39743 10.1371/journal.pone.0039743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang T, Cai G, Qiu Y, Fei N, Zhang M, et al. (2012) Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 6:320–329 10.1038/ismej.2011.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, et al. (2012) Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22:292–298 10.1101/gr.126573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, et al. (2013) Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 105:1907–11 10.1093/jnci/djt300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, et al. (1991) Identification and characterization of the familial adenomatous polyposis coli gene. Cell 66: 589–600. [DOI] [PubMed] [Google Scholar]
- 8. Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, et al. (1991) Identification of FAP locus genes from chromosome 5q21. Science 253: 661–665. [DOI] [PubMed] [Google Scholar]
- 9. Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, et al. (1991) Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science 253: 665–669. [DOI] [PubMed] [Google Scholar]
- 10. Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, et al. (1992) APC mutations occur early during colorectal tumorigenesis. Nature 359: 235–237. [DOI] [PubMed] [Google Scholar]
- 11. Kinzler KW, Vogelstein B (1996) Lessons from hereditary colorectal cancer. Cell 87: 159–170. [DOI] [PubMed] [Google Scholar]
- 12. Senda T, Iizuka-Kogo A, Onouchi T, Shimomura A (2007) Adenomatous polyposis coli (APC) plays multiple roles in the intestinal and colorectal epithelia. Med Mol Morphol 40: 68–81. [DOI] [PubMed] [Google Scholar]
- 13. Fearon ER (2011) Molecular genetics of colorectal cancer. Annu Rev Pathol 6: 479–507. 10.1146/annurev-pathol-011110-130235 [DOI] [PubMed] [Google Scholar]
- 14. Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, et al. (1992) Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 256: 668–670. [DOI] [PubMed] [Google Scholar]
- 15. Dove WF, Clipson L, Gould KA, Luongo C, Marshall DJ, et al. (1997) Intestinal neoplasia in the ApcMin mouse: independence from the microbial and natural killer (beige locus) status. Cancer Res 57: 812–814. [PubMed] [Google Scholar]
- 16. Li Y, Kundu P, Seow SW, de Matos CT, Aronsson L, et al. (2012) Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APCMin/+ mice. Carcinogenesis 33: 1231–1238. 10.1093/carcin/bgs137 [DOI] [PubMed] [Google Scholar]
- 17. Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, et al. (2014) Gut microbial metabolism drives transformation of msh2-deficient colon epithelial cells. Cell 158: 288–299. 10.1016/j.cell.2014.04.051 [DOI] [PubMed] [Google Scholar]
- 18. Mackenzie GG, Sun Y, Huang L, Xie G, Ouyang N, et al. (2010) Phospho-sulindac (OXT-328), a novel sulindac derivative, is safe and effective in colon cancer prevention in mice. Gastroenterology. 139:1320–32. 10.1053/j.gastro.2010.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sweetser DA, Hauft SM, Hoppe PC, Birkenmeier EH, Gordon JI (1988) Transgenic mice containing intestinal fatty acid-binding protein-human growth hormone fusion genes exhibit correct regional and cell-specific expression of the reporter gene in their small intestine. Proc Natl Acad Sci U S A 85: 9611–9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paulsen JE1, Namork E, Steffensen IL, Eide TJ, Alexander J. (2000) Identification and quantification of aberrant crypt foci in the colon of Min mice—a murine model of familial adenomatous polyposis. Scand J Gastroenterol. 2000;35:534–9. [DOI] [PubMed] [Google Scholar]
- 21. Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A. (2004) Development of an extensive set of 16S rDNA-targeted primer for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 97:1166–1177. [DOI] [PubMed] [Google Scholar]
- 22. Maeda H, Fujimoto C, Haruki Y, Maeda T, Kokequchi S, et al. (2003) Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol Med Microbiol. 39:81–86. [DOI] [PubMed] [Google Scholar]
- 23. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, et al. (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 104:13780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li E, Hamm CM, Gulati AS, Sartor RB, Chen H, et al. (2012) Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS One.7:e26284 10.1371/journal.pone.0026284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hara N, Alkanani AK, Ir D, Robertson CE, Wagner BD, et al. (2012) Prevention of virus-induced type 1 diabetes with antibiotic therapy. J Immunol 189: 3805–3814. 10.4049/jimmunol.1201257 [DOI] [PubMed] [Google Scholar]
- 26. Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, et al. (2013) Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339: 1084–1088. 10.1126/science.1233521 [DOI] [PubMed] [Google Scholar]
- 27. Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194. [PubMed] [Google Scholar]
- 28. Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185. [DOI] [PubMed] [Google Scholar]
- 29. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schloss PD, Westcott SL (2011) Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol 77: 3219–3226. 10.1128/AEM.02810-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pruesse E, Peplies J, Glockner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. 10.1093/bioinformatics/bts252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, et al. (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sokal RR, Rohlf FJ. (1994) Biometry: the principles and practice of statistics in biological research. Freeman, New York. [Google Scholar]
- 34. Robertson CE, Harris JK, Wagner BD, Granger D, Browne K, et al. (2013) Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics 29: 3100–01. 10.1093/bioinformatics/btt526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McMurdie PJ, Holmes S. (2014) Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 10: e1003531 10.1371/journal.pcbi.1003531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paulson JN, Stine OC, Bravo HC, Pop M. (2013) Differential abundance analysis for microbial marker-gene surveys. Nat Methods 10: 1200–02 10.1038/nmeth.2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Benjamini Y, Hochberg Y (1995) Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 57: 289–300. [Google Scholar]
- 38.James M. Curran (2013). Hotelling: Hotelling's t-squared test and variants. R package version 1.0–2. Available: http://cran.r-project.org/package=hotelling
- 39.Oksanen, J, Kindt R, Legendre P, O’Hara B, Simpson GL, et al. (2008) Vegan: Community Ecology Package. R package version 1.15–1. Available: http://cran.r-project.org/, http://vegan.r-forge.r-project.org.
- 40. Ouyang N, Zhu C, Zhou D, Nie T, Go MF, et al. (2012) MC-12, an annexin A1-based peptide, is effective in the treatment of experimental colitis. PLoS One. 2:e41585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, et al. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liao Y, Smyth GK, Shi W. (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–30. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 43. Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang T, DeSimone RA, Jiao X, Rohlf FJ, Zhu W, et al. (2012) Host genes related to paneth cells and xenobiotic metabolism are associated with shifts in human ileum-associated microbial composition. PLoS One. 7:e30044 10.1371/journal.pone.0030044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skaug H, Fournier D, Bolker B, Magnusson A and Nielsen A (2015-01-25). Generalized Linear Mixed Models using AD Model Builder. R package version 0.8.1.
- 46. McAlpine CA, Barak Y, Matise I, Cormier RT. (2006) Intestinal-specific PPARγ deficiency enhances tumorigenesis in ApcMin/+ mice. Int. J. Cancer 119: 2339–46. [DOI] [PubMed] [Google Scholar]
- 47. Zackular JP1, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, et al. (2013) The gut microbiome modulates colon tumorigenesis. MBio. 4:e00692–13. 10.1128/mBio.00692-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Serino M, Luche E, Gres S, Baylac A, Berge M, et al. (2012) Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut 61: 543–53. 10.1136/gutjnl-2011-301012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wasan HS, Novelli M, Bee J, Bodmer WF. (1997) Dietary fat influences on polyp phenotype in multiple intestinal neoplasia mice. Proc Natl Acad Sci U S A. 94:3308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Day SD, Enos RT, McClellan JL, Steiner JL, Velázquez KT, et al. (2013) Linking inflammation to tumorigenesis in a mouse model of high-fat-diet-enhanced colon cancer. Cytokine. 64:454–62. 10.1016/j.cyto.2013.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gu S, Chen D, Zhang JN, Lv X, Wang K, et al. (2013) Bacterial community mapping of the mouse gastrointestinal tract. PLoS One 8: e74957 10.1371/journal.pone.0074957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nagalingam NA1, Kao JY, Young VB. (2011) Microbial ecology of the murine gut associated with the development of dextran sodium sulfate-induced colitis. Inflamm Bowel Dis. 17:917–26. 10.1002/ibd.21462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Campbell JH1, Foster CM, Vishnivetskaya T, Campbell AG, Yang ZK, et al. (2012) Host genetic and environmental effects on mouse intestinal microbiota. ISME J. 6:2033–44. 10.1038/ismej.2012.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Robertson SJ, Zhou JY, Geddes K, Rubino SJ, Cho JH, et al. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes. 2013;4: 222–31 10.4161/gmic.24373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen LC1, Hao CY, Chiu YS, Wong P, Melnick JS, et al. (2004) Alteration of gene expression in normal-appearing colon mucosa of APC(min) mice and human cancer patients. Cancer Res. 64:3694–700. [DOI] [PubMed] [Google Scholar]
- 56. De Palma G1, Nadal I, Medina M, Donat E, Ribes-Koninckx C, et al. (2010) Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 10:63 10.1186/1471-2180-10-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nadal I, Santacruz A, Marcos A, Warnberg J, Garagorri JM, et al. (2009) Shifts in clostridia, bacteroides and immunoglobulin-coating fecal bacteria associated with weight loss in obese adolescents. 33:758–67 [DOI] [PubMed] [Google Scholar]
- 58. Brocardo M1, Lei Y, Tighe A, Taylor SS, Mok MT, et al. (2008) Mitochondrial targeting of adenomatous polyposis coli protein is stimulated by truncating cancer mutations: regulation of Bcl-2 and implications for cell survival. J. Biol Chem. 283: 5950–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper. Raw 16S rRNA gene sequence data are deposited in NCBI Short Read Archive under BioProject Assession Number: PRJNA270112 (www.ncbi.nlm.nih.gov/bioproject/PRJNA270112). The RNA-Sequence data are deposited in NCBI's Gene Expression Omnibus database with accession number GSE67634.