Abstract
The recent explosion of RNA-seq studies has resulted in a newfound appreciation for the importance of riboregulatory RNAs in the posttranscriptional control of eukaryotic and prokaryotic genetic networks. The current review will explore the role of trans-riboregulatory RNAs in various adaptive responses of host and pathogen in the oral cavity.
Keywords: small RNA, noncoding RNA, microRNA, host pathogen interactions, gene expression regulation
1. Introduction
Recent improvements in sequencing technology have revealed a surprising diversity of noncoding RNAs expressed in nearly all cell types that have been examined thus far. In bacteria, noncoding RNAs constitute at least 10 – 20% of all expressed genes [86], yet despite their ubiquity and abundance, only a minute fraction of these molecules actually have assigned functions. Similarly, most of what is currently known about bacterial noncoding RNAs has been derived from studies in just a handful of species. The situation is substantially more daunting when considering human noncoding RNAs. Nearly 15,000 unique long noncoding RNA (lncRNA) transcripts have been recently annotated from the human genome [23]. As this number does not account for any of the plethora of <200 nt noncoding RNA species, such as microRNAs (miRNA) or small-interfering RNAs (siRNA), the true breadth of the human noncoding transcriptome must be astounding. However, as more attention has been focused upon deciphering the roles of noncoding RNAs, it is becoming increasingly evident that many of these molecules are likely to be riboregulatory RNAs involved in posttranscriptional control of gene expression [15]. Within the next decade, it would not be surprising to discover that riboregulatory RNAs play an even greater role than transcription factors for the regulation of genetic networks. Posttranscriptional mechanisms also have a variety of characteristics that make them particularly suited for highly dynamic genetic pathways like many of the major cellular adaptive responses. This includes the regulation of accessory genes and virulence factors in pathogens [29, 34, 54, 79, 86] as well as the corresponding immune responses of their infected hosts [5, 75, 105]. Posttranscriptional mechanisms offer a faster response time at a reduced energetic cost compared to most transcriptional mechanisms [34]. Perhaps of even greater importance is the fact that posttranscriptional mechanisms also provide the option of directly overriding existing genetic programs in response to environmental signals [34]. For example, preexisting mRNA pools transcribed during a previous growth condition can be rapidly inhibited from further translation or even selectively degraded in response to new environmental stimuli. This will prevent these mRNAs from yielding proteins that would otherwise provide little or no utility in the current environment. This is a key distinction from transcriptional mechanisms, which are clearly essential for the production of new transcripts, but are typically incapable of inactivating them once they are transcribed.
There are several basic mechanisms of posttranscriptional control employed by both bacterial and human cells that can be broadly classified via control by either cis elements within mRNAs, trans riboregulators, or via sequestration of regulatory RNAs/proteins. Frequently, genes are regulated using a combination of these mechanisms as well. Regulation in cis often involves mRNA secondary structures within the 5’ and/or 3’ untranslated regions (UTRs) of mRNAs [8, 34, 35, 91]. These structures ultimately influence the translation efficiency and mRNA stability of the molecules to which they are attached. In contrast, trans riboregulators perform a similar function, but do so via direct hybridization (seed pairing) to heterologous target mRNAs [4, 27, 97]. Since trans riboregulation typically occurs through imperfect complementarity between the regulator and target, a single riboregulator may have numerous targets as part of a larger posttranscriptional regulon [78]. Posttranscriptional regulation by sequestration is an indirect mechanism by which an RNA molecule serves as a sink to titrate other regulatory RNAs or proteins away from target mRNAs [1, 6, 25, 32]. Such RNAs are commonly referred to as “decoys” or “sponges”. A substantial body of recent research in both human and bacterial cells exists for each of these aforementioned regulatory mechanisms. Therefore, due to space limitations, this review will be specifically focused upon a comparison of the recent advances related to trans riboregulation in the host and bacterial pathogen with an emphasis on the human oral cavity. Although not considered here, we would also like to highlight the importance of microRNAs that play crucial roles in viral pathogenesis. Viral modulation of the host miRNA machinery can promote viral replication, while the expression of viral miRNAs in host cells may play critical roles in viral pathogenesis. The reader is referred to several comprehensive reviews for additional information on the subject [36, 31, 103, 41].
2. Trans-riboregulatory control of the host immune response
2.1 Sources of host riboregulatory molecules
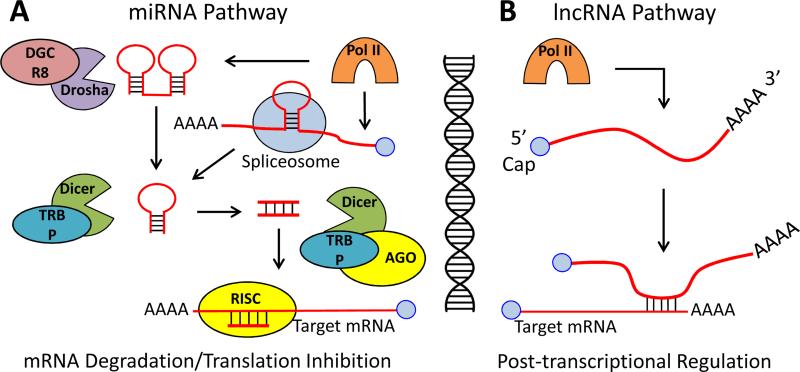
Recently, there has been considerable interest in the central roles of microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) for the control of many diverse processes such as oncogenesis [118], cell differentiation [44, 93], and more recently the immune response [105]. For miRNAs, mature riboregulators generally range in size between 20 – 22 nt in length and are typically derived from larger immature transcripts produced by RNA polymerase II [37, 119]. The immature primary miRNA transcripts are converted into miRNAs via a variety of pathways, but the two best-studied mechanisms are via the canonical miRNA pathway and the mirtron pathway (Fig. 1A). The canonical pathway begins with the production of the primary miRNA transcript, which forms a hairpin structure of about 55 – 70 nt. [37, 88, 119]. This hairpin is processed in the nucleus by the Drosha enzyme in complex with the RNA binding protein DGCR8 [37, 88, 119]. Drosha is an RNase III-type endoribonuclease that is specific for double stranded RNA. The processed hairpin is then exported to the cytoplasm via the exportin-5 protein and further processed by another RNase III enzyme (Dicer) in complex with the RNA binding protein TRBP [37, 88, 119]. The second processing step removes the single stranded loop of the hairpin to yield an RNA duplex that can be loaded onto either of four argonaute (AGO) proteins [37, 63, 88, 119]. One strand of the small RNA duplex is subsequently degraded to yield the mature miRNA, which together with an AGO protein ultimately forms the core of the RNA-induced silencing complex (RISC) [63]. The mirton pathway uses most of these same processing steps to form the RISC. However, mirton transcripts do not require Drosha processing, since they are created through RNA splicing mechanisms that typically yield hairpins directly recognizable by exportin-5 [20, 116]. Thus, mirtons are exclusively derived from intron sequences.
Fig 1. General overview of riboregulation by miRNA and lncRNA.
A) RNA polymerase II transcripts of primary miRNAs are processed by the RNase III family enzyme Drosha, whereas immature mirtrons are derived from intron splicing. After transport to the cytoplasm, the RNAs are further processed by the RNase III family enzyme Dicer in conjunction with accessory proteins required for cellular localization and mRNA targeting (TRBP and AGO). Seed pairing with miRNAs occurs in the 3’ UTR of target mRNAs leading to translation inhibition and mRNA degradation. B) lncRNAs are transcribed by RNA polymerase II and are capped and polyadenylated similar to mRNAs. Posttranscriptional regulation by lncRNAs includes regulation of translation, RNA turnover, and mRNA splicing. No additional proteins are required for the regulatory function of lncRNAs, but lncRNAs may interact with other regulatory proteins.
In general, much less is known about lncRNAs compared to miRNAs. lncRNAs are loosely defined based upon their size (>200 nt) [48] and like miRNAs, lncRNAs are generally transcribed by RNA polymerase II [64, 67, 123] (Fig. 1B). However, lncRNAs exhibit a much greater resemblance to mRNAs. They are capped, polyadenylated, and often contain introns that are spliced by the same mechanisms used to generate mature mRNAs [64, 67, 82, 123]. Current evidence suggests that lncRNA expression is considerably lower than typical mRNA expression [23].
2.2 Mechanisms of riboregulatory control of human gene expression
Both miRNAs and lncRNAs are components of posttranscriptional regulatory networks in humans. Many lncRNAs also regulate gene expression through a variety of other mechanisms including epigenetic control and transcriptional regulation [64, 67], but these are beyond the scope of this review. As described above, the major mediator of miRNA function is the RISC. There are two mechanisms by which the RISC can affect gene expression posttranscriptionally. By far, the most common mechanism involves seed pairing interactions between nucleotides 2 – 8 of the miRNA and complementary sequences found in the 3’ UTR of target mRNAs [14]. Successful seed pairing interactions result in translation repression at the level of either initiation or post-initiation. The mechanisms involved in miRNA translation repression are still an area of active investigation and the process is not fully understood. However, current evidence suggests that miRNAs interfere with the formation of the closed loop structure of mRNAs, which are normally required to stimulate translation. Maintenance of mRNA loop structures occurs via the interaction of the eIF4F complex located at the 5’ cap and the PABPC protein located at the poly-A tail [46]. These protein interactions are critical to prevent deadenylation of the poly-A tail and subsequent decapping, which would otherwise expose an mRNA to degradation by the 5’ – 3’ exoribonuclease XRN1 [46]. There is still active debate regarding the relative contribution of translation repression vs. transcript degradation for the posttranscriptional control of gene expression, but recent results point towards a greater role for transcript degradation [46]. In instances in which the miRNA has perfect complementarity with its target, the RISC may also directly degrade target mRNAs, as certain AGO proteins like AGO2 are catalytically active ribonucleases [8, 14, 46, 63]. Thus far, this mechanism has only been observed in rare instances, presumably because miRNAs typically exhibit imperfect complementarity and only a subset of AGO proteins is catalytically active. In contrast to miRNAs, lncRNAs do not seem to require interactions with an analogous structure to the RISC to control gene expression. Similar to many bacterial non-coding small RNAs (sRNAs), seed pairing interactions with lncRNAs can directly stimulate or inhibit translation as well as modify mRNA target accessibility to ribonucleases [64, 67, 82, 123]. However, unlike prokaryotic sRNAs, lncRNAs also have the additional option of posttranscriptional control via modulation of mRNA splicing patterns [64, 67, 82, 123]. While each of these regulatory mechanisms has been observed with lncRNA, at present there are only a few examples in the literature, likely due to the dearth of information regarding lncRNAs in general. Therefore, it is still unclear whether they are major sources of posttranscriptional riboregulators. A survey of human antisense transcriptomes demonstrated a vast potential reservoir of antisense RNAs, which could conceivably function in posttranscriptional regulation [42, 64]. Therefore, it will be interesting to see in the coming years whether this is indeed the case.
2.3 Trans-riboregulators of the immune response
To date, studies of viruses have produced much of what is known about the host riboregulatory molecules triggered by pathogen infection [18]. However, research in recent years has demonstrated that bacterial pathogens are also potent stimuli for the production of numerous miRNAs that play critical regulatory roles in a variety of immune response pathways [26, 62]. Studies of the role of lncRNA in the immune response are currently in the earliest stages. Therefore, it is still unclear whether these molecules are truly key players in the host response to infection. Given the extreme diversity of lncRNAs overall as well as the presence of numerous lymphocyte-specific lncRNAs [77], it seems inevitable that they too will be discovered to be central regulators of the immune response. It is also worth noting that the stimulation of immune cells results in a profound effect upon the lncRNA transcriptome [13, 58].
2.4 Clinical studies of miRNA responses to oral inflammatory diseases
There are currently only a handful of clinical studies that have examined the human miRNA responses to oral diseases [16, 57, 76, 80, 96, 117, 124] and nothing is yet known about the corresponding lncRNA responses. Despite this, some interesting patterns can be found among the available data suggesting the regulatory RNA responses triggered by periodontitis and endodontic infections are quite distinct. Table 1 lists the differentially regulated miRNAs that have been reported in at least two or more miRNA profiling studies using clinical samples of health vs. disease. It should also be noted that there are numerous miRNAs uniquely regulated in each of these studies, so the list of shared miRNA responses is likely to expand dramatically as additional profiling studies are published. Furthermore, when comparing these data, it is important to consider that clinical specimens are a collection of multiple cell types, unlike what would normally be analyzed in in vitro studies. Therefore, transcriptional responses are the net averages of the sample as a whole, rather than any single cell type, which probably contributes to variability between studies. Even so, one can already identify a sizeable shared core set of differentially regulated miRNAs among the current studies. From this core set of miRNAs, it is apparent that most of the reported miRNA responses in the periodontitis studies are quite similar to each other [57, 76, 80, 96, 117], while the same is true of the endodontic miRNA responses [16, 124]. However, when comparing the results of periodontitis vs. endodontic infections, nearly the entire overlapping set of miRNAs curiously exhibits an inverse relationship (Table 1). The only exceptions are mir-199a-5p, mir-214*, and mir-766. Both mir199a-5p and mir-214* are lower expressed in disease samples of both periodontitis and inflamed pulp, while mir-766 is higher expressed in both [76, 117, 124]. In ovarian epithelial tumor cells and hepatocellular cancer cells, the primary miRNAs for mir-199a and mir-214 are down-regulated together as a cluster and thus their expression is linked [24, 122]. Furthermore, NF-κB activation by either the unfolded protein response or LPS represses transcription of this cluster [24]. Therefore, it is conceivable that the lower expression of mir199a-5p and mir-214* in both periodontitis and endodontic infections could be similarly attributable to inflammation and the concomitant activation of NF-κB. Mir-766 was one of only several miRNAs reported to have significantly increased expression in inflamed pulps and it was induced in inflamed gingiva as well [76, 124]. Recently, this miRNA was found to play a key role in suppressing the production of sirtuin 6 (SIRT6) in human dermal fibroblasts [92]. SIRT6 plays an important role in the differentiation of odontoblasts [99], fibroblasts [92, 104], and osteoclasts [55]. It also exerts anti-inflammatory effects by interfering with certain NF-κB signaling pathways [55, 104, 111]. Therefore, it would be interesting to determine whether mir-766 knockdown and/or SIRT6 overexpression could reduce the immunopathogenesis of periodontitis and pulpitis in experimental models. When comparing the miRNA responses of periodontits and endodontic infections, it is also striking that multiple members of the mir-181 family exhibit opposing expression patterns. This family of miRNAs all contains similar seed sequences and inhibits numerous components of inflammatory pathways [100] in addition to inhibiting myeloid differentiation of HL-60 human promyelocytic leukemia cells and CD34+ hematopoietic stem cells [100]. In endodontic infections, nearly this entire group of miRNA is lower expressed, which is indicative of active inflammation [100]. Counterintuitively, most of these same miRNA are higher expressed in periodontitis samples, despite ongoing chronic inflammation in the gingiva. A similar phenomenon can be observed among the let-7 family of miRNAs as well. There are 12 distinct let-7 miRNAs expressed in human cells that are associated with a diverse range of regulatory pathways including the production of multiple cytokines [95, 98]. Studies of let-7 expression in human cells exposed to a variety of bacterial pathogens consistently report a repressive effect upon their expression [26, 62, 95]. Consistent with these studies, two let-7 miRNAs were repressed in periapical lesions. However, four separate let-7 miRNAs were higher expressed in inflamed gingiva. In addition, miRNAs mir-101, mir-142-3p, and mir-223 were solely up-regulated in periodontitis samples with mir-142-3p and mir-223 also being among the most significantly affected [57, 76, 80, 96, 117]. Mir-101, mir-142-3p, and mir-223 were all recently found to be higher expressed in a mouse model of inflammatory bowel disease (IBD) [89], which, like periodontitis, is a chronic inflammatory disease caused by an abnormal immune response to the flora [17, 45]. Mir-101 is induced by a variety of innate immune activators via multiple toll-like receptors (TLR). In murine macrophages, mir-101 inhibits the translation of MAPK phosphatase-1 (MKP-1), which normally acts downstream of TLRs to dampen the innate immune response [126]. Thus, mir-101 can amplify activation of innate immune pathways in response to TLR activation by inhibiting MKP-1. Mir-142-3p was one of the few miRNAs found to be differentially regulated in three separate periodontitis studies. Its expression was previously demonstrated to be specific to cells of the hematopoietic lineage [53] and it functions in the differentiation and proliferation of immune cells as well the production of cytokines [28, 51, 102, 112, 125]. Since a similar increase in mir-142-3p expression was not observed in pulpitis and periapical disease, the higher mir-142-3p expression in gingival samples is likely as a result of a genuine differential regulation in periodontitis, rather than just an increased number of immune cells within the samples. Like mir-142-3p, mir-223 was also one of the few miRNAs identified independently in more than two periodontitis studies. In the aforementioned mouse model of IBD, mir-223 upregulation increases IL-17 production leading to subsequent IL-17-mediated immunopathogenesis [89]. Considering the central role of IL-17 in the pathogenesis of periodontal disease [40], mir-223 overexpression may be a similarly important riboregulatory pathway in periodontitis.
Table 1.
miRNA expression during periodontal disease and endodontic pulp and periapical infections.
| miRNA periodontal disease | miRNA pulp infection | miRNA periapical infection | |||||
|---|---|---|---|---|---|---|---|
| miRNA ref. |
[96] | [117] | [80] | [57] | [76] | [124] | [16] |
| hsa-let-7a | ↑ | ↑ | |||||
| hsa-let-7c | ↑ | ↓ | ↓ | ||||
| hsa-let-7f | ↑ | ↑ | ↓ | ||||
| hsa-let-7i | ↑ | ↑ | |||||
| hsa-mir-23b | ↑ | ↓ | |||||
| hsa-mir-27b | ↑ | ↓ | |||||
| hsa-mir-29a | ↑ | ↑ | ↓ | ||||
| hsa-mir-29c | ↑ | ↓ | |||||
| hsa-mir-30a | ↑ | ↓ | ↓ | ||||
| hsa-mir-30a* | ↓ | ↓ | |||||
| hsa-mir-30b | ↑ | ↓ | |||||
| hsa-mir-30c | ↑ | ↓ | |||||
| hsa-mir-30d | ↑ | ↑ | ↓ | ||||
| hsa-mir-30e | ↓ | ↑ | ↑ | ↑ | ↓ | ||
| hsa-mir-31* | ↓ | ↑ | |||||
| hsa-mir-33a | ↓ | ↑ | |||||
| hsa-mir-34a | ↑ | ↓ | |||||
| hsa-mir-34c-5p | ↑ | ↑ | |||||
| hsa-mir-95 | ↑ | ↑ | ↓ | ↓ | |||
| hsa-mir-98 | ↑ | ↓ | |||||
| hsa-mir-99a | ↓ | ↓ | |||||
| hsa-mir-101 | ↑ | ↑ | |||||
| hsa-mir-106b | ↑ | ↑ | |||||
| hsa-mir-125b | ↑ | ↓ | |||||
| hsa-mir-126 | ↑ | ↑ | |||||
| hsa-mir-130a | ↑ | ↑ | |||||
| hsa-mir-140-3p | ↑ | ↓ | |||||
| hsa-140-5p | ↑ | ↓ | |||||
| hsa-mir-141 | ↓ | ↑ | |||||
| hsa-mir-142-3p | ↑ | ↑ | ↑ | ||||
| hsa-mir-144* | ↑ | ↑ | |||||
| hsa-mir-148a | ↑ | ↓ | |||||
| hsa-mir-155 | ↑ | ↓ | |||||
| hsa-mir-181a | ↑ | ↓ | |||||
| hsa-mir-181a* | ↓ | ↓ | |||||
| hsa-mir-181a-2* | ↓ | ↓ | |||||
| hsa-mir-181b | ↑ | ↓ | ↓ | ||||
| hsa.mir-181c | ↑ | ↑ | ↓ | ↓ | |||
| hsa-mir-181d | ↑ | ↓ | ↓ | ||||
| hsa-mir-185 | ↑ | ↑ | |||||
| hsa-mir-186 | ↑ | ↓ | |||||
| hsa-mir-199a-5p | ↓ | ↓ | |||||
| hsa-mir-203 | ↓ | ↑ | ↑ | ||||
| hsa-mir-205 | ↓ | ↓ | |||||
| hsa-mir-205* | ↓ | ↓ | |||||
| hsa-mir-214* | ↓ | ↓ | ↓ | ||||
| hsa-mir-218 | ↑ | ↓ | |||||
| hsa-mir-223 | ↑ | ↑ | ↑ | ||||
| hsa-mir-301a | ↑ | ↑ | |||||
| hsa-mir-338-3p | ↑ | ↓ | |||||
| hsa-mir-363 | ↑ | ↑ | |||||
| hsa-mir-374a | ↑ | ↓ | |||||
| hsa-mir-379 | ↑ | ↓ | |||||
| hsa-mir-381 | ↑ | ↓ | |||||
| hsa-mir-451 | ↑ | ↓ | |||||
| hsa-mir-455-3p | ↑ | ↓ | ↓ | ||||
| hsa-mir-455-5p | ↓ | ↓ | |||||
| hsa-mir-497 | ↑ | ↑ | |||||
| hsa-mir-513c | ↓ | ↓ | |||||
| hsa-mir-671-5p | ↑ | ↓ | |||||
| hsa-mir-766 | ↓ | ↓ | |||||
| hsa-mir-1274b v16.0 | ↓ | ↓ | |||||
With the largely distinct miRNA expression patterns observed in periodontitis and endodontic infections, it will be interesting to see whether this trend continues as more miRNA profiling studies are published in the future. Since only a subset of people is prone to developing periodontitis, perhaps the distinct miRNA expression patterns in periodontitis samples is a reflection of the aberrant oral immune responses exhibited among this population [9, 38]. If so, it may be possible to identify a periodontitis-specific miRNA signature that could be used as a risk assessment to predict individual susceptibility. Likewise, particular dysregulated miRNAs might also serve as potential therapeutic targets to reduce periodontitis immunopathogenesis.
3. Trans-riboregulatory control of bacterial adaptive responses
The regulatory pathways controlled by sRNA in oral bacteria are diverse and include the control of biofilm formation, toxin-antitoxin modules, and virulence factors in addition to the other important cellular processes highlighted in the following sections.
3.1 Sources of prokaryotic riboregulatory molecules
The main source for bacterial sRNAs is the chromosome, but sRNAs were initially discovered on plasmids, phages, and transposons [11] indicating that the accessible pool of regulatory sRNAs might be much larger, especially for bacterial communities engaged in frequent horizontal gene transfer like the oral biofilm [85]. It has been estimated that 10 – 20% of all genes within a bacterial genome code for regulatory RNAs [86] and an average of 100 to 200 sRNAs have been discovered per single genome [11]. sRNAs generally range in size from 40 nt to 500 nt, which is considerably larger than the analogous microRNA (miRNA) riboregulators found in eukaryotes [121, 52]. Most characterized sRNAs are encoded within intergenic regions, lack protein coding sequences, and contain independent promoters and terminators [109]. However, there is also a rare subset classified as dual-function sRNAs that contain small translated open reading frames [47, 60]. The most prominent example is RNAIII from Staphylococcus aureus, which encodes a 26-codon hemolysin in addition to its pleiotropic riboregulatory function [68]. Conversely, larger protein-encoding genes can contain internal promoter sequences for sRNAs as well [90]. In some cases, sRNAs are also generated through the processing of larger transcripts. This was initially discovered for the Escherichia coli 6S RNA, a small, noncoding RNA involved in transcriptional control of gene expression [114, 113]. Transcription of the 6S RNA gene ssrS is growth phase dependent and regulated by two promoters, ssrS P1 and ssrS P2 giving rise to two possible transcripts. The longer transcript originating from P1 is processed by RNase E and G and the smaller transcript by RNase E [56]. Other mechanisms of sRNA generation include the 5’ and 3’ UTRs of mRNAs, which may be processed by RNases to produce functional sRNAs [109]. Likewise, transcription attenuation mechanisms can lead to functional sRNAs created from prematurely terminated mRNAs [109]. A new mechanism of sRNA-like riboregulatory control was recently identified in Streptococcus mutans, where the 5’ UTR of the irvA mRNA was demonstrated to function as a trans-riboregulator of gbpC mRNA. ([59] and see below). Heteroduplex formation between irvA and gbpC mRNAs serves as the key regulatory step allowing gbpC mRNA to be translated during environmental stress. This novel riboregulatory function for a protein-encoding mRNA suggests that previously identified “strictly” protein-encoding mRNAs might require reexamination for potential roles as trans-acting riboregulators [59].
3.2 Mechanisms of riboregulatory control of bacterial gene expression
Most mechanistic models of trans riboregulatory control of bacterial gene expression have been identified in E. coli [33], Salmonella [108] and S. aureus [30] and as of today only two mechanisms have been described in oral bacteria, both in S. mutans [59, 49]. In the following paragraphs the basic principles of trans riboregulation are introduced [12, 11, 65, 66], which represent the core set of prokaryotic riboregulatory posttranscriptional mechanisms.
3.2.1 Role of RNA chaperones and other ribonucleoprotein complexes
Ribonucleoprotein complexes are essential regulatory components in bacteria. Although sRNA interactions with mRNA are mediated via complementary seed pairing, RNA duplex formation commonly requires an additional protein component to function as an RNA chaperone catalyzing the interaction. One of the best-characterized RNA chaperones is Hfq (host factor required for phage Q β RNA replication, also known as host factor 1) [110, 22]. In many enteric bacteria, Hfq is crucial for remodeling RNA secondary structures and enabling seed pairing between sRNAs and their targets [110, 52]. Hfq is also able to stabilize sRNAs by protecting them from RNase degradation. Interestingly, only about 50% of sequenced species encode obvious Hfq orthologs and it seems to be largely dispensable for heteroduplex formation in numerous species, particularly among Gram positive bacteria [101, 106]. Currently, Listeria monocytogenes is the only Gram positive species shown to require Hfq and even this is only for a small subset of its sRNA interactions [73]. Interestingly, no Hfq orthologs have been identified in any species of the Streptococcus genus [65]. Thus, it is still unclear whether Gram positive species truly require an RNA chaperone for seed pairing interactions or they have evolved alternate mechanisms [86]. Given the diverse roles of Hfq in enteric bacteria [110], it is expected that most bacteria would presumably utilize some type of RNA chaperone-dependent mechanism for sRNA riboregulatory interactions.
3.2.2 Riboregulatory control of translation
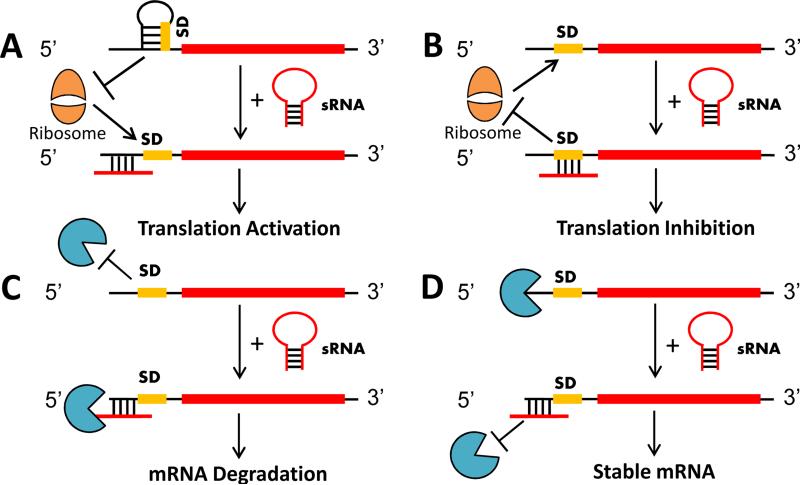
The majority of characterized sRNA seed pairing interactions target the 5’ UTR of mRNAs frequently occurring in or around the Shine-Dalgarno (SD) sequence (Fig. 2). Proximal or distal sRNA binding to the SD sequence can either directly or indirectly influence the binding of the 30S ribosomal subunit, thus influencing translation efficiency. In some instances, a naturally occurring secondary structure within the 5’ UTR can occlude the SD sequence leading to poor translation initiation. However, distal sRNA hybridization in the UTR can destabilize this secondary structure freeing the SD sequence and ultimately increasing the yield of protein [11, 66]. Heteroduplex formation between an sRNA and its target sequence can also mask the SD sequence of a transcript that would otherwise be freely accessible. As a consequence, such seed pairing interactions inhibit translation initiation. Alternatively, sRNA interactions upstream or downstream of the SD sequence may also induce inhibitory secondary structures to occlude the SD sequence and reduce translation of the target mRNA [11, 65, 66].
Fig. 2. Mechanisms of sRNA-mediated posttranscriptional regulation in bacteria.
A) sRNA interacts with the 5’ UTR of the transcript disrupting intra-molecular base pairing and allowing access to the SD sequence for translation initiation B) sRNA interacts directly with the ribosome binding site (SD sequence) preventing translation initiation C) sRNA guides a ribonuclease to the target mRNA to initiate mRNA degradation D) sRNA interacts with the target mRNA to occlude an endogenous RNase cleavage site in the mRNA
3.2.3 Regulation of mRNA stability
In addition to modulating translation efficiency, sRNA seed pairing interactions can have a substantial negative or positive effect upon mRNA stability (Fig. 2). Similar to the aforementioned debate of the relative contribution of translation repression vs. transcript degradation by miRNAs, translation and mRNA stability are often intimately connected in prokaryotes, since mRNAs not protected by ribosomes are generally more accessible to ribonuclease degradation [120]. Therefore, sRNA interactions that inhibit translation tend to reduce mRNA stability, whereas mRNA stability is generally higher in heavily translated mRNAs. Seed pairing can also directly promote mRNA degradation by creating substrates for double stranded RNA specific ribonucleases, such as RNase III. In some cases, ribonucleoprotein complexes with RNA chaperones, such as Hfq serve to actively recruit RNases to a target via protein-protein interactions. sRNA recognition and guided RNase degradation of mRNA occurs most frequently via interactions in the 5’ UTR, but interactions in the coding region have also been described [11, 65, 66]. There are two mechanisms by which sRNAs can modulate RNase cleavage of a target to increase its stability. First, seed pairing within the 5’ UTR may recruit ribonucleases that process the UTR to remove inhibitory secondary structures occluding the SD sequence. In this case, increased mRNA stability is a consequence of stimulating translation. Alternatively, it was recently demonstrated that sRNA interactions are capable of directly stabilizing target mRNAs by occluding endogenous RNase cleavage sites within target mRNAs [11, 65, 66].
3.3 Trans-riboregulators of pathogen adaptive responses
3.3.1 Global sRNA expression during host infection
Our understanding of oral bacterial sRNAs has increased substantially just within the past few years. Thus far, the primary focus has been to identify novel sRNAs using bioinformatics and high-throughput sequencing approaches [65]. Surprisingly little is currently known about the molecular mechanisms of oral bacterial sRNA-target interactions and their role in host-pathogen interactions. Most of what is known has been derived from studies of Streptococcus pyogenes and Streptococcus pneumoniae, which both reside in the oral cavity, but are not significant members of the dental biofilm. Examples from these species are included in the next sections to demonstrate the breadth of sRNA dependent modulation of host-pathogen interactions and because they belong to the genus Streptococcus, which constitutes about 80% of the initial dental biofilm [87].
S. pneumoniae colonizes the nasopharynx in over 40% of healthy individuals, but can also cause numerous diseases like pneumonia, otitis media, sepsis, and meningitis [61]. A recent analysis of the TIGR4 strain identified 89 putative sRNAs that each have >90% conservation in other sequenced S. pneumoniae strains. Over half of these have been confirmed as being expressed by either Northern blot analysis or RT-qPCR and a definite role in pathogenesis was established for eight of the sRNAs within a murine model of sepsis. Subsequent fitness profiling of S. pneumoniae suggests that sRNAs play a much more significant role during systemic infection as well as for tissue specific tropism [61]. 27 sRNA mutants had reduced virulence during lung infection. In the nasopharynx, 19 sRNAs appeared essential for infection, while 5 sRNA mutants exhibited attenuated virulence. Within blood, 5 sRNAs were found to be essential and an additional 11 were attenuated for virulence. Interestingly, the roles of sRNAs in these different host niches were also specific with only one sRNA, SN46, playing a role in all three host-specific tissues [61]. This suggests that sRNA-dependent control of pathogenesis can be restricted to specific host tissues.
A recent study with Aggregatibacter actinomycetemcomitans used a similar approach with a murine model of infection to detect changes in the expression of non-coding (nc) RNAs, which are functional RNAs containing no coding sequences, such as tRNAs, rRNAs, and most sRNAs. Besides being the proposed causative agent of localized aggressive periodontitis [94], A. actinomycetemcomitans can cause extraoral infections, such as brain abscesses [83]. Jorth et al. used a murine abscess model to compare the in vivo transcriptome with that of in vitro grown biofilms. The study identified 210 ncRNAs, including 75 sRNAs. 80 out of the 210 ncRNAs were differently expressed in vivo when compared to in vitro culture, with 39 up-regulated and 41 down-regulated. Notably, hfq was specifically higher expressed in vivo. Considering the critical RNA chaperone function for Hfq in other bacteria, it will be interesting to determine whether Hfq plays a similar role in A. actinomycetemcomitans pathogenesis.
3.3.2 Riboregulation of pathogen adaptive responses and in vivo relevance
A more detailed understanding of sRNA-mediated regulation of bacterial pathogenesis is available for the sRNA FasX from S. pyogenes (Group A streptococcus, GAS) [66], which is a human-specific pathogen that causes a variety of diseases ranging from superficial infection to life-threatening diseases [19]. FasX (size 205 nt) is part of the fasBCAX operon encoding a putative two-component system with two membrane spanning histidine kinases and a DNA binding response regulator controlling fasX expression [50]. The expression of fasX increases during exponential growth, but decreases in the stationary phase [66]. Trans-riboregulation by FasX targets two important virulence factors streptokinase (Ska) and Cpa, which is required for pilus biogenesis. Streptokinase is a critical secreted virulence factor promoting S. pyogenes dissemination through its activation of host plasminogen into plasmin, a protease that dissolves fibrin present in blood clots after injury. FasX expression increases streptokinase production 10-fold due to its stabilization of ska mRNA, which occurs via seed pairing located 24 to 32 nucleotides upstream of the start codon [66, 84]. This is an unusual location for a positive interaction, as most sRNA interactions occurring near the start codon inhibit target translation. In contrast to ska mRNA, FasX seed pairing with cpa mRNA results in decreased cpa translation and mRNA stability by occluding the cpa SD sequence. Consequently, this interaction inhibits pilus production. In general, pilus formation promotes adherence to host cells and facilitates biofilm formation. The ability of FasX to simultaneously enhance thrombolytic activity and inhibit pilus mediated adherence and biofilm formation ultimately favors pathogen dissemination over colonization [84, 66]. The importance of this opposing effect upon virulence factor production was recently confirmed in a murine model of bacteremia. Mice infected with a fasX mutant survived significantly longer, thus confirming fasX as a major checkpoint regulator in the adaptive responses of S. pyogenes [21].
3.3.3 In vitro mechanistic studies of oral bacterial adaptive responses
A combination of bioinformatic tools was used to identify and predict the respective mRNA targets of a group of iron- and Fur-regulated sRNAs in the periodontopathogen A. actinomycetemcomitans [2]. Potential binding sites for the iron-dependent transcriptional regulator Fur were identified in the promoter regions of all four sRNAs, consistent with their predicted iron-dependent expression. Deletion of the fur ortholog increased expression of the tested sRNAs as expected. However, in contrast to expectations, the fur mutation did not alter biofilm development under the tested conditions, despite a known regulation of biofilm associated genes by iron [3]. Even more surprising, an overproduction of one of the identified sRNAs, designated JA03, diminished biofilm formation almost completely. These results point to a much more complex posttranscriptional regulatory network for A. actinomycetemcomitans biofilm formation than previously assumed.
Iron availability in the form of hemin was also recently demonstrated to regulate a large group of sRNAs in the periodontopathogen Porphyromonas gingivalis. Out of 37 putative sRNAs identified, 24 were found to be differentially regulated in response to hemin depletion [81]. Since free iron is an essential element that is typically in extremely low abundance in the oral cavity, it is not surprising that P. gingivalis has evolved an intricate posttranscriptional regulatory network to maintain iron homeostasis. Presumably this would also be true of many other oral bacteria, especially for periodontal pathogens.
Another class of sRNAs referred to as antisense RNAs (aRNA) was recently demonstrated to control a toxin-antitoxin (TA) module expressed by the cariogenic species S. mutans [49]. TA modules are implicated in cell growth arrest and may even lead to cell death under certain adverse growth conditions. However, in the absence of environmental stress, the toxin component of the module is efficiently inactivated by the antitoxin component. The functionality of TA modules parallels those of plasmid addiction modules: the antitoxin is produced in abundance, but is highly labile, whereas the toxin is much more stable [115]. During impaired growth conditions, TA module gene expression is impacted resulting in a net accumulation of the toxin component [107]. In S. mutans, the Fst-like toxin encoding a 32 amino acid peptide and its cis-encoded antisense sRNA antitoxin both exhibit the expected differences in stabilities. The fst transcript half-life is 90 min., whereas the antitoxin RNA has a 30 min. half-life. Analysis of the TA module locus further demonstrated that the toxin and the antitoxin were transcribed across direct tandem repeats. This provides a region of perfect complementarity for both RNAs that facilitates seed pairing inhibition of toxin translation. As expected, overexpression of the Fst toxin component led to toxicity in S. mutans and co-expression of the antitoxin inhibited toxicity, which confirmed the functionality of the TA module [49].
A novel mechanism of trans-acting riboregulatory control was also recently discovered in S. mutans [59]. A protein-encoding dual-function mRNA, irvA, was demonstrated to function both in the production of the IrvA protein, while also serving as a trans riboregulator via sRNA-like seed pairing interactions. The irvA gene is encoded on the chromosome adjacent to its key regulator irvR, which encodes a LexA-like self-cleaving transcription repressor [74]. IrvR is a potent repressor of irvA expression under normal growth conditions. However, this repression is relieved when the cell encounters a variety of environmental stresses. As a consequence of irvA induction, S. mutans abolishes its natural competence ability, represses bacteriocin production, and strongly upregulates the production of a surface exposed lectin called GbpC [7]. Even though gbpC is constitutively expressed, its mRNA is exceptionally unstable and is normally degraded before any appreciable translation can occur on gbpC transcripts. However, when irvA repression is relieved by environmental stress, hybrid duplexes form between irvA and gbpC mRNAs due to seed pairing interactions between the irvA 5’ UTR and the gbpC coding sequence. This results in a tremendous increase in gbpC mRNA stability and a concomitant increase in GbpC protein production. Since the seed region in the irvA 5’ UTR was shown to overlap with an endogenous RNase J2 cleavage site in gbpC mRNA, it was proposed that hybrid duplex formation between the two mRNAs directly inhibits RNase J2-mediated degradation of gbpC mRNA during episodes of environmental stress. In this interaction, the increase in gbpC mRNA stability was also shown to be largely independent of gbpC translation, which demonstrates that mRNA stability and translation are not always directly correlated. It is still unclear why GbpC production would be regulated by environmental stress, but it was suggested that the surface lectin activity of GbpC could be used to bolster biofilm integrity under adverse growth conditions [59].
While our understanding of riboregulatory control of gene expression in oral bacteria has increased substantially in recent years, it is still very rudimentary. With the tremendous improvements in next-generation sequencing technologies, we can expect future meta-transcriptomic studies of pathogenic oral biofilms to provide unprecedented insights into the posttranscriptional landscape of microbial communities. Further mechanistic studies will be vital for interpreting the implications of these data. Such studies should revolutionize our understanding of riboregulatory control of key pathways important for virulence, adaptation, and persistence in the oral cavity.
4. Perspective/Outlook
Riboregulatory molecules are increasingly being recognized for their central role in controlling host immune response pathways and pathogen virulence factor production. To date, much of what is known about the miRNA responses to bacterial pathogens has come from studies of a relatively small number of species. Among these studies, there are several miRNAs that are commonly differentially regulated even when using multiple in vitro cell culture systems and animal models, mainly the let-7 family of miRNAs, mir-146, and mir-155 [26, 62]. Overall, this trend has generally held true for studies of oral bacteria as well [10, 43, 69, 71, 72]. Despite this synergy among studies using experimental model systems, these commonly observed miRNA responses have not been particularly reflective of the miRNA responses found in oral clinical samples (Table 1). For example, while a variety of let-7 miRNAs were identified in periodontitis samples, they exhibited opposite expression patterns to what has been observed in in vitro and animal model studies [26, 62, 95]. The question then is whether this difference is simply a reflection of the limited number of available clinical datasets or if there is truly a fundamental difference in the miRNA profiles of clinical disease samples vs. experimental models. It is also curious that the miRNA responses in periodontitis have thus far exhibited minimal similarities to endodontic infections [16, 57, 76, 80, 96, 117, 124]. Of course, one would expect a certain amount of distinction between the two simply due to the different cell types involved in both diseases. However, both diseases are also characterized by inflammation [39, 70]. Presumably, this in itself should result in a shared inflammation-specific miRNA profile, yet this has not been the case thus far. Are these differences also attributable to the small number of available clinical datasets or are they suggestive of fundamental differences in the inflammatory responses to the oral flora? Likewise, the tissue-specific sRNA responses of S. pneumoniae suggest that this organism and probably many other pathogens utilize specific posttranscriptional riboregulators to rapidly adapt to different niches and growth conditions within the host. For oral biofilm bacteria, one can predict that a similarly distinct sRNA profile would be observable for bacteria residing in different locations in the oral cavity or for organisms residing in sites of variable health status. The next major breakthroughs in our understanding of the role of riboregulation in oral disease are likely to arise from correlation studies of both host miRNA responses and bacterial sRNA responses in samples of health and disease. The relative accessibility of such samples presents a tremendous opportunity for the oral research community to be at the forefront of both miRNA and bacterial sRNA research.
Acknowledgements
This work was supported by an NIH-NIDCR grant DE021726 to J.K. and NIH-NIDCR grants DE018893 and DE022083 to JM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Almeida MI, Reis RM, Calin GA. Decoy activity through microRNAs: the therapeutic implications. Expert opinion on biological therapy. 2012;12:1153–9. doi: 10.1517/14712598.2012.693470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amarasinghe JJ, Connell TD, Scannapieco FA, Haase EM. Novel iron-regulated and Fur-regulated small regulatory RNAs in Aggregatibacter actinomycetemcomitans. Molecular oral microbiology. 2012;27:327–49. doi: 10.1111/j.2041-1014.2012.00645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amarasinghe JJ, Scannapieco FA, Haase EM. Transcriptional and translational analysis of biofilm determinants of Aggregatibacter actinomycetemcomitans in response to environmental perturbation. Infection and immunity. 2009;77:2896–907. doi: 10.1128/IAI.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nature reviews. Molecular cell biology. 2013;14:475–88. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 5.Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nature reviews. Immunology. 2010;10:24–35. doi: 10.1038/nri2685. [DOI] [PubMed] [Google Scholar]
- 6.Bak RO, Mikkelsen JG. miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley interdisciplinary reviews. RNA. 2014;5:317–33. doi: 10.1002/wrna.1213. [DOI] [PubMed] [Google Scholar]
- 7.Banas JA, Vickerman MM. Glucan-binding proteins of the oral streptococci. Critical reviews in oral biology and medicine : an official publication of the American Association of Oral Biologists. 2003;14:89–99. doi: 10.1177/154411130301400203. [DOI] [PubMed] [Google Scholar]
- 8.Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cellular and molecular life sciences : CMLS. 2012;69:3613–34. doi: 10.1007/s00018-012-0990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartold PM, Van Dyke TE. Periodontitis: a host-mediated disruption of microbial homeostasis. Unlearning learned concepts. Periodontology. 2013;62:203–17. doi: 10.1111/j.1600-0757.2012.00450.x. 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benakanakere MR, Li Q, Eskan MA, Singh AV, Zhao J, Galicia JC, et al. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. The Journal of biological chemistry. 2009;284:23107–15. doi: 10.1074/jbc.M109.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brantl S, Bruckner R. Small regulatory RNAs from low-GC Gram-positive bacteria. RNA biology. 2014;11:443–56. doi: 10.4161/rna.28036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caldelari I, Chao Y, Romby P, Vogel J. RNA-mediated regulation in pathogenic bacteria. Cold Spring Harbor perspectives in medicine. 2013;3:a010298. doi: 10.1101/cshperspect.a010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–92. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Chan LT, Zhong S, Naqvi AR, Self-Fordham J, Nares S, Bair E, et al. MicroRNAs: new insights into the pathogenesis of endodontic periapical disease. Journal of endodontics. 2013;39:1498–503. doi: 10.1016/j.joen.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nature reviews. Immunology. 2008;8:458–66. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 18.Cullen BR. Viruses and microRNAs: RISCy interactions with serious consequences. Genes & development. 2011;25:1881–94. doi: 10.1101/gad.17352611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cunningham MW. Pathogenesis of group A streptococcal infections. Clinical microbiology reviews. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curtis HJ, Sibley CR, Wood MJ. Mirtrons, an emerging class of atypical miRNA. Wiley interdisciplinary reviews. RNA. 2012;3:617–32. doi: 10.1002/wrna.1122. [DOI] [PubMed] [Google Scholar]
- 21.Danger JL, Cao TN, Cao TH, Sarkar P, Trevino J, Pflughoeft KJ, et al. The small regulatory RNA FasX enhances group A Streptococcus virulence and inhibits pilus expression via serotype-specific targets. Molecular microbiology. 2015 doi: 10.1111/mmi.12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. The Journal of biological chemistry. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan Q, Wang X, Gong W, Ni L, Chen C, He X, et al. ER stress negatively modulates the expression of the miR-199a/214 cluster to regulates tumor survival and progression in human hepatocellular cancer. PloS one. 2012;7:e31518. doi: 10.1371/journal.pone.0031518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Current biology : CB. 2010;20:R858–61. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eulalio A, Schulte L, Vogel J. The mammalian microRNA response to bacterial infections. RNA biology. 2012;9:742–50. doi: 10.4161/rna.20018. [DOI] [PubMed] [Google Scholar]
- 27.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annual review of biochemistry. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 28.Fan HB, Liu YJ, Wang L, Du TT, Dong M, Gao L, et al. miR-142-3p acts as an essential modulator of neutrophil development in zebrafish. Blood. 2014;124:1320–30. doi: 10.1182/blood-2013-12-545012. [DOI] [PubMed] [Google Scholar]
- 29.Fechter P, Caldelari I, Lioliou E, Romby P. Novel aspects of RNA regulation in Staphylococcus aureus. FEBS letters. 2014;588:2523–9. doi: 10.1016/j.febslet.2014.05.037. [DOI] [PubMed] [Google Scholar]
- 30.Felden B, Vandenesch F, Bouloc P, Romby P. The Staphylococcus aureus RNome and its commitment to virulence. PLoS pathogens. 2011;7:e1002006. doi: 10.1371/journal.ppat.1002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frappier L. Regulation of Herpesvirus Reactivation by Host MicroRNAs. Journal of virology. 2015;89:2456–8. doi: 10.1128/JVI.03413-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gopel Y, Gorke B. Lies and deception in bacterial gene regulation: the roles of nucleic acid decoys. Molecular microbiology. 2014;92:641–7. doi: 10.1111/mmi.12604. [DOI] [PubMed] [Google Scholar]
- 33.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annual review of microbiology. 2004;58:303–28. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 34.Gripenland J, Netterling S, Loh E, Tiensuu T, Toledo-Arana A, Johansson J. RNAs: regulators of bacterial virulence. Nature reviews. 2010;8:857–66. doi: 10.1038/nrmicro2457. [DOI] [PubMed] [Google Scholar]
- 35.Grzybowska EA, Wilczynska A, Siedlecki JA. Regulatory functions of 3'UTRs. Biochemical and biophysical research communications. 2001;288:291–5. doi: 10.1006/bbrc.2001.5738. [DOI] [PubMed] [Google Scholar]
- 36.Guo YE, Steitz JA. Virus meets host microRNA: the destroyer, the booster, the hijacker. Molecular and cellular biology. 2014;34:3780–7. doi: 10.1128/MCB.00871-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ha M, Kim VN. Regulation of microRNA biogenesis. Nature reviews. Molecular cell biology. 2014;15:509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 38.Hajishengallis G. Too old to fight? Aging and its toll on innate immunity. Molecular oral microbiology. 2010;25:25–37. doi: 10.1111/j.2041-1014.2009.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nature reviews. Immunology. 2015;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hajishengallis G, Chavakis T, Hajishengallis E, Lambris JD. Neutrophil homeostasis and inflammation: novel paradigms from studying periodontitis. Journal of leukocyte biology. 2014 doi: 10.1189/jlb.3VMR1014-468R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris JF, Micheva-Viteva S, Li N, Hong-Geller E. Small RNA-mediated regulation of host-pathogen interactions. Virulence. 2013;4:785–95. doi: 10.4161/viru.26119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–7. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Honda T, Takahashi N, Miyauchi S, Yamazaki K. Porphyromonas gingivalis lipopolysaccharide induces miR-146a without altering the production of inflammatory cytokines. Biochemical and biophysical research communications. 2012;420:918–25. doi: 10.1016/j.bbrc.2012.03.102. [DOI] [PubMed] [Google Scholar]
- 44.Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO reports. 2012;13:971–83. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huibregtse IL, van Lent AU, van Deventer SJ. Immunopathogenesis of IBD: insufficient suppressor function in the gut? Gut. 2007;56:584–92. doi: 10.1136/gut.2006.103523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature reviews. Genetics. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 47.Kaito C, Saito Y, Ikuo M, Omae Y, Mao H, Nagano G, et al. Mobile genetic element SCCmec-encoded psm-mec RNA suppresses translation of agrA and attenuates MRSA virulence. PLoS pathogens. 2013;9:e1003269. doi: 10.1371/journal.ppat.1003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 49.Koyanagi S, Levesque CM. Characterization of a Streptococcus mutans intergenic region containing a small toxic peptide and its cis-encoded antisense small RNA antitoxin. PloS one. 2013;8:e54291. doi: 10.1371/journal.pone.0054291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kreikemeyer B, Boyle MD, Buttaro BA, Heinemann M, Podbielski A. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Molecular microbiology. 2001;39:392–406. doi: 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- 51.Lagrange B, Martin RZ, Droin N, Aucagne R, Paggetti J, Largeot A, et al. A role for miR-142-3p in colony-stimulating factor 1-induced monocyte differentiation into macrophages. Biochimica et biophysica acta. 2013;1833:1936–46. doi: 10.1016/j.bbamcr.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 52.Lalaouna D, Simoneau-Roy M, Lafontaine D, Masse E. Regulatory RNAs and target mRNA decay in prokaryotes. Biochimica et biophysica acta. 2013;1829:742–7. doi: 10.1016/j.bbagrm.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 53.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le Rhun A, Charpentier E. Small RNAs in streptococci. RNA biology. 2012;9:414–26. doi: 10.4161/rna.20104. [DOI] [PubMed] [Google Scholar]
- 55.Lee HS, Ka SO, Lee SM, Lee SI, Park JW, Park BH. Overexpression of sirtuin 6 suppresses inflammatory responses and bone destruction in mice with collagen-induced arthritis. Arthritis and rheumatism. 2013;65:1776–85. doi: 10.1002/art.37963. [DOI] [PubMed] [Google Scholar]
- 56.Lee JY, Park H, Bak G, Kim KS, Lee Y. Regulation of transcription from two ssrS promoters in 6S RNA biogenesis. Molecules and cells. 2013;36:227–34. doi: 10.1007/s10059-013-0082-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee YH, Na HS, Jeong SY, Jeong SH, Park HR, Chung J. Comparison of inflammatory microRNA expression in healthy and periodontitis tissues. Biocell : official journal of the Sociedades Latinoamericanas de Microscopia Electronica. 2011;35:43–9. [PubMed] [Google Scholar]
- 58.Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1002–7. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu N, Niu G, Xie Z, Chen Z, Itzek A, Kreth J, et al. The Streptococcus mutans irvA Gene Encodes a trans-Acting Riboregulatory mRNA. Molecular cell. 2015;57:179–90. doi: 10.1016/j.molcel.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mangold M, Siller M, Roppenser B, Vlaminckx BJ, Penfound TA, Klein R, et al. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Molecular microbiology. 2004;53:1515–27. doi: 10.1111/j.1365-2958.2004.04222.x. [DOI] [PubMed] [Google Scholar]
- 61.Mann B, van Opijnen T, Wang J, Obert C, Wang YD, Carter R, et al. Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS pathogens. 2012;8:e1002788. doi: 10.1371/journal.ppat.1002788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maudet C, Mano M, Eulalio A. MicroRNAs in the interaction between host and bacterial pathogens. FEBS letters. 2014;588:4140–7. doi: 10.1016/j.febslet.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 63.Meister G. Argonaute proteins: functional insights and emerging roles. Nature reviews. Genetics. 2013;14:447–59. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 64.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature reviews. Genetics. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 65.Merritt J, Chen Z, Liu N, Kreth J. Posttranscriptional regulation of oral bacterial adaptive responses. Current oral health reports. 2014;1:50–8. doi: 10.1007/s40496-013-0005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller EW, Cao TN, Pflughoeft KJ, Sumby P. RNA-mediated regulation in Gram-positive pathogens: an overview punctuated with examples from the group a Streptococcus. Molecular microbiology. 2014;94:9–20. doi: 10.1111/mmi.12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic acids research. 2012;40:6391–400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. The EMBO journal. 1995;14:4569–77. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nahid MA, Rivera M, Lucas A, Chan EK, Kesavalu L. Polymicrobial infection with periodontal pathogens specifically enhances microRNA miR-146a in ApoE−/− mice during experimental periodontal disease. Infection and immunity. 2011;79:1597–605. doi: 10.1128/IAI.01062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nair PN. Pathogenesis of apical periodontitis and the causes of endodontic failures. Critical reviews in oral biology and medicine : an official publication of the American Association of Oral Biologists. 2004;15:348–81. doi: 10.1177/154411130401500604. [DOI] [PubMed] [Google Scholar]
- 71.Naqvi AR, Fordham JB, Khan A, Nares S. MicroRNAs responsive to Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis LPS modulate expression of genes regulating innate immunity in human macrophages. Innate immunity. 2013;20:540–51. doi: 10.1177/1753425913501914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neiva KG, Calderon NL, Alonso TR, Panagakos F, Wallet SM. Type 1 diabetes-associated TLR responsiveness of oral epithelial cells. Journal of dental research. 2014;93:169–74. doi: 10.1177/0022034513516345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nielsen JS, Lei LK, Ebersbach T, Olsen AS, Klitgaard JK, Valentin-Hansen P, et al. Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic acids research. 2010;38:907–19. doi: 10.1093/nar/gkp1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Niu G, Okinaga T, Qi F, Merritt J. The Streptococcus mutans IrvR repressor is a CI-like regulator that functions through autocleavage and Clp-dependent proteolysis. Journal of bacteriology. 2010;192:1586–95. doi: 10.1128/JB.01261-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annual review of immunology. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 76.Ogata Y, Matsui S, Kato A, Zhou L, Nakayama Y, Takai H. MicroRNA expression in inflamed and noninflamed gingival tissues from Japanese patients. Journal of oral science. 2014;56:253–60. doi: 10.2334/josnusd.56.253. [DOI] [PubMed] [Google Scholar]
- 77.Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. Journal of immunology. 2009;182:7738–48. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- 78.Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Research in microbiology. 2009;160:278–87. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 79.Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell host & microbe. 2010;8:116–27. doi: 10.1016/j.chom.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 80.Perri R, Nares S, Zhang S, Barros SP, Offenbacher S. MicroRNA modulation in obesity and periodontitis. Journal of dental research. 2012;91:33–8. doi: 10.1177/0022034511425045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Phillips P, Progulske-Fox A, Grieshaber S, Grieshaber N. Expression of Porphyromonas gingivalis small RNA in response to hemin availability identified using microarray and RNA-seq analysis. FEMS microbiology letters. 2014;351:202–8. doi: 10.1111/1574-6968.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Picardi E, D'Erchia AM, Gallo A, Montalvo A, Pesole G. Uncovering RNA Editing Sites in Long Non-Coding RNAs. Frontiers in bioengineering and biotechnology. 2014;2:64. doi: 10.3389/fbioe.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rahamat-Langendoen JC, van Vonderen MG, Engstrom LJ, Manson WL, van Winkelhoff AJ, Mooi-Kokenberg EA. Brain abscess associated with Aggregatibacter actinomycetemcomitans: case report and review of literature. Journal of clinical periodontology. 2011;38:702–6. doi: 10.1111/j.1600-051X.2011.01737.x. [DOI] [PubMed] [Google Scholar]
- 84.Ramirez-Pena E, Trevino J, Liu Z, Perez N, Sumby P. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Molecular microbiology. 2010;78:1332–47. doi: 10.1111/j.1365-2958.2010.07427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roberts AP, Kreth J. The impact of horizontal gene transfer on the adaptive ability of the human oral microbiome. Frontiers in cellular and infection microbiology. 2014;4:124. doi: 10.3389/fcimb.2014.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Romby P, Charpentier E. An overview of RNAs with regulatory functions in gram-positive bacteria. Cellular and molecular life sciences : CMLS. 2010;67:217–37. doi: 10.1007/s00018-009-0162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rosan B, Lamont RJ. Dental plaque formation. Microbes and infection / Institut Pasteur. 2000;2:1599–607. doi: 10.1016/s1286-4579(00)01316-2. [DOI] [PubMed] [Google Scholar]
- 88.Sand M. The pathway of miRNA maturation. Methods in molecular biology. 2014;1095:3–10. doi: 10.1007/978-1-62703-703-7_1. [DOI] [PubMed] [Google Scholar]
- 89.Schaefer JS, Montufar-Solis D, Vigneswaran N, Klein JR. Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10−/− mice precedes expression in the colon. Journal of immunology. 2011;187:5834–41. doi: 10.4049/jimmunol.1100922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schluter JP, Reinkensmeier J, Daschkey S, Evguenieva-Hackenberg E, Janssen S, Janicke S, et al. A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing alpha- proteobacterium Sinorhizobium meliloti. BMC genomics. 2010;11:245. doi: 10.1186/1471-2164-11-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sharma A, Diecke S, Zhang WY, Lan F, He C, Mordwinkin NM, et al. The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. The Journal of biological chemistry. 2013;288:18439–47. doi: 10.1074/jbc.M112.405928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nature reviews. Molecular cell biology. 2014;15:565–76. doi: 10.1038/nrm3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Slots J, Reynolds HS, Genco RJ. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infection and immunity. 1980;29:1013–20. doi: 10.1128/iai.29.3.1013-1020.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Staedel C, Darfeuille F. MicroRNAs and bacterial infection. Cellular microbiology. 2013;15:1496–507. doi: 10.1111/cmi.12159. [DOI] [PubMed] [Google Scholar]
- 96.Stoecklin-Wasmer C, Guarnieri P, Celenti R, Demmer RT, Kebschull M, Papapanou PN. MicroRNAs and their target genes in gingival tissues. Journal of dental research. 2012;91:934–40. doi: 10.1177/0022034512456551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Molecular cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Su JL, Chen PS, Johansson G, Kuo ML. Function and regulation of let-7 family microRNAs. MicroRNA. 2012;1:34–9. doi: 10.2174/2211536611201010034. [DOI] [PubMed] [Google Scholar]
- 99.Sun HL, Wu YR, Huang C, Wang JW, Fu DJ, Liu YC. The effect of SIRT6 on the odontoblastic potential of human dental pulp cells. Journal of endodontics. 2014;40:393–8. doi: 10.1016/j.joen.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 100.Sun X, Sit A, Feinberg MW. Role of miR-181 family in regulating vascular inflammation and immunity. Trends in cardiovascular medicine. 2014;24:105–12. doi: 10.1016/j.tcm.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic acids research. 2002;30:3662–71. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun Y, Varambally S, Maher CA, Cao Q, Chockley P, Toubai T, et al. Targeting of microRNA-142-3p in dendritic cells regulates endotoxin-induced mortality. Blood. 2011;117:6172–83. doi: 10.1182/blood-2010-12-325647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Swaminathan G, Martin-Garcia J, Navas-Martin S. RNA viruses and microRNAs: challenging discoveries for the 21st century. Physiological genomics. 2013;45:1035–48. doi: 10.1152/physiolgenomics.00112.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tian K, Liu Z, Wang J, Xu S, You T, Liu P. Sirtuin-6 inhibits cardiac fibroblasts differentiation into myofibroblasts via inactivation of nuclear factor kappaB signaling. Translational research : the journal of laboratory and clinical medicine. 2014 doi: 10.1016/j.trsl.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 105.Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nature immunology. 2014;15:484–91. doi: 10.1038/ni.2887. [DOI] [PubMed] [Google Scholar]
- 106.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Molecular microbiology. 2004;51:1525–33. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 107.Van Melderen L. Toxin-antitoxin systems: why so many, what for? Current opinion in microbiology. 2010;13:781–5. doi: 10.1016/j.mib.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 108.Vogel J. A rough guide to the non-coding RNA world of Salmonella. Molecular microbiology. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- 109.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, et al. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic acids research. 2003;31:6435–43. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nature reviews. 2011;9:578–89. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wan F, Lenardo MJ. The nuclear signaling of NF-kappaB: current knowledge, new insights, and future perspectives. Cell research. 2010;20:24–33. doi: 10.1038/cr.2009.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang XS, Gong JN, Yu J, Wang F, Zhang XH, Yin XL, et al. MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood. 2012;119:4992–5004. doi: 10.1182/blood-2011-10-385716. [DOI] [PubMed] [Google Scholar]
- 113.Wassarman KM. 6S RNA: a regulator of transcription. Molecular microbiology. 2007;65:1425–31. doi: 10.1111/j.1365-2958.2007.05894.x. [DOI] [PubMed] [Google Scholar]
- 114.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–23. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 115.Wen J, Fozo EM. sRNA antitoxins: more than one way to repress a toxin. Toxins. 2014;6:2310–35. doi: 10.3390/toxins6082310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Westholm JO, Lai EC. Mirtrons: microRNA biogenesis via splicing. Biochimie. 2011;93:1897–904. doi: 10.1016/j.biochi.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xie YF, Shu R, Jiang SY, Liu DL, Zhang XL. Comparison of microRNA profiles of human periodontal diseased and healthy gingival tissues. International journal of oral science. 2011;3:125–34. doi: 10.4248/IJOS11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xue B, He L. An expanding universe of the non-coding genome in cancer biology. Carcinogenesis. 2014;35:1209–16. doi: 10.1093/carcin/bgu099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Molecular cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yarchuk O, Iost I, Dreyfus M. The relation between translation and mRNA degradation in the lacZ gene. Biochimie. 1991;73:1533–41. doi: 10.1016/0300-9084(91)90188-7. [DOI] [PubMed] [Google Scholar]
- 121.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–9. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 122.Yin G, Chen R, Alvero AB, Fu HH, Holmberg J, Glackin C, et al. TWISTing stemness, inflammation and proliferation of epithelial ovarian cancer cells through MIR199A2/214. Oncogene. 2010;29:3545–53. doi: 10.1038/onc.2010.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. Journal of molecular biology. 2013;425:3723–30. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhong S, Zhang S, Bair E, Nares S, Khan AA. Differential expression of microRNAs in normal and inflamed human pulps. Journal of endodontics. 2012;38:746–52. doi: 10.1016/j.joen.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 125.Zhou Q, Haupt S, Prots I, Thummler K, Kremmer E, Lipsky PE, et al. miR-142-3p is involved in CD25+ CD4 T cell proliferation by targeting the expression of glycoprotein A repetitions predominant. Journal of immunology. 2013;190:6579–88. doi: 10.4049/jimmunol.1202993. [DOI] [PubMed] [Google Scholar]
- 126.Zhu QY, Liu Q, Chen JX, Lan K, Ge BX. MicroRNA-101 targets MAPK phosphatase-1 to regulate the activation of MAPKs in macrophages. Journal of immunology. 2010;185:7435–42. doi: 10.4049/jimmunol.1000798. [DOI] [PubMed] [Google Scholar]