Abstract
Different sites within the oropharynx harbour unique microbial communities. Co-evolution of microbes and host has resulted in complex interkingdom circuitries. Metabolic signalling is crucial to these processes, and novel microbial communication factors are progressively being discovered. Resolving interkingdom networks will lead to better understanding of oral health or disease aetiology.
Keywords: Biofilms, Quorum sensing, Microbial communities, Candida albicans, Streptococcus, Porphyromonas
1. Introduction
Omes, beyond the originally coined genome, have become fashionable for depicting wholeness in the data-rich biological sciences: proteome, lipidome, interactome etc. Much has been researched and written about the oral microbiome. At least 2000 different microbial taxa have been detected in the human oral cavity and more than 350 genomes have been fully sequenced. Recent systematic biomolecular analyses of dental calculus from adult human skeletons c. 950–1200 CE shows that the oral cavity has long served as a reservoir for microbes implicated in oral and systemic diseases [1]. Despite changes in lifestyle, diet and oral hygiene over a millennium, DNA and proteins from Porphyromonas gingivalis, Tannerella forsythia and Treponema denticola, bacteria that are associated with periodontal disease, were abundant in ancient dental calculus samples. The presence of other potential pathogens such as Streptococcus mutans, Filifactor alocis, Olsenella uli, Streptococcus pneumoniae, Streptococcus pyogenes, and Neisseria meningitidis, shows that the oral cavity has long harboured bacteria associated with oral infections, bacteraemia and cardiovascular disease. The functional profiles of human proteins present within modern or ancient calculus are also highly similar [1].
Despite such commonalities, microbial diversity in communities colonizing tooth enamel, mucosal surfaces etc. is individual specific and site specific [2]. There is a conserved oral community in healthy mouths at the genus level. Abundant groups include Streptococcus, Granulicatella, Gemella, Rothia, Neisseria, Haemophilus, Lautrophia and Prevotella. On the other hand, there is evidence that a wide range of polymicrobial consortia can be associated with periodontal disease. Deep sequencing has confirmed decades of evidence from traditional laboratory culture studies that Gram-negative bacteria (e.g. Selenomonas, Prevotella, P. gingivalis, T. denticola, T. forsythia, Catonella, etc.) are enriched in periodontal disease samples [3].
Oligotyping provides a higher taxonomic resolution of communities in different habitats and across individuals [4]. Instead of simply recognizing the existence of a taxon, it is possible to assign sequence variants within a taxon with differential localization. Perhaps surprisingly, oligotypes characteristic of subgingival plaque tend to be detected also in tonsillar crypts, suggesting that the tonsils provide a habitat for oral anaerobes [4]. Making use of the invaluable Human Oral Microbiome Database (HOMD; www.homd.org), oligotypes were related to named species such that co-occurrence data were obtained at both the oligotype and species levels. Distinct oral site distributions of Streptococcus gordonii and Streptococcus salivarius were revealed, confirming that the latter organism is only found at mucosal sites, and not present in supra- or sub-gingival plaque. Conversely, Streptococcus infantis and Streptococcus mitis oligotypes were found in all oral samples taken from 9 different sites, including plaque [4]. Veillonellae were found at all sites, but could be divided into two oligotypes, differing by a single nucleotide, one oligotype being found in plaque and the other on mucosal surfaces.
Complementing these molecular-based detection methods, visualization of species within oral microbial communities can be performed by CLASI-FISH (combinatorial labelling and spectral imaging - fluorescence in situ hybridization) [5]. This can distinguish multiple differentially-labelled microbes in oral microbial communities with proximity analyses revealing co-occurrence. In dispersed dental plaque, Streptococcus, Veillonella, Prevotella and Actinomyces predominated, the two latter genera showing the most inter-species associations [5].
These various metagenomic and co-occurrence maps provide essential information about who goes where, but not much about what they get up to. From a functional viewpoint we would like to determine: how different species recognize each other and what mediates their physical interactions; the metabolic networks that are crucial to community stability; the ecological benefits for interspecies connections; molecular to cellular effects on the host; and how the host responds. The communities in the oral cavity are more biologically complex than current views might indicate. For example, fungi interact with bacteria, viruses that are exogenous or indigenous interact with bacteria and host, and microbes are preyed upon by protozoa. Different taxonomic kingdoms have co-evolved in generating the human oral ecosystem, and components of all kingdoms interact with the host, in sickness or in health. Studies of interkingdom communication are in their infancy, but it is quickly becoming evident that signals and responses between members of different kingdoms are intricately involved in shaping the oral microbiome.
2. Deconstructing the oral microbiome
Communication between bacteria is fundamental to social evolution theories for microbial communities, so-named sociomicrobiology [6]. Cooperation between cells will be favoured when the individuals have a shared selfish interest in doing so. Evolution of cooperation can occur when the waste product of one individual provides a nutrient for another. A well cited example of this in oral microbial communities is the physical interaction of Veillonella with Streptococcus, wherein lactic acid produced by the latter is a carbon source for the veillonellae [7]. Benefits can flow in both directions too, known as cross-feeding, and this can occur between different variants of the same species, or between different species. For example, cooperation between two early colonizers of dental plaque Streptococcus oralis and Actinomyces oris can be mutually beneficial [8].
These kinds of metabolic interactions are only just beginning to be dissected and they probably account, in large part, for the inability to cultivate in pure culture a significant proportion of the oral microbiota identified as a result of metagenomic analyses. Metabolic dependency is likely rife within the microbial communities, and new approaches to isolating oral microbiota components that depend on others for growth, or that are mutually dependent, involve utilizing specific enrichments (mini-trap), single cell cultivation and nutritionally deprived growth media [9]. Recently, these sorts of approaches have led to the cultivation of a TM7 phylum bacterium, which up until now has been recalcitrant to cultivation [10]. The bacteria are reported as small cocci (about 300 nm diam.) and are obligate ectosymbionts living in tight surface and metabolic associations with a strain of Actinomyces odontolyticus [10]. The TM7 genome is small (705 kb) and reveals complete deficiency in amino acid synthetic capacity. Transcriptional analyses suggest that signalling occurs between these two phyla, and that inflammatory cytokine tumour necrosis factor-alpha (TNF-α) production in macrophages, induced by A. odontolyticus, is repressed in the presence of TM7. It seems likely that other obligate epibiotic interactions, episymbiotic, and endosymbiotic associations between microorganisms will be discovered in the future.
3. Bacterial wires and mobiles
3.1. Cell-cell contact
Studies of microbial coaggregation between subgingival organisms have begun to identify the mechanistic basis of these interactions. For example, fusobacteria are thought to bridge the early and later colonizers within dental plaque [11]. Fusobacterium nucleatum expresses at least one adhesin that recognizes streptococci, and a galactose-specific lectin that interacts with P. gingivalis. These consortia are locked in metabolic communication that can be considered as signalling, because the organisms sense and respond to specific molecules. Contact-dependent signalling has been demonstrated in P. gingivalis following interaction of the short fimbrial adhesin (Mfa) with S. gordonii SspA/B adhesins. This initiates a signalling cascade in P. gingivalis that prepares the organisms for community biofilm living [12].
One of the best characterized examples of contact-dependent communication is bacterial conjugation mediated by sex pili assembled by type IV secretion systems. It has recently been demonstrated that type IV-like pili [13] are induced by S. pneumoniae undergoing transformation in response to the competence-stimulating peptide signal. The pili appear to be involved directly in extracellular DNA (eDNA) binding [14] and can be also critical for biofilm formation by oropharyngeal microorganisms [15]. The discovery that some kinds of pili, denoted nanowires, can propagate charge [16] suggests that there may be much more to learn about pilus function in oral cavity intermicrobial communication and host cell signalling. Of course bacteria-host cell contact is fundamental to adhesion, internalization and intracellular signalling processes that are dealt with in other papers as part of this special collection.
3.2. Cell-cell signalling
Through the exchange of small chemical signals, such as acyl homoserine lactones (AHLs) or oligopeptides, bacteria are able to monitor population density and regulate gene expression by the process of quorum sensing (QS). There has been little evidence for AHL-mediated signalling in oral microbial communities, although a Pseudomonas species producing AHL has recently been isolated from the tongue surface [17]. AHLs have been shown to be chemoattractants for neutrophils, alter barrier integrity of epithelial cells, and increase macrophage phagocytic activity [18]. However, another QS molecule named autoinducer-2 (AI-2) appears to be well utilized in oral microbial communities, and the luxS gene encoding the synthase enzyme for AI-2 has been widely detected [8]. Although QS evolved as a means for bacteria to coordinate behaviour, evidence is emerging that QS may provide a means for bacteria to directly interact with eukaryotes. AHL-based signalling molecules are able to modulate inflammatory responses and induce apoptosis, possibly through G-protein coupled receptors [19]. P. aeruginosa produces AHL that kills hyphal filaments produced by the fungus Candida albicans, but not the yeast forms [20], and secretes phenazines that inhibit fungal respiratory metabolism [21].
Bacterial cell wall peptidoglycan fragments, or muropeptides, have long been known to serve as signals between bacteria and eukaryotic organisms. Muropepetides, and N-acetyl-D-glucosamine derived from peptidoglycan, stimulate C. albicans to undergo yeast to hyphal filament transition [22]. Peptidoglycan is also a key stimulant of immune responses. Pneumococcal (S. pneumoniae) peptidoglycan triggers the production of interferon-γ (IFN-γ) and interleukin-17 (IL-17) from T helper cells, contributing to acquired immunity to pneumococcal infections [23]. Recruitment of macrophages required for pneumococcal clearance is enhanced by peptidoglycan sensing [24], while the resident microbiota generates peptidoglycan fragments that systemically prime the innate immune system [25]. NOD1 and NOD2 (Nucleotide-binding Oligomerization Domain proteins) are cytosolic pattern-recognition receptor proteins that respond to intracellular peptidoglycan fragments. NOD1 is expressed in a range of cell types, while NOD2 production is more restricted [26]. NOD1 detects D-glutamyl-meso-diaminopimelic acid (iE-DAP), a dipeptide found in Gram-negative bacteria and some Gram-positive bacteria. NOD2 detects muramyl dipeptide (MDP) that is always present in peptidoglycan [26]. Activation of NODs drives innate inflammatory responses through the nuclear-factor (NF)-κB and mitogen-activated protein kinase (MAPK) signalling pathways. This leads to expression of pro-inflammatory immune factors such as TNF-α, interleukin-6 (IL-6), CC-chemokine ligand 2 (CCL2), interleukin-8 (IL-8), and defensins. These help drive recruitment and priming of innate immune cells, including neutrophils and monocytes, in response to both bacterial and viral infections.
Oxidized fatty acids, or oxylipins, are signalling molecules that are transmitted and received by organisms from different kingdoms. Fungi found in the oral microbiome, such as C. albicans, Candida dubliniensis and Candida glabrata, can produce prostaglandin E2 (PGE2) from arachidonic acid supplied by the host. PGE2 modulates inflammation, pulmonary function, fertility and gastric mucosal integrity. C. albicans also produces eicosanoids, one of these being resolvin E1, which is anti-inflammatory and attenuates neutrophil migration [27]. Low levels of resolvin E1 produced by C. albicans could dampen the adaptive immune response and protect the fungus from immune recognition. Some of the signals covered in this section and their identities are summarized in Table 1.
Table 1.
Some of the small-molecule signals produced by oral microorganisms and received by human cells or vice versa.
| Microbial products influencing microbiome and host | Host products influencing microbiome | |||
|---|---|---|---|---|
| Compound | Example of producer |
Effects | Compound | Effects |
| Acyl-homoserine lactones (AHLs) | Pseudomonas | QS in bacteria, kill C. albicans hyphal filaments, neutrophil chemoattractants, stimulate phagocytes, actin remodelling in epithelial cells | Estradiol | Stimulates growth of C. albicans, impairs dendritic cell function and Th17 response |
| Muropeptides | S. pneumoniae | Promote C. albicans yeast to hypha transition, trigger production of IFN-γ, IL-17 | Catecholamines | Stimulate Actinomyces, Eikenella, Campylobacter, inhibit P. gingivalis |
| iE-DAP | P. gingivalis | Activates NOD, stimulates production of TNF-α, IL-6, CCL2, IL-8, and defensins (e.g. HBD) | Hydrogen peroxide | Induces oxidative stress in bacteria, triggers C. albcians filamentation, kills susceptible microorganisms |
| Farnesol | C. albicans | Inhibits C. albicans filamentation, upregulates epithelial TLR2, increases IL-6 and HBD production | Antimicrobial peptides | Various species-or strain-specific inhibitory effects on bacteria and fungi |
| Indole | Fusobacterium, T. denticola | Inhibits AHL-mediated QS, promotes apoptosis, down-regulates IL-8, upregulates Il-10 | ||
| Hydrogen peroxide | Streptococcus, Lactobacillus | Induces oxidative stress in bacteria and fungi, triggers C. albicans filamentation, kills susceptible microorganisms | ||
| Oxylipins | C. albicans | Attenuate neutrophil migration, affect mucosal integrity | ||
| Neuroactive compounds | Lactobacillus | Anti-inflammatory | ||
| Butyrate | P. gingivalis | Reactivation of HIV-1, apoptosis, increases HBD production, causes cell cycle arrest in fibroblasts | ||
4. Host emissions
Microbial endocrinology is a relatively new research area that focuses on the ability of microorganisms to sense host-associated chemicals such as hormones. Prokaryotic responsiveness to catecholamine hormones (e.g. noradrenaline, adrenaline, dopamine) is widespread, and there is evidence for enhancement of growth (and virulence) of bacteria colonizing the gastrointestinal tract, skin and oral cavity [28]. The fact that some oral bacteria implicated in periodontitis e.g. Eikenella, Campylobacter, are especially stimulated by catecholamines (stress hormones) [29] has been linked to the knowledge that stress is a known risk-factor for periodontal disease. The QseC sensor kinase is a bacterial receptor for adrenaline/noradrenaline in Escherichia coli and activates virulence genes in response to the adrenergic signals [30]. It is not clear at present if QseC orthologs found in other Gram-negative bacteria, e.g. Aggregatibacter actinomycetemcomitans, are generally responsive to catecholamine hormones or have different cognate signals.
Noncatecholamine mammalian hormones, such as estradiol, are found in saliva [31], and this hormone enhances C. albicans infectivity by promoting the yeast to hypha transition and growth [32]. Estradiol also impairs dendritic cell function, such that the cells are less efficient at up-regulating antigen-presenting pathways, interleukin-23 (IL-23), and Th17 immune response, thus rendering the host more susceptible to C. albicans infection [33].
5. Cool reception
Quorum sensing molecules produced by bacteria and fungi can directly modulate the behaviour of human cells, as mentioned briefly above [18]. AHLs have been shown to be taken up into epithelial cells where they affect the distribution of a GTPase-activating protein and phosphorylation of Rac1 and Cdc42, which are upstream effectors of actin remodelling [34]. However, there is a vast array of molecules produced by microorganisms that affect host cells and only a few pertinent to the oral microbiome will be considered here. Moreover the complexity of the signals produced will depend upon the complexity of the microbial community.
An interesting area of investigation is into the effects of neuroactive compounds, produced by the microbiota, which can influence components of the host nervous system and ultimately the brain. Production of γ-aminobutyric acid (GABA), the primary inhibitory neurotransmitter of the mammalian brain, by microorganisms has been known for decades, but this is only one of numerous neuroactive signals produced including dopamine, acetylcholine and agmatine [35]. The pathways used by the microbes to produce these substances are exactly the same as the host’s pathways. Neuroendocrine hormones secreted by the microbiota exert anti-inflammatory activities. Lactobacillus spp. can produce milligram quantities of GABA and this could be a mechanism for prophylaxis of inflammatory conditions by administration of probiotics. Moreover there is now evidence that bacteria secreting neuroendocrine hormones can influence host behaviour. Since common synthesis pathways exist, it is possible that the microorganisms utilize neurochemicals for intercellular communication, and that the brain can therefore influence the prevalence of certain microbial species [35].
Other microbial signals that are immunomodulatory include 2-amino acetophenone produced by P. aeruginosa [36], and indole, which is produced by many oral bacterial species including Fusobacterium and T. denticola. Indole elicits an epithelial cell response that strengthens host cell-barrier properties [37], down-regulates IL-8 and increases anti-inflammatory cytokine interleukin-10 (IL-10) production. Commensal oral bacteria F. nucleatum and S. gordonii perturb the gingival transcriptiome much less than does P. gingivalis [38], and it is suggested that indole acts as a beneficial signal and is not recognized as a pathogenic signal.
Farnesol is a QS molecule that regulates virulence and morphogenesis in C. albicans. Farnesol controls hyphal filament formation by blocking the degradation of Nrg1, the major repressor of hyphal development [39]. In addition, farnesol promotes epithelial cell defence against C. albicans by up-regulating Toll-like receptor 2 (TLR2) expression and increasing production of IL-6 and human β-defensin 2 [40]. These various examples of microbial production of signals that cool host immune responses relate to the development of an oral microbiome that in health lives in relative harmony with the host.
The transition of a relatively benign commensal oral cavity microbiota to a dysbiotic periodontal microbiota can result in deregulated inflammation of gingival soft tissues. This occurs through elevated neutrophil transmigration into the tissues, and activation of bone-resorbing osteoclasts through RANK ligand expression by CD4+ T cells. Recent studies in mice have suggested that P. gingivalis, even in low abundance, can trigger dysbiosis of the community structure leading to development of periodontal disease [41]. A current hypothesis is that local tissue damage, for example initiated by P. gingivalis, leads to accumulation of commensal bacteria that secrete NOD1-stimulating ligands e.g. iE-DAP (see above). This results in chronic inflammatory stimulation with NOD1-mediated neutrophil recruitment to induce bone loss [42]. To add further microbial complexity to the periodontal disease process, the presence of both fungi (e.g. C. albicans) and of protozoa (e.g. Entamoeba gingivalis) has been correlated with periodontitis [43, 44].
6. Going viral
It has been suggested that viruses play a role in various types of destructive periodontal disease. Human cytomegalovirus (HCMV), Epstein-Barr virus (EBV) and other herpesviruses have been closely associated with active periodontitis [45]. Synergism between specific periodontal pathogens (P. gingivalis, Prevotella, T. denticola, A. actinomycetemcomitans) and herpesviruses might therefore contribute to increased aggressiveness in disease pathogenesis. It is speculated that the HCMV latent genome is carried into the inflamed periodontium by infected macrophages and T cells. Subsequent CMV activation infects other cell types, triggers release of IL-1β and TNF-I, and results in increased susceptibility to tissue breakdown and loss of alveolar bone [46].
It is well known of course that immunocompromised subjects with human immunodeficiency virus type 1 (HIV-1) infection are more susceptible to a number of opportunistic microbial pathogens. However, it has not been so well appreciated that P. gingivalis can induce HIV-1 reactivation through production of butyric acid and thus aid progression of HIV-1 [47]. In addition, HIV-1 particles trapped on the P. gingivalis cell surface can become internalized when bacteria invade CD4− epithelial cells [48]. These various synergies between virus, bacterium and host are only just becoming understood, and show clearly that interkingdom interactions play major roles in shaping health or disease. Even the beneficial effects of vaccination may be subject to interkingdom influences, and unintentional consequences, for the live attenuated influenza virus vaccine can enhance oropharyngeal carriage of S. pneumoniae [49].
7. A mushrooming field
7.1. Fungi in the oral cavity
Fungi represent a small but significant component of the oral microbiome, and it is worth noting that many, if not most, microbiome studies have not included probes for fungi. Even some of the more recent metagenomics approaches do not incorporate fungal genome assemblies into data for periodontal disease microbiomes, despite evidence for Candida spp. being present in subgingival plaque. Fungi, especially Candida spp., are carried orally by everybody at one time or other in life, and approximately 40% of humans are continuously colonized. C. albicans now represents one of the most common microorganisms in hospital-acquired infections, with candidemia having a mortality rate of approximately 50%. Opportunisitc Candida infections occur when the host’s immune defences are compromised, from restricted salivary flow associated with ill-fitting dentures (denture stomatitis) to HIV-1 infection or immune suppression following surgery. Since most infections appear to arise from indigenous colonizing fungi, reduction in carriage levels might reduce incidence of disease.
7.2. Signalling in fungi
Similar to bacteria, fungi produce a range of QS molecules that provide a check on population density and control virulence factor expression. Farnesol (see above) is produced by growing populations of C. albicans and inhibits the formation of hyphae and biofilms [39]. The receptor for farnesol has not been identified, but the histidine kinase Chk1 and Ras-Cyr1 (adenylyl cyclase) pathway is activated and Tup1, transcriptional cofactor repressor of filament formation, is upregulated [50]. Farnesol also acts as an interspecies QS molecule with, for example, C. tropicalis, acts intergenerically with Aspergillus, and impacts the host (see above). Tyrosol, an aromatic alcohol produced by C. albicans, acts in opposition to farnesol by inducing hyphal filament formation. As described above, C. albicans produces oxylipins that act as QS molecules, increasing filamentation under some conditions. Fungi are also responsive to carbon dioxide (CO2) and the yeast to hyphae morphological transition associated with pathogenesis in C. albicans is triggered by elevated CO2 [51]. This is sensed by Cyr1, activating the catalytic domain, increasing cAMP synthesis, and activating protein kinase A (PKA) [52]. Cyr1 also senses pH and temperature, and is directly activated by bacterial peptidoglycan (MDP, see above) binding to the LRR (leucine-rich repeat) domain [52].
7.3. Signalling between fungi and bacteria
Fungal-bacterial interactions are implicated in promoting colonization, biofilm formation, and virulence. Cross-kingdom signalling seems to have become an intense area of research over recent years. A range of small molecules with signalling or QS functions, produced by fungi or bacteria, modulate the collective behaviour of mixed species communities. When species of oral streptococci form biofilms with C. albicans, both bacteria and fungi seem to benefit in that dual species biofilms are more luxurious in growth than the single species biofilm counterparts [53–56]. C. albicans is able to lower O2 tension, and may also provide growth stimulatory factors for S. gordonii. In turn, growth with S. gordonii enables C. albicans to persist under conditions <pH 4.5, and S. gordonii produces nutrient by-products that are stimulatory to C. albicans, enhancing the length of hyphal filaments [53]. A number of interactive signals are proposed for these phenomena. S. gordonii appears to block, at least in part, the inhibitory effects of farnesol [53]. There is also evidence that AI-2 [53], peptidoglycan fragments [22, 52] or hydrogen peroxide [57] produced by these catalase-negative bacteria could enhance filamentation. There may be additional growth factors or catabolites that are produced by the interkingdom partnership that promote synergy, such as enabling increased efficiency of energy production (new carbon sources or provision of amino acids).
In S. mutans dual species biofilms with C. albicans, the streptococcal QS system for development of competence for DNA uptake is turned on by C. albicans [55], as are at least two genes associated with virulence [56]. Co-infection resulted in enhanced development of carious lesions over and above those caused by S. mutans alone [56]. Paradoxically, the competence-stimulating peptides released by S. mutans [58] and by S. gordonii [59] both modulate biofilm formation by C. albicans, suggesting that C. albicans can sense these signals produced by streptococci just as the bacteria can sense farnesol, and perhaps other QS molecules produced by fungi. Trans-2-decenoic acid formed by S. mutans [60] is also inhibitory to hypha formation.
7.4. Mediators of fungi-bacteria communication
It has long been known that a variety of oral streptococci are able to coaggregate with C. albicans and that the interactions occur principally with hyphal filaments [53], although C. albicans cells primed for hypha formation are also bound by oral streptococci. Several cell-wall anchored proteins e.g. SspA, SspB and CshA, on the surface of S. gordonii, all appear to be involved in the binding of C. albicans by the bacteria. More specifically, the SspB polypeptide has been shown to interact with a hyphal cell wall protein designated Als3 [61]. This is a member of the Als protein family in C. albicans that comprises glycosylphosphatidylinositol (GPI)-modified proteins linked to the fungal cell wall. Als3 is the only member of the family to be expressed uniquely on hyphae, while Als1 is expressed at the site of initial hyphal filament extension [62]. The interaction of SspB protein on S. gordonii with the N-terminal region of Als3 [63] is thought to drive, at least in part, the formation of dual species biofilms of these organisms. Als3 and other cell wall proteins are glycosylated, and early stage O-mannosylation of these proteins is critical for activation of hyphal adhesin functions [64]. Als3 is also a receptor for Staphylococcus aureus binding to C. albicans [65], which facilitates staphylococcal invasion into host tissues leading to systemic infection.
7.5. Consequences of communication between fungi and bacteria
In the oral cavity, C. albicans has the potential to associate with hundreds of different species of bacteria, and so it is likely that there are novel physical and chemical interactions to be discovered. However, one consequence of the interactions of C. albicans with streptococci, in addition to providing metabolic synergy, might be that persistence of the fungus in the oral cavity is enhanced. The virulence of coaggregating bacteria or fungi may also be promoted. Colonization of the oral cavity by S. oralis in experimental animals was augmented in the presence of C. albicans [66]. Co-infection also resulted in enlarged oral thrush lesions, upregulated the mucosal inflammatory responses, and promoted deep organ dissemination of C. albicans. This is a form of pathogenic synergy, also exemplified in S. aureus co-infections with C. albicans [65], which can turn a non-lethal monomicrobial infection into an infection with high mortality rate. Clearly these kinds of co-infections have serious implications for clinical management of patients, so better understanding of the mechanisms of these interkingdom networks should impact on improving patient treatment outcomes.
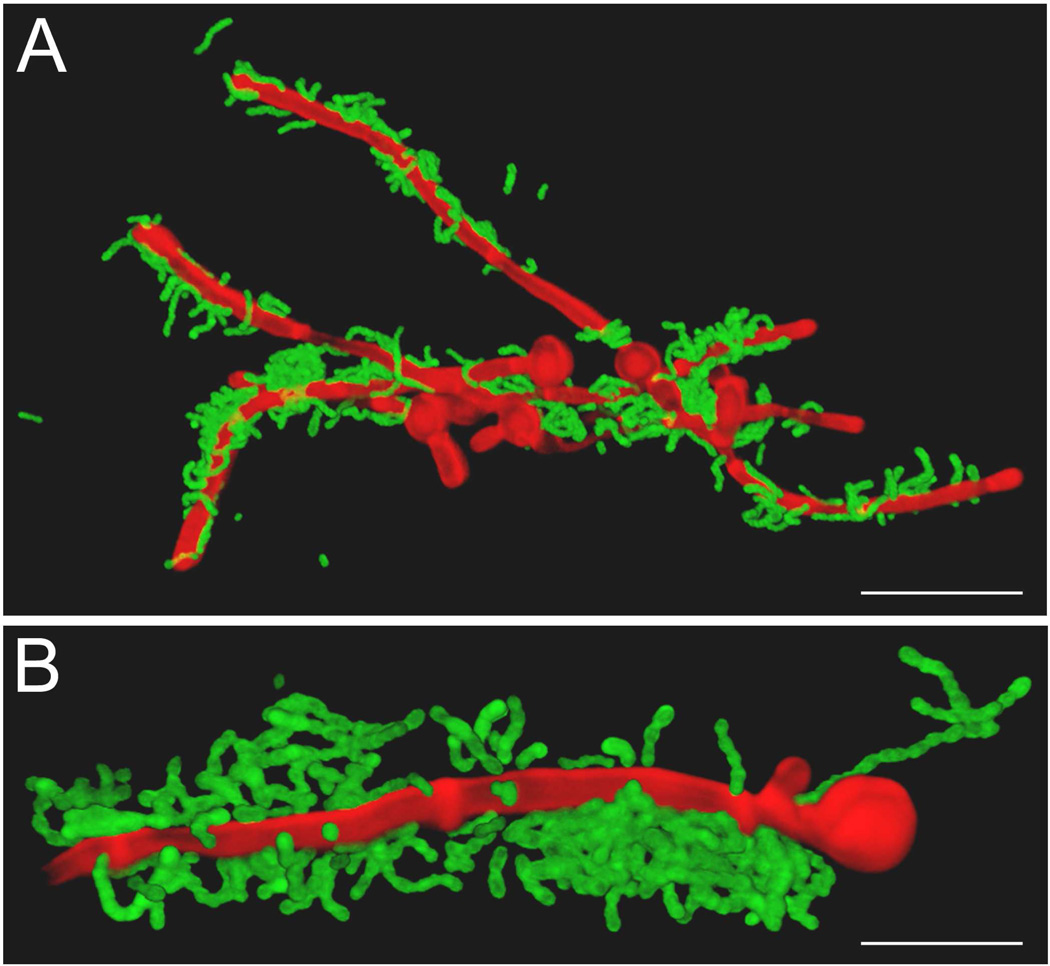
The relations between oral bacteria and C. albicans have been largely focused on extracellular associations (Fig. 1) in which streptococci adhere to hyphal filaments, sometimes in specific regions [65], and then form clusters or microsocieties surrounding or enveloping the hyphal filaments. This we believe represents an upregulation of streptococcal surface proteins as a result of adherence to the hyphae, or receiving a chemical signal from the fungi, and the intercellular aggregation of streptococci to form clumps (Fig. 1). In S. pneumoniae, development of competence is accompanied by clumping of cells, effected by the release of eDNA. A similar process may occur in the eDNA-mediated interactions of S. gordonii with C. albicans in biofilms [59].
Fig. 1.
Fluorescence confocal scanning laser micrographs of interactions of oral streptococci with Candida albicans. Panel A: Streptococcus gordonii DL1 cells interacting with C. albicans hyphae. Bacterial cells were labelled with fluorescein isothiocyanate (FITC) [64] and incubated with hypha-forming cells of C. albicans for 2 h at 37°C. C. albicans cells were stained with calcofluor white [64]. Note that streptococci are attached only to hyphae, they form micro-societies in some areas, and are able to cross-link the C. albicans hyphae. Panel B: Streptococcus oralis 34 cells interacting with C. albicans hyphae under similar conditions to panel A. Large aggregates of streptococci have formed around sites of contact with hyphae, suggesting a process of adherence followed by aggregation and accumulation. Note a chain of streptococci in intimate contact with the hyphal filament close to the mother cell.
Associations of Helicobacter pylori with C. albicans may be even more intimate. Candida spp. isolated from the oral cavity of dyspeptic patients were found to contain bacteria-like bodies inside their vacuoles. These contained H. pylori-specific 16S rRNA and H. pylori proteins such as urease, vacuolating toxin (VacA), periredoxin and thiolperoxidase [67]. Fluorescence microscopy revealed H. pylori cells inside the mildly-acidic vacuoles of mother and daughter cells, suggesting that the endobacteria could multiply and transmit. The accommodation of H. pylori in endosymbiosis with Candida that grow on the oral cavity mucosa could contribute to persistence of H. pylori in the gastrointestinal tract. It would be interesting to determine in future if members of the resident oral microbiota (e.g. streptococci) are also able to interact with C. albicans in this way. This would have implications for persistence of oral bacteria outside of the oral cavity environment, and could provide mechanisms for resisting effects of antibacterial agents or hiding from immune system components.
8. Messages for home
Interkingdom networking of free living microorganisms and the host in the oral ecosystem provides a fascinating view of how cell population regulatory molecules have been shared or adapted. Interactions between viruses, bacteria, fungi and the host that have been discussed in this article are summarized in Fig. 2. We are just at the tip of the iceberg in understanding the multitude of potential cross-kingdom interactions that go to determine health or disease for the host, and life or death for the microbes. The immense biological data set for the oral cavity ecosystem will provide new understanding of the ways in which human microbial communities develop, and will be mined for new molecules that might be utilized for influencing community development or host response. Many of these microorganisms may also have evolved unique biological factors significant for our wellbeing. Thus compounds biosynthesized by microbiota components may have potential therapeutic applications. An inhibitor of C. albicans biofilm formation, designated mutanobactin, has been isolated from S. mutans, and is roughly 300 times more potent than farnesol [68]. The human microbiome could also be a promising source of new antibiotics. The biosynthetic gene clusters for a class of antibiotics, thiopeptides, are widely distributed in metagenomes of the human microbiota. One of these, lactocillin, is produced by the vaginal microbiota and has potent activity against a range of Gram-positive vaginal pathogens [69]. It is evident then that interkingdom signalling molecules that are utilized by microbes to manipulate populations could also be used by us, the host, to manipulate our microbiota. New molecules will likely also be found that will impact directly on host cells, hopefully as a means of controlling unwanted inflammatory responses, apoptotic processes, or haemostasis.
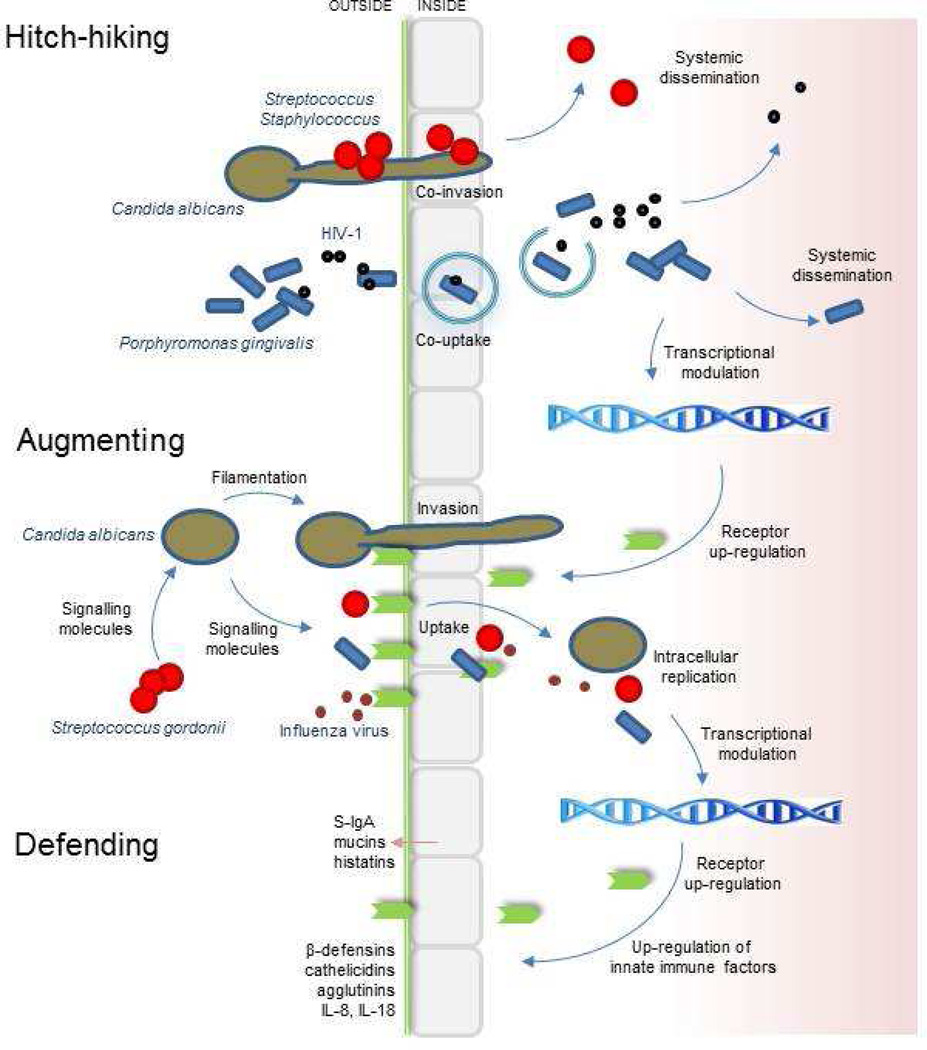
Fig. 2.
Summary of interkingdom interactions associated with the oral microbiome and epithelial barrier. Hitch-hiking involves the carriage of one organism by another across the epithelial cell barrier, as exemplified in the diagram by interactions of fungi-bacteria and bacteria-virus. Bacteria-bacteria hitch-hiking is also documented [70] but is not shown. Hitch-hiking extends the repertoire of microbe-host cell interactions. Internalization may lead to degradation through autophagy pathways, intracellular replication, uptake by macrophages and systemic spread. Upregulation of surface receptors (e.g. E-cadherin, proteoglycans, integrins) as a result of microbial adhesion or invasion promotes secondary interactions e.g. influenza-Streptococcus pneumoniae. Signalling molecules produced by bacteria (e.g. AHLs) are able to modulate C. albicans hyphal morphogenesis and epithelial cells, while signalling molecules produced by fungi (e.g. farnesol) may affect bacteria and host cells. All of these various interactions may augment the virulence and pathogenesis of the oral microbiome. Host epithelial cell small-molecule responses to microbes include upregulation of antimicrobial peptides (e.g. β-defensins) and cytokines (not specifically addressed in this review paper). Agglutinins (mucin-like proteins) such as gp340 are macromolecular defences secreted by epithelial cells. Other macromolecular defence factors (e.g. mucins) are secreted by specialized cells. The reader is directed to the many recent specialist reviews on antimicrobial peptides, mucins, and other elements of innate immunity for further information.
Acknowledgements
We would like to thank colleagues, including Rich Lamont, Aras Kadioglu and Mark Ramsdale for their insightful discussions, Lindsay Dutton for providing images, and NIH (NIDCR) for current research funding (R01-DE016690) related to work reviewed in this article.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors have no conflicts of interest.
References
- 1.Warinner C, Rodrigues JFM, Vyas R, Trachsel C, Shved N, Grossmann J, et al. Pathogens and host immunity in the ancient human oral cavity. Nature Genet. 2014;446:336–344. doi: 10.1038/ng.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One. 2012;7:e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eren AR, Borisy GG, Huse SM, Welch LJM. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA. 2014;111:E2875–E2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valm AM, Welch JLM, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, et al. Systems-level analysis of microbial community organization through combinatorial labelling and spectral imaging. Proc Natl Acad Sci USA. 2011;108:4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nature Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 7.Chalmers NI, Palmer RJ, Jr, Cisar JO, Kolenbrander PE. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J Bacteriol. 2008;190:145–154. doi: 10.1128/JB.00983-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakubovics NS. Talk of the town: interspecies communication in oral biofilms. Mol Oral Microbiol. 2010;25:4–14. doi: 10.1111/j.2041-1014.2009.00563.x. [DOI] [PubMed] [Google Scholar]
- 9.Sizova MV, Hohmann T, Hazen A, Paster BJ, Halem SR, Murphy CM, et al. New approaches for isolation of previously uncultivated oral bacteria. Appl Environ Microbiol. 2012;78:194–203. doi: 10.1128/AEM.06813-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu SY, et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci USA. 2015;112:244–249. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenkinson HF. Beyond the oral microbiome. Environ Microbiol. 2011;13:3077–3087. doi: 10.1111/j.1462-2920.2011.02573.x. [DOI] [PubMed] [Google Scholar]
- 12.Kuboniwa M, Lamont RJ. Subgingival biofilm formation. Periodontol 2000. 2010;52:38–52. doi: 10.1111/j.1600-0757.2009.00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melville S, Craig L. Type IV pili in Gram-positive bacteria. Microbiol Mol Biol Rev. 2013;77:323–341. doi: 10.1128/MMBR.00063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laurebceau R, Péhau-Arnaudet G, Baconnais S, Gault J, Malosse C, Dujeancourt A, et al. A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLoS Pathog. 2013;9:e1003473. doi: 10.1371/journal.ppat.1003473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konto-Ghiorghi Y, Mairey E, Mallet A, Duménil G, Caliot E, Trieu-Cuot P, et al. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 2009;5:e1000422. doi: 10.1371/journal.ppat.1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boesen T, Nielsen LP. Molecular dissection of bacterial nanowires. mBio. 2013;4:e00270–e00213. doi: 10.1128/mBio.00270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen JW, Chin S, Tee KK, Yin WF, Choo YM, Chan KG. N-acyl homoserine lactone-producing Pseudomonas putida strain T2-2 from human tongue surface. Sensors (Basel) 2013;13:13192–13203. doi: 10.3390/s131013192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rumbaugh KP, Kaufmann GF. Exploitation of host signaling pathways by microbial quorum sensing signals. Curr Opin Microbiol. 2012;15:162–168. doi: 10.1016/j.mib.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Shiner EK, Terentyev D, Bryan A, Sennoune S, Martinez-Zaguilan R, Li G, et al. Pseudomonas aeruginosa autoinducer modulates host cell responses through calcium signalling. Cell Microbiol. 2006;8:1601–1610. doi: 10.1111/j.1462-5822.2006.00734.x. [DOI] [PubMed] [Google Scholar]
- 20.Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004;54:1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 21.Morales DK, Grahl N, Okegbe C, Dietrich LE, Jacobs NJ, Hogan DA. Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. mBio. 2013;4:e00526–e00512. doi: 10.1128/mBio.00526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu XL, Lee RT, Fang HM, Wang YM, Li R, Zou H, et al. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe. 2008;4:28–39. doi: 10.1016/j.chom.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Olliver M, Hiew J, Mellroth P, Henriques-Normark B, Bergman P. Human monocytes promote Th1 and Th17 responses to Streptococcus pneumoniae. Infect Immun. 2011;79:4210–4217. doi: 10.1128/IAI.05286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis KM, Nakamura S, Weiser JN. Nod2 sensing of lysozyme-digested peptidoglycan promotes macrophage recruitment and clearance of S. pneumoniae colonization in mice. J Clin Invest. 2011;121:3666–3676. doi: 10.1172/JCI57761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vlarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol. 2014;14:9–23. doi: 10.1038/nri3565. [DOI] [PubMed] [Google Scholar]
- 27.Haas-Stapleton EJ, Lu Y, Hong S, Arita M, Favoreto S, Nigam S, et al. Candida albicans modulates host defense by biosynthesizing the pro-resolving mediator resolvin E1. PLoS One. 2007;2:e1316. doi: 10.1371/journal.pone.0001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freestone P. Communication between bacteria and their hosts. Scientifica. 2013;2013:361073. doi: 10.1155/2013/361073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts A, Matthews JB, Socransky SS, Freestone PP, Williams PH, Chapple IL. Stress and the periodontal diseases: growth responses of periodontal bacteria to Escherichia coli stress-associated autoinducer and exogenous Fe. Oral Microbiol Immunol. 2005;20:147–153. doi: 10.1111/j.1399-302X.2004.00196.x. [DOI] [PubMed] [Google Scholar]
- 30.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci USA. 2006;103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stefan-van Staden R-I, Gugoaşă LA, Calenic B, Legler J. Pattern recognition of estradiol, testosterone and dihydrotestosterone in children’s saliva samples using stochastic microsensors. Sci Rep. 2014;4:5579. doi: 10.1038/srep05579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Essmann M, Burt ET, Larsen B. Estrogen effects on Candida albicans: a potential virulence-regulating mechanism. J Infect Dis. 2000;181:1441–1446. doi: 10.1086/315406. [DOI] [PubMed] [Google Scholar]
- 33.Relloso M, Aragoneses-Fenoll L, Lasarte S, Bourgeois C, Romera G, Kuchler K, et al. Estradiol impairs the Th17 immune response against Candida albicans. J Leukoc Biol. 2012;91:159–165. doi: 10.1189/jlb.1110645. [DOI] [PubMed] [Google Scholar]
- 34.Karlsson T, Turkina MV, Yakymenko O, Magnusson K-E, Vikström E. The Pseudomonas aeruginosa N-acylhomoserine lactone quorum sensing molecules target IQGAP1 and modulate epithelial cell migration. PLoS Pathog. 2012;8:e1002953. doi: 10.1371/journal.ppat.1002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyte M. Microbial endocrinology: host-microbiota neuroendocrine interactions influencing brain behaviour. Gut Microbes. 2014;5:381–389. doi: 10.4161/gmic.28682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzika AA, Constantinou C, Bandyopadhaya A, Psychogios N, Lee S, Mindrinos M, et al. A small volatile bacterial molecule triggers mitochondrial dysfunction in murine skeletal muscle. PLoS One. 2013;8:e74528. doi: 10.1371/journal.pone.0074528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell-tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci USA. 2010;107:228–233. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasegawa Y, Mans JJ, Mao S, Lopez MC, Baker HV, Handfield M, et al. Gingival epithelial cell transcriptional responses to commensal and opportunistic oral microbial species. Infect Immun. 2007;75:2540–2547. doi: 10.1128/IAI.01957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Y, Su C, Unjoe O, Liu H. Quorum sensing controls hyphal initiation in Candida albicans through Ubr1-mediated protein degradation. Proc Natl Acad Sci USA. 2014;111:1975–1980. doi: 10.1073/pnas.1318690111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Decanis N, Savignac K, Rouabhia M. Farnesol promotes epithelial cell defense against Candida albicans through Toll-like receptor 2 expression, interleukin-6 human β-defensin 2 production. Cytokine. 2009;45:132–140. doi: 10.1016/j.cyto.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 41.Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2014;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiao Y, Darzi Y, Tawaratsumida K, Marchesan JT, Hasegawa M, Moon H, et al. Induction of bone loss by pathobiont-mediated Nod1 signaling in the oral cavity. Cell Host Microbe. 2013;13:595–601. doi: 10.1016/j.chom.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McManus BA, Maguire R, Cashin PJ, Claffey N, Flint S, Abdulrahim MH, et al. Enrichment of multilocus sequence typing clade 1 with oral Candida albicans isolates in patients with untreated periodontitis. J Clin Microbiol. 2012;50:3335–3344. doi: 10.1128/JCM.01532-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonner M, Amard V, Bar-Pinatel C, Charpentier F, Chatard J-M, Desmuyck Y, et al. Detection of the amoeba Entamoeba gingivalis in periodontal pockets. Parasite. 2014;21:30. doi: 10.1051/parasite/2014029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slots J. Herpesviral-bacterial synergy in the pathogenesis of human periodontitis. Curr Opin Infect Dis. 2007;20:278–283. doi: 10.1097/QCO.0b013e3280964da0. [DOI] [PubMed] [Google Scholar]
- 46.Contreras A, Botero JE, Slots J. Biology and pathogenesis of cytomegalovirus in periodontal disease. Periodontol 2000. 2014;64:40–56. doi: 10.1111/j.1600-0757.2012.00448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imai K, Victoriano AF, Ochiai K, Okamoto T. Microbial interaction of periodontopathic bacterium Porphyromonas gingivalis and HIV- possible causal link of periodontal disease to AIDS progression. Curr HIV Res. 2012;10:238–244. doi: 10.2174/157016212800618183. [DOI] [PubMed] [Google Scholar]
- 48.Mantri CK, Chen C, Dong X, Goodwin JS, Xie H. Porphyromonas gingivalis-mediated epithelial cell entry of HIV-1. J Dent Res. 2014;29:794–800. doi: 10.1177/0022034514537647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mina MJ, McCullers JA, Klugman KP. Live attenuated influenza vaccine enhances colonization of Streptococcus pneumoniae and Staphylococcus aureus in mice. mBio. 2014;5:e01040–e01013. doi: 10.1128/mBio.01040-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davis-Hanna A, Piispanen AE, Stateva LI, Hogan DA. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol Microbiol. 2008;67:47–62. doi: 10.1111/j.1365-2958.2007.06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hall RA, De Sordi L, MacCallum DM, Topal H, Eaton R, Bloor JW, et al. CO2 acts as a signalling molecule in populations of the fungal pathogen Candida albicans. PLoS Pathog. 2010;6:e1001193. doi: 10.1371/journal.ppat.1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L. Fungal adenylyl cylase acts as a signal sensor and integrator and plays a central role in interaction with bacteria. PLoS Pathog. 2013;9:e1003612. doi: 10.1371/journal.ppat.1003612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bamford CV, d’Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. 2009;77:3696–3704. doi: 10.1128/IAI.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diaz PI, Xie Z, Sobue T, Thohpson A, Biyikoglu B, Ricker A, et al. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun. 2012;80:620–632. doi: 10.1128/IAI.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sztajer H, Szafranski SP, Tomasch J, Reck M, Nimtz M, Rohde M, et al. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 2014;8:2256–2271. doi: 10.1038/ismej.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai CH, et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 2014;82:1968–1981. doi: 10.1128/IAI.00087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nasution O, Srinivasa K, Kim M, Kim YJ, Kim W, Jeong W, et al. Hydrogen peroxide induces hyphal differentiation in Candida albicans. Eukaryot Cell. 2008;7:2008–2011. doi: 10.1128/EC.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jarosz LM, Deng DM, van der Mei HC, Crielaard W, Krom BP. Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot Cell. 2009;8:1658–1664. doi: 10.1128/EC.00070-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jack AA, Daniels DE, Jepson MA, Vickerman MM, Lamont RJ, Jenkinson HF, et al. The Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology. 2015;161:411–421. doi: 10.1099/mic.0.000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vilchez R, Lemme A, Ballhausen B, Thiel V, Schulz S, Jansen R, et al. Streptococcus mutans inhibits Candida albicans hyphal formation by the fatty acid signalling molecule trans-2-decenoic acid (SDSF) Chembiochem. 2010;11:1552–1562. doi: 10.1002/cbic.201000086. [DOI] [PubMed] [Google Scholar]
- 61.Silverman RJ, Nobbs AH, Vickermann MM, Barbour ME, Jenkinson HF. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun. 2010;78:4644–4652. doi: 10.1128/IAI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coleman DA, Oh SH, Zhao X, Hoyer LL. Heterogeneous distribution of Candida albicans cell-surface antigens demonstrated with an Als1-specific monoclonal antibody. Microbiology. 2010;156:3645–3659. doi: 10.1099/mic.0.043851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bamford CV, Nobbs AH, Barbour ME, Lamont RJ, Jenkinson HF. Functional regions of Candida albicans hyphal cell wall protein Als3 that determine interaction with the oral bacterium Streptococcus gordonii. Microbiology. 2015;161:18–29. doi: 10.1099/mic.0.083378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dutton LC, Nobbs AH, Jepson K, Jepson MA, Vickerman MM, Aqeel Alawfi S, et al. O-mannosylation in Candida albicans enables development of interkingdom biofilm communities. mBio. 2014;5:e00911. doi: 10.1128/mBio.00911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schlecht LM, Peters BM, Krom BP, Freiberg JA, Hänsch GM, Filler SG, et al. Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiology. 2015;161:168–181. doi: 10.1099/mic.0.083485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu H, Sobue T, Thompson A, Xie Z, Poon K, Ricker A, et al. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 2014;16:214–231. doi: 10.1111/cmi.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siavoshi F, Saniee P. Vacuoles of Candida yeast as a specialized niche for Helicobacter pylori. World J Gastroenterol. 2014;20:5263–5273. doi: 10.3748/wjg.v20.i18.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X, Du L, You J, King JB, Cichewicz RH. Fungal biofilm inhibitors from a human oral microbiome-derived bacterium. Org Biomol Chem. 2012;20:2044–2050. doi: 10.1039/c2ob06856g. [DOI] [PubMed] [Google Scholar]
- 69.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edwards AM, Grossman TJ, Rudney JD. Fusobacterium nucleatum transports noninvasive Streptococus cristatus into human epithelial cells. Infect Immun. 2006;74:654–662. doi: 10.1128/IAI.74.1.654-662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]