Abstract
Apicomplexan parasites cause some of the most severe human diseases including malaria (caused by Plasmodium), toxoplasmosis, and cryptosporidiosis. Treatments are limited by lack of effective drugs and development of resistance to available agents. By exploiting novel features of protein kinases in these parasites, it may be possible to develop new treatments. We summarize here recent advances in identifying small molecule inhibitors against a novel family of plant-like, calcium-dependent kinases that are uniquely expanded in apicomplexan parasites. Analysis of the 3-D structure, activation mechanism, and sensitivity to small molecules have identified a number of attractive chemical scaffolds that are potent and selective inhibitors of these parasite kinases. Further optimization of these leads may yield promising new drugs for treatment of these parasitic infections.
Keywords: Serine - threonine protein kinases, gatekeeper, ATP-binding pocket, orthogonal inhibitors, chemotherapy, parasites
Targeting essential kinases in apicomplexan parasites
The phylum Apicomplexa contains several important human pathogens including Plasmodium spp., the causative agents of malaria 1, Cryptosporidium parvum (and C. hominis), an important cause of diarrheal disease in young children in Africa 2, and Toxoplasma gondii, and important opportunistic pathogen that causes disease in immunocompromised patients 3, and due to congenital infection 4. Globally, these parasites cause acute and chronic infections in many individuals and lead to severe morbidity and mortality. Treatments for such infections are limited by few drug choices, inability to completely cure chronic infections, and development of resistance. For example, in the case of C. parvum, the only approved treatment for adults Nitazoxanide is not effective in young children or immunocompromised patients 5. Although therapies for toxoplasmosis can control acute infection, they do not eradicate the chronic stages 6. Finally, drug resistance is an acute problem in P. falciparum: resistance to chloroquine and sulfa compounds is widespread in many areas of the world, and more recently, resistance is emerging to artemisinin, especially in South East Asia 7, 8. As such, there is a need to identify new targets that are essential and druggable and to develop new lead compounds if we are to ultimately achieve better therapeutic interventions.
Eukaryotic protein kinases control a number of essential pathways and they are expanded in many lineages, including humans that have more than 500 members 9. Although apicomplexan parasites lack some common kinases such as protein kinase C, they contain members of most major classes of protein kinases including AGC (named for protein kinase A, protein kinase G and protein kinase C), CMGC (named for cyclin dependent linases (CDK), MAP kinases (MAPK), glycogen synthase kinase 3 (GSK3) and cdc-like kinases (CLK)), calmodulin kinases (CaMK), and casein kinase 1 (CK1) groups, as well as tyrosine kinases-like (TKL) kinases 10, 11. However, apicomplexans lack conventional tyrosine kinases (TK) and have reduced or absent MAPK family members 10, 11. However, apicomplexans lack conventional TKs and have reduced or absent MAPK family members 10, 11. They also contain several expanded families, notably the FIKK kinases that are exported by P. falciparum into the infected red blood cell 12, 13 and the rhoptry (ROP) kinase family 14, 15, implicated in virulence of T. gondii 16. Genome-wide approaches to defining kinase function by gene disruption techniques have been reported for P. berghei 17 and in P. falciparum 18, thus defining a number of essential kinases.
Given the key role of kinases in controlling signaling and the frequency of activating mutations in cancer, more than 15 specific inhibitors that target several important human kinases have been approved for cancer chemotherapy 19. The conservation of kinase active sites makes it even more challenging to develop treatments for parasitic infection due to the need to identify unique structural or regulatory properties that can be selectively targeted by inhibitors that do not also inhibit host enzymes 20. Recent studies have identified a family of plant-like, calcium-dependent serine-threonine (S/T) protein kinases (CPDKs) that are conserved in apicomplexans 21. CDPKs fulfill the three central requirements for identifying new drug targets: essentiality, druggability, and unique structural or regulatory features. This review summarizes the CDPK family, highlights their biology and unique structural-regulatory features, and summarizes efforts to design selective inhibitors against these enzymes.
Biology of CDPKs
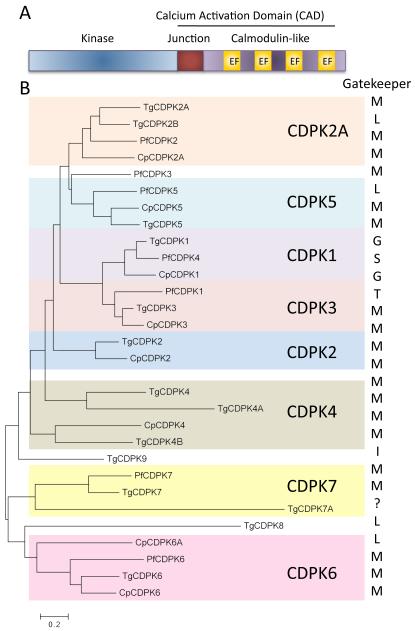
CDPKs are a unique family of S/T kinases found in protists, oomycetes, green algae, and higher plants, but lacking in animal cells 22. CDPKs typically consist of an N-terminal kinase domain, a junctional domain, and a series of calcium binding domains known as EF hands (Figure 1A). The combination of the junctional and calmodulin-like regions is referred to as the calcium activation domain (CAD) 23. It has been suggested that CDPKs arose by fusion of a CamK domain with calmodulin 24, 25, which subsequently diversified separately in protists 26 and plants 22. The best-studied parasite CPDKs contain this canonical domain structure (Figure 1A); however, some members contain N-terminal extensions (i.e. CDPK2, CDPK6, CDPK5), variable numbers of EF hands (i.e. CDPK4), EF hands located N-terminal to the kinase domain (i.e. CDPK6, CDPK7), or contain a PH domain (i.e. CDPK7) 21. Collectively, Plasmodium and Cryptosporidium contain 7 CDPKs while there are 14 genes in Toxoplasma (Figure 1B) 21, 27. Phylogenetic analysis groups these CDPKs into major clades many of which have orthologs across the three species, while others occur only in one group (i.e. TgCDK8, TgCDPK9, and PfCDPK3) (Figure 1B). Several CDPKs are modified by N-terminal myristoylation and/or palmitoylation, as a means of targeting them to membranes 21, and this location may influence substrate choice, given their similar preferred motifs for phosphorylation 28, 29. Genetic disruption of CDPKs has shown they control a wide range of phenotypes in T. gondii or Plasmodium spp. including egress (PfCDPK5 30, TgCDPK1 28 and TgCDPK3 29, 31, 32), microneme secretion (TgCDPK1 29, PfCDPK1 33), motility (TgCDPK1 28, PbCDPK3 34), development (PbCDPK1 35, PbCDPK4 36,), or cell division (TgCDPK7 37, PfCDPK7 38). Here we will focus on the enzymes that are either essential and/or that have been the target of efforts to identify small molecule inhibitors.
Figure 1.
CDPK domain structure and phylogeny. A) Typical domain structure of a canonical CDPK. EF hand named for the domain structure first identified in the calcium-binding domain of parvalbumin. B) Phylogenetic representation of the CDPKs in Plasmodium falciparum (Pf), Toxoplasma gondii (Tg) and Cryptosporidium parvum (Cp). Gatekeeper residues are indicated to the right in single letter amino acid code. The gatekeeper for TgCDPK7A could not be determined due to a Ser-rich insert in this portion of the protein. The numbering schemes of CDPKs among different parasites are also not identical, largely for historical reasons 21. Protein names from 21 with inclusion of two new genes from T. gondii: TgDPK4B (TgME49_292055) and TgCDPK7A (TgME49_237860) (http://ToxoDB.org). Tree was generated using MEGA (6.06 MAC) 96 using a Maximum Likelihood algorithm.
The essentiality of several CDPKs has been demonstrated using genetic approaches. For example, TgCDPK1 controls the release of micronemes, which are discharged at the anterior end of the cell to release adhesive proteins, and several of the downstream targets of this kinase have been indentified 39. Because micronemal proteins participate in cell motility, host cell invasion, and egress, TgCDPK1 is essential and conditional suppression results in significant impairment of growth 28. In contrast, the ortholog of TgCDPK1 in P. berghei (known as PbCDPK4) controls xanthurenic acid-induced calcium release and development of male gametes during sexual reproduction in the mosquito 36. In vitro biochemical studies reveal that the junctional domain that links the kinase domain to the C-terminal calmodulin domain regulates the activity of PfCDPK4 40, as previously suggested for plant CDPKs 24, 25. Given the large evolutionary distances among apicomplexans, which span ~ 400 mya 41, it is perhaps not surprising that CDPK orthologs do not necessarily perform conserved functions.
Not surprisingly given the conservation, some of the functions of CDPKs are partially overlapping. For example, the functions of TgCDPK3 partially mimic those of TgCDPK1 and they can also be compensated for by activation of protein kinase G (PKG), a distinct kinase that is also required for both egress and entry of T. gondii 29, 42. Recent studies implicate TgCDPK3 in control of calcium homeostasis, suggesting that it acts upstream of TgCDPK1 43. Consistent with this finding, over-expression of TgCDPK1 can partially rescue the egress phenotype of Δcdpk3 mutants 43. The degree to which substrates overlap vs. being specific to individual kinases is uncertain, but the simultaneous requirement for three kinases in stimulated egress of T. gondii (i.e. CDPK1, CDPK3, and PKG) attests to the importance of this process in the intracellular cycle of T. gondii. By contrast only CDPK5 has been shown essential for egress in P. falciparum, although PKG is also required for this process 30, 44. Recently it was suggested that PKG lies upstream, controlling production of phosphoinositides that activate calcium release from intracellular stores 45. Hence, cyclic nucleotide pools, PKG, and intracellular calcium signaling appear to be intricately linked in apicomplexans.
The ortholog of TgCDPK3 in Plasmodium is known as CDPK1, which is expressed in sexual stages, sporozoites, and during asexual replication in red blood cells where it is expressed late in the cycle during schizogony 46. PfCDPK1 has been shown to phosphorylate members of the motor complex involved in gliding motility (e.g. myosin light chain called MTIP and GAP45) in vitro 47. Although PfCDPK1 has long been considered a candidate for regulating this complex, confirmation that these modifications are important in vivo has been lacking 48. Moreover, recent studies that regulated the expression PfCDPK using a degradation fusion protein 49, or disrupted the gene for PbCDPK1 50, indicate it is not essential during asexual development. Nonetheless, phosphorylation of motor complex proteins is detected in vivo 18, 51, although the responsible kinases and essentiality of these changes remain uncertain. In contrast to its non-essential role in asexual stages, genetic studies have revealed a clear role for PbCDPK1 in activating translation of repressed mRNAs during sexual development in the mosquito 35.
Collectively, the above studies identify several CDPKs that are essential, during at least one stage in the life cycle, hence identifying potential drug targets. Before discussing the unique structural and regulatory features of CDPKs, we will briefly review what is known about protein kinases from mammalian systems where there is a wealth of knowledge on structure, function, and chemical inhibitors to draw on for comparison.
Structure-function and inhibition of eukaryotic protein kinases
Eukaryotic protein kinases have a conserved structure consisting of globular N- and C-lobes linked by a hinge region that collectively define 12 conserved subdomains 52. The N-lobe is dominated by a series of beta strands and a single alpha helical region αC that makes functional interactions with the C-lobe. The C-lobe is comprised of a series of alpha helical regions including the F helix that defines its hydrophobic core. The pocket between the N- and C-lobes is involved in binding ATP and together they coordinate transfer the γ phosphoryl group from ATP to the hydroxyl side chain on serine (S), theronine (T) or tyrosine (Y) residues in the target 52.
In the 25 years since PKA was first crystallized, X-ray crystal structures have been solved for a large number of kinases 53. Comparison of these structures reveals major conformational changes that accompany activation 54. In addition to their catalytic activity, kinases may impart signaling function by virtue of this bi-molecular switch 55. Structural studies have revealed the functions of key catalytic residues in the catalytic triad (Lys in domain II (β3 strand), Asp in the DFG loop, and Asp in the catalytic loop) as well as a Gly-rich loop, flanked by β1-β2 strands, which positions the γ-phosphate of the ATP. As part of the catalytic cycle, the kinase closes down on the bound nucleotide and repositions the DFG loop, so that the Asp residue coordinates a critical Mg2+ ion that is common to the active site of most protein kinases.
Although earlier studies emphasized differences in closed vs. open conformations, more recent studies have defined two hydrophobic spines, referred to as the C, or catalytic spine, and the R, or regulatory spine, in controlling activation 56, 57. Recognized not by sequence conservation but rather by their local spatial patterns of connectivity, they control conformational changes between inactive and active forms 56, 58, 59. This transition is associated with repositioning of the Phe residue of the DFG loop to complete the R spine, which is formed from residues of both the N- and C-lobes 56, 57. In contrast, the C spine does not undergo dramatic repositioning, but rather the adenine base of the ATP completes this hydrophobic structure. In active kinases where the R spine is complete, the position of the DFG motif is often described as DFG-in, while in inactive forms it is in a DFG-out conformation, breaking the hydrophobicity of the R spine 56, 57. This revised structural model explains many of the features of the activation mechanism of diverse kinases, including the allosteric interactions that allow pseudokinases to activate their partner enzymes 60.
Unique structural and regulatory features of CDPKs
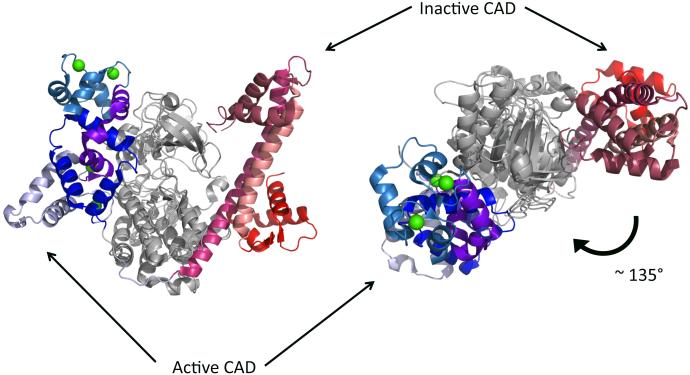
A number of parasite CDPKs have been crystallized either with calcium bound (e.g. PDB: 3HX4, 3IGO, 3LIJ) or without calcium (e.g. PDB: 3HZT, 3KU2, 4RGJ). The resulting crystallographic structures show what these protein kinases are auto-inhibited in the absence of calcium and they become activated in its presence. Super-imposition of the active and inactive structures of TgCDPK1 reveals how calcium-binding results in rearrangement and translocation the CAD to a different conformation and position (with respect to the kinase domain) (Figure 2). In particular, the top view shows the CAD is displaced by approximately 135° - perhaps largest movement of a regulatory domain ever observed in a kinase domain. When bound to calcium, TgCDPK1 has all the known features of an active kinase domain. As shown in Figure 3, this includes the positioning of the αC helix close to the ATP-binding pocket, allowing it to extend E99 to contribute to coordination of ATP. The activation loop is also well-structured and oriented so as to position the DFG triad to interact with ATP. Finally, the two hydrophobic spines are intact in the active structure, while the R spine is disoriented and the αC helix lies away from the ATP-binding pocket in the inactive structure (Figure 3).
Figure 2.
Activation of CDPKs results in a significant rearrangement of the CAD. Active (calcium bound) and inactive structures of TgCDPK1 are superposed on each other to illustrate the magnitude of the translation of the CAD. Side view on the left. Top view on the right. Green sphere, calcium ions.
Figure 3.
Structure of TgCDPK1 in different states (only kinase domain shown): (A) Calcium-bound TgCDPK1 (PDB: 3HX4; CAD hidden)contains all elements of an active kinase: αC helix (blue helix) is positioned close to the ATP pocket, extending E99 in that direction. Activation loop (blue strand) is well structured and oriented away from the ATP pocket so as to position DFG triad for interaction with ATP. The hydrophobic r-spine (yellow) is intact, as is the hydrophobic c-spine (green, two portions linked by the adenosine moiety of the AMPPNP in this structure). (B) Calcium-free TgCDPK1 shows elements of inactive kinase (PDB: 3KU2; CAD hidden): αC helix (red helix) is positioned farther from the ATP pocket. The hydrophobic r-spine is bent. The activation loop (red strand) is oriented toward the ATP pocket. (C) Binding 3-BrB-PP1 (PDB: 3MA6; only KD crystallized) stabilizes TgCDPK1 in the inactive conformation, similar to Fig. 3B
Inhibition of eukaryotic protein kinases
Because protein kinases undergo dramatic conformational reorganization, as well as being catalytically active, the precise architecture of these structural changes is important in the function of inhibitors. So called type I inhibitors bind competitively with ATP to the hinge region, displacing ATP, and preventing catalysis. In contrast, type II inhibitors such as imatinib (Gleevec), stabilize the DFG-out conformation 61. Many type I inhibitors, originally discovered by screening of small molecules, have also been converted to type II inhibitors through an understanding of their structural interactions with the ATP-binding pocket and medicinal chemistry 61. Notably, resistance to Type II inhibitors can be acquired by various mutations, in either the gatekeeper or activation domain that favor the DFG-in or active state 62. More recent efforts to develop type III (proximal to the nucleotide binding pocket) and type IV (targeting other allosteric sites) inhibitors offer greater potential for selectivity, with a trade off in potency 63. Although protein kinases within parasites often show conservation of structure and function, their evolutionary distance from humans 64 also means these enzymes are highly divergent, hence improving the chances for discovery of potent inhibitors that act orthogonally to their human counterparts 20.
The gatekeeper and analog sensitive kinase inhibitors
Lying between the C and R spines, the gatekeeper residue forms part of the ATP-binding pocket and the side chain at this residue can influence the activity of inhibitors. Most mammalian and yeast kinases have large hydrophobic gatekeeper residues 9, a feature that is important in stabilizing the C and R spines. However, it is often possible to mutate this reside to a smaller side chain without loosing catalytic activity, and this change has facilitated the development of analog-sensitive (AS) kinases 65, 66. AS kinases are sensitive to bulky inhibitors that mimic the structure of ATP but which are excluded by large gatekeeper kinases. The foundation for this approach was based on early work defining the sensitivity of c-Src (T-238 gatekeeper) to a compound called PP1 (containing a p-tolyl derivative at the C3 position of the scaffold pyrazolo [3,4-d] pyrimidine), which was shown to be a competitive inhibitor of ATP binding to kinases with small gatekeeper residues (i.e. T, V, A, G) 67, 68. Kinases with larger bulky gatekeeper residues can be made sensitive to PP1 by altering the gatekeeper to a smaller residue (typically A or G), while sensitive kinases can be rendered insensitive by substitution of larger gatekeeper residues 69. Further modification of PP1 to include larger C3 substituents (henceforth referred to as R1 to facilitate comparison to other scaffolds) results in greater discrimination between large and small gatekeeper containing kinases 70. Correspondingly, the modification of kinases to include a larger hydrophobic pocket formed by the small side chain of Gly (hydrogen) allow for selective inhibition of AS kinases using bulky ATP mimetics, thus revealing their function in complex biology of intact or semi permeable cells 71. A parallel strategy has also been developed for orthogonal labeling of substrates of AS kinases using bulky analogs of ATP-γ-S 72, 73. As described below, the natural occurrence of small gatekeepers within several parasite kinases has been exploited to take advantage of chemical scaffolds such as PP1 derivatives that are excluded by the normal bulky gatekeepers of mammalian kinases.
Progress in designing selective inhibitors of CDPKs
The fact that CDPKs are restricted to plants and apicomplexan parasites makes them potential targets of selective drug candidates. Although this premise is somewhat weakened by their similarities to human CaMKs in the ATP-binding pocket, a solution has been inspired by the observation that some CDPKs have Thr, Ser or other small residues in the gatekeeper position (Figure 1B). This feature is not conserved in human CaMKs, and is seen only in tyrosine kinases and MAPKs, both of which are absent in apicomplexan genomes. Moreover, several CDPKs are unique in having Gly in the gatekeeper, a residue not seen in any mammalian protein kinase 9. The subset of small gatekeeper-containing CDPKs includes PfCDPK1 (Thr), TgCDPK1 (Gly), CpCDPK1 (Gly) and PfCDPK4 (Ser) (Figure 1B). The first three have been the targets of a significant portion of medicinal chemistry efforts invested on parasite kinases 74-80.
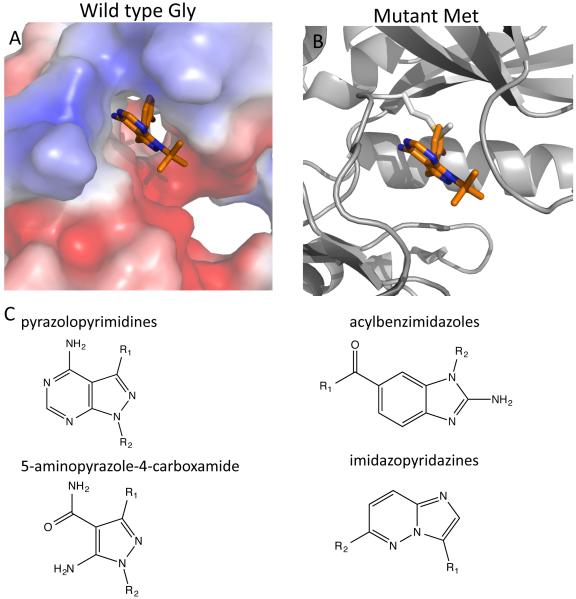
The presence of bulky substituents in the C3 position (i.e. R1) of the pyrazole in the pyrazolopyrimidine scaffold, such as that found in 3-methyl-benzyl (MB)-PP1 or 3-bromo-benzyl (BrB)-PP1, makes them potent inhibitors of TgCDPK1 and CpCDPK1 28, 79. This property was utilized to demonstrate the essentiality of CDPK1 in T. gondii to confirm the specificity of these compounds in vivo using a chemical genetics approach 28. A co-structure of TgCDPK1 with 3BrB-PP1 (Figure 4A) shows how the 3-bromo-benzyl moiety occupies the hydrophobic pocket created by the Gly gatekeeper (G128), while the 4-aminopyrimidine ring interacts with the hinge region in the ATP-binding pocket of the kinase. The same compound superposed on the structure of mutant TgCDPK1 with Met replacing G128 shows how it would clash with a gatekeeper residue with a longer side-chain (Figure 4B). Extension of this principle led to a series of compounds that extend the bulky substituent in the R1 position of PP1, including analogs that have naphthyl and larger heterocycle derivatives 81. Further modification of this scaffold by addition of cyclic groups at R2 also improved selectivity over the human kinase Src due to a favorable interaction with the ribose-binding pocket 82. For example, substituting 6-alkoxy-2-naphthyl at the R1 position and 4-piperidine methylene at the R2 position (a compound referred to as BKI-1), results in markedly improved selectivity over host Src 82. An alternative strategy extended the R1 region of the PP1 scaffold by addition of halogen containing benzyl moieties that are connected via a methylene linking rather than a direct aryl linkage 83. The added flexibility of this linkage may improve binding to the hinge region while allowing the bulky substiuent to occupy the hydrophobic pocket created by the small gatekeeper 83. These later compounds also show improved stability to microsome degradation, thus providing more favorable in vivo properties 83. Although early PP1 analogs such as 1-NM-PP1 provided little benefit for treatment of toxoplasmosis in mice 84, more potent derivatives such as 3-MB-PP1 or 3-ClB-PP1, or modifications that improve microsome stability, lead to greater efficacy in controlling both acute infection and reducing chronic cyst burdens in mice 83. Further modification of BKI-1 by addition of a N-methyl group to yield compound 1294 also showed potency against acute toxoplasmosis in the mouse model 85. Importantly, although the primary target of PP1 inhibitors in T. gondii is CDPK1 28, 29, studies using long-term treatment to generate resistance mutants also identified a MAPK homolog as a potential secondary target 86. Although such off-target activity may be desirable in reducing the risk of resistance developing, it also suggests that use of some PP1-like inhibitors may lead to activity against host kinases.
Figure 4.
Example of bulky PP1-like inhibitors interacting with the small gatekeeper in TgCDPK1. A) 3-BrB-PP1 interacting with TgCPDK1, which has a Gly gatekeeper (PDB: 3MA6) creating a deep hydrophobic pocket that the 3-bromo-benzyl ring points backward into. (B) Superposition of 3-BrB-PP1 from Fig. 4A on the structure of mutant TgCDPK1(G128M) shows steric clash with Met gatekeeper. (C) Compounds that inhibit CDPKs with small gatekeepers are derivatives of pyrazolopyrimidines, acylbenzimidazoles, 5-aminopyrazole-4-carboxamide and imidazopyridazines. The substituent mostly likely to occupy the hydrophobic pocket next to the small gatekeeper is denoted as R1 in each case.
TgCDPK1 and CpCDPK1 share 70% sequence identity in the kinase domain and consequently they have been found to have highly correlated chemical sensitivities 78. Compound 1294 was also reported to have activity against CpCDPK1 in vitro and to reduce the burden of oocyst shedding in immunodeficient SCID-beige mice 87. Neospora caninum, an apicomplexan parasite that causes abortion in cattle, is also susceptible to compound 1294 in a surrogate rodent model of infection 88. PP1 derivatives also have some activity against Plasmodium CDPKs that have small gatekeepers, although none have Gly at this residue. For example, 1294 and NA-PP1 also inhibit PfCDPK4 (Ser gatekeeper) in vitro and prevent male exflagellation of transgenic P. berghei parasites expressing PfCDPK4 89. Moreover, compound 1294 shows potency against PfCDPK1 in vitro 90 and blocks male gametocyte exflagellation in P. falciparum 91. Notably this later study examined the effects of 1294 on host enzymes and detected an undesirable hERG activity, which was abrogated by modification of the R2 group to a pyran 91. Collectively, these studies reveal that PP1 derivatives provide attractive leads for targeting several essential CDPKs in apicomplexan parasites.
Acylbenzimidazoles (Figure 4C) are also potent inhibitors of small gatekeeper kinases especially TgCDPK1 82. These compounds use the benzimidazole scaffold to interact with the hinge while extending a phenone group to occupy the hydrophobic pocket next to the Gly gatekeeper (e.g. PDB: 3UPX). Unlike pyrazolopyrimidines, these compounds do not inhibit tyrosine kinases such as Src and Abl 82, making them potentially more selective drug candidates. A third series of potent inhibitors of TgCDPK1 and CpCDPK1 is based on the 5-aminopyrazole-4-carboxamide scaffold 80 (Figure 4C). In these compounds, the carboxamide interacts with the hinge of the CDPKs while a bulky substituent in the 3-position of the aminopyrazole is ensconced in the cavity next to the Gly gatekeeper (e.g. PDB: 4M84).
A number of pyrazolopyrimidines, acylbenzimidazoles, and aminopyrazole carboxamides have been co-crystallized with either TgCDPK1 or CpCDPK1 (Table 1). Interestingly, in each case, interaction with the inhibitor stabilizes the protein in the DFG-out position. A perusal of the PDB suggests that this is also common of co-structures of human tyrosine kinases with gatekeeper-exploiting inhibitors. An illustrative example is shown in Figure 4C in the TgCDPK1 co-crystal structure with 3-BrB-PP. Whether this configuration is due to stabilization of the DFG-out conformation due to the way the compound interacts with the ATP binding pocket or simply a consequence of crystal packing is uncertain. The PP1 scaffold has been adapted to bind to Src in a type II DFG-out conformation 92, suggesting these modifications may be beneficial to screen against CDPKs.
Table 1.
Co-crystal structures of inhibitors with parasite CDPKs
| Protein | Pyrazolopyrimidines | Acylbenzimidazoles | Aminopyrazole carboxamides |
Others |
|---|---|---|---|---|
| TgCDPK1 | 3I7B, 3I7C, 3MA6, 3NCG, 3N51, 3SXF, 3T3U, 3T3V, 3UPZ, 3UQF, 3UQG, 3V51, 3V5P, 3V5T, 4JBV, 4TZR, 4WG3, 4WG4 |
3UPX | 4M84, 4ONA, 4WG5 |
3NYV, 4XLL |
| CpCDPK1 | 2WEI, 3MWU, 3NCG |
The search for antimalarial kinase inhibitors has focused particularly around PfCDPK1, largely because this kinase has long been considered essential 46, despite some uncertainty about its precise biological role in vivo. An in vitro enzyme activity screen against ~20,000 compounds resulted in identification of a series of trisubstituted purines that were potent in vitro inhibitors 93. One of the most potent of these called purfalcamine blocked late stage schizogony 93, although the specific target of this inhibitor in the parasite remains uncertain. Using similar in vitro enzyme assays for small molecule screens, a number of other chemical series have emerged as potential inhibitors of PfCDPK1, including pyrazolopyrimidines, azabenzimididazoles, and imidazopyridazines 74, 75, 77. The small Thr gatekeeper of PfCDPK1 was important in its susceptibility to these inhibitors in vitro; however, the potency of enzyme inhibition in vitro was not correlated with inhibition of parasite growth in red blood cells 74. Furthermore, the imidazopyridazine series was independently identified in a whole cell screen using a kinase inhibitor library, with specific analogs found to be effective in vivo 94. Synthesis of additional imidazopyridazine analogs was used to improve their ADME properties, for example by reducing Log P (Log D), enhancing microsome stability, and improving selectivity over human kinases 74-77. Despite these improvements, treatment of mice infected with P. berghei with some of the most potent derivatives only resulted in modest efficacy, despite achieving what should have been therapeutic serum levels 95. The failure to achieve better control may be explained by more recent data showing that PbCDPK1 is in fact not essential for asexual growth 33, but rather plays an important role in sexual development 35. Nonetheless, the potency of some of the imidazopyridazine inhibitors on asexual parasite growth of P. falciparum suggests that there is another essential target(s) in the parasite. Identifying the actual target of these inhibitors will be important to extend the SAR studies and thus improve potency and selectivity against what would appear to be an essential target in the parasite.
Concluding remarks
CDPKs have a number of features that make them attractive targets for development of new drugs to treat infections with apicomplexan parasites. CDPKs are found in plants and apicomplexan parasites, but not humans. CDPKs have a novel mechanism of activation not shared by other kinases. In some cases, CDPKs control essential features of the biology. CDPKs have a preponderance of small gatekeeper residues that makes them uniquely exploitable using bulky analogs of ATP mimetics as inhibitors. Remaining challenges are to optimize SAR to provide greater selectivity while retaining potency, improving bioavailability, and resisting metabolism and/or excretion to achieve levels sufficient for in vivo efficacy. An additional challenge is that the biological niche exploited by each of these three parasites differs widely from the CNS (toxoplasmosis), liver or blood (malaria), or the gut (cryptosporidiosis). Hence, designing drugs with optimal bioavailability and PK will require tailoring compounds for these distinct tissue environments. Although these are formidable challenges, they are worth undertaking given the limitations of existing treatments for these important human diseases.
Highlights.
Apicomplexan parasites contain a family of plant-like calcium-dependent serine / threonine protein kinases (CDPKs) that control a variety of essential functions.
CDPKs contain an unusual domain architecture and use a unique activation mechanism that is based on an auto-inhibitory calmodulin-like domain fused to the C-terminus of the kinase domain.
Several CDPKs have been validated as potential drug targets based on genetic experiments demonstrating that they are essential for parasite growth or survival.
Unique chemical scaffolds have been exploited to develop selective inhibitors of essential CDPKs in parasites offering promise as new therapeutic leads.
Acknowledgments
We are grateful to Oliver Billker, Sebastian Lourido, Amy Wernimont, Flora Rutaganira, and Kevan Shokat for helpful comments and Shaojun Long for assistance with curating the gene list for phylogenetic analysis. Supported in part by a grant from the NIH (AI094098).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miller LH, et al. Malaria biology and disease pathogenesis: insights for new treatments. Nat Med. 2013;19:156–167. doi: 10.1038/nm.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotloff KL, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 3.Saadatnia G, Golkar M. A review on human toxoplasmosis. Scand J Infect Dis. 2012;44:805–814. doi: 10.3109/00365548.2012.693197. [DOI] [PubMed] [Google Scholar]
- 4.Torgerson PR, Mastroiacovo P. The global burden of congenital toxoplasmosis: a systematic review. Bull World Health Organ. 2013;91:501–508. doi: 10.2471/BLT.12.111732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Checkley W, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis. 2014 doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCabe RE. Antitoxoplasma chemotherapy. In: Joynson DHM, Wreghitt TG, editors. Toxoplasmosis: a comprehensive clinical guide. Cambridge Univ. Press; 2001. pp. 319–359. [Google Scholar]
- 7.Gamo FJ. Antimalarial drug resistance: new treatments options for Plasmodium. Drug Discov Today Technol. 2014;11:81–88. doi: 10.1016/j.ddtec.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Sibley CH. Tracking artemisinin resistance in Plasmodium falciparum. Lancet Infect Dis. 2013;13:999–1000. doi: 10.1016/S1473-3099(13)70260-3. [DOI] [PubMed] [Google Scholar]
- 9.Manning G, et al. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 10.Miranda-Saavedra D, et al. The kinomes of apicomplexan parasites. Microbes Infect. 2012;14:796–810. doi: 10.1016/j.micinf.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Talevich E, et al. An evolutionary perspective on the kinome of malaria parasites. Philos Trans R Soc Lond B Biol Sci. 2012;367:2607–2618. doi: 10.1098/rstb.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nunes M, et al. A novel protein kinase family in Plasmodium falciparum is differentially transcribed and secreted to various cellular compartments of the host cell. Molec. Micro. 2006;63:391–403. doi: 10.1111/j.1365-2958.2006.05521.x. [DOI] [PubMed] [Google Scholar]
- 13.Schneider AG, Mercereau-Puijalon O. A new Apicomplexa-specific protein kinase family: multiple members in Plasmodium falciparum, all with an export signature. BMC Genomics. 2005;6:30. doi: 10.1186/1471-2164-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peixoto L, et al. Integrative genomics approaches highlight a family of parasite-specific kinases that regulate host responses. Cell Host Microbe. 2010;8:208–218. doi: 10.1016/j.chom.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talevich E, Kannan N. Structural and evolutionary adaptation of rhoptry kinases and pseudokinases, a family of coccidian virulence factors. BMC Evol Biol. 2013;13:117. doi: 10.1186/1471-2148-13-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter CA, Sibley LD. Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat Rev Microbiol. 2012;10:766–778. doi: 10.1038/nrmicro2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tewari R, et al. The systematic functional analysis of Plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe. 2010;8:377–387. doi: 10.1016/j.chom.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solyakov L, et al. Global kinomic and phospho-proteomic analyses of the human malaria parasite Plasmodium falciparum. Nat Commun. 2011;2:565. doi: 10.1038/ncomms1558. [DOI] [PubMed] [Google Scholar]
- 19.Bamborough P. System-based drug discovery within the human kinome. Expert Opin Drug Discov. 2012;7:1053–1070. doi: 10.1517/17460441.2012.724056. [DOI] [PubMed] [Google Scholar]
- 20.Lucet IS, et al. Plasmodium kinases as targets for new-generation antimalarials. Future Med Chem. 2012;4:2295–2310. doi: 10.4155/fmc.12.183. [DOI] [PubMed] [Google Scholar]
- 21.Billker O, et al. Calcium-dependent signaling and kinases in apicomplexan parasites. Cell Host Microbe. 2009;5:612–622. doi: 10.1016/j.chom.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valmonte GR, et al. Calcium-dependent protein kinases in plants: evolution, expression and function. Plant Cell Physiol. 2014;55:551–569. doi: 10.1093/pcp/pct200. [DOI] [PubMed] [Google Scholar]
- 23.Wernimont AK, et al. Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat. Struct. Molec. Biol. 2010;17:596–601. doi: 10.1038/nsmb.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harmon AC, et al. CDPKs - a kinase for every Ca2+ signal? Trends Plant Sci. 2000;5:154–159. doi: 10.1016/s1360-1385(00)01577-6. [DOI] [PubMed] [Google Scholar]
- 25.Harper JF, Harmon AC. Plants, symbiosis and parasites: a calcium signalling connection. Nat. Rev. Molec. Cell Biol. 2005;6:555–566. doi: 10.1038/nrm1679. [DOI] [PubMed] [Google Scholar]
- 26.Nagamune K, et al. Calcium regulation and signaling in apicomplexan parasites. Subcell. Biochem. 2008;47:70–81. doi: 10.1007/978-0-387-78267-6_5. [DOI] [PubMed] [Google Scholar]
- 27.Artz JD, et al. The Cryptosporidium parvum kinome. BMC Genomics. 2011;12:478. doi: 10.1186/1471-2164-12-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lourido S, et al. Calcium-dependent protein kinase 1 is an essential regulator of exocytosis in Toxoplasma. Nature. 2010;465:359–362. doi: 10.1038/nature09022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lourido S, et al. Distinct signalling pathways control Toxoplasma egress and host-cell invasion. EMBO J. 2012;31:4524–4534. doi: 10.1038/emboj.2012.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dvorin JD, et al. A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science. 2010;328:910–912. doi: 10.1126/science.1188191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garrison E, et al. A forward genetic screen reveals that calcium-dependent protein kinase 3 regulates egress in Toxoplasma. PLoS Pathog. 2012;8:e1003049. doi: 10.1371/journal.ppat.1003049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCoy JM, et al. TgCDPK3 regulates calcium-dependent egress of Toxoplasma gondii from host cells. PLoS Pathog. 2012;8:e1003066. doi: 10.1371/journal.ppat.1003066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bansal A, et al. Characterization of Plasmodium falciparum calcium-dependent protein kinase 1 (PfCDPK1) and its role in microneme secretion during erythrocyte invasion. J Biol Chem. 2013;288:1590–1602. doi: 10.1074/jbc.M112.411934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siden-Kiamos I, et al. Plasmodium berghei calcium-dependent protein kinase 3 is required for ookinate gliding and mosquito midgut invasion. Mol. Microbiol. 2006;60:1355–1363. doi: 10.1111/j.1365-2958.2006.05189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sebastian S, et al. A Plasmodium calcium-dependent protein kinase controls zygote development and transmission by translationally activating repressed mRNAs. Cell Host Microbe. 2012;12:9–19. doi: 10.1016/j.chom.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Billker O, et al. Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell. 2004;117:503–514. doi: 10.1016/s0092-8674(04)00449-0. [DOI] [PubMed] [Google Scholar]
- 37.Morlon-Guyot J, et al. The Toxoplasma gondii calcium-dependent protein kinase 7 is involved in early steps of parasite division and is crucial for parasite survival. Cell Microbiol. 2014;16:95–114. doi: 10.1111/cmi.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar P, et al. Regulation of Plasmodium falciparum development by Calcium-Dependent Protein Kinase 7 (PfCDPK7) J Biol Chem. 2014 doi: 10.1074/jbc.M114.561670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lourido S, et al. Exploiting the unique ATP-binding pocket of Toxoplasma calcium-dependent protein kinase 1 to identify its substrates. ACS Chem Biol. 2013 doi: 10.1021/cb400115y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranjan R, et al. Dissection of mechanisms involved in the regulation of Plasmodium falciparum calcium dependent protein kinase 4 (PfCDPK4) J. Biol. Chem. 2009 doi: 10.1074/jbc.M900656200. 10.1074/jbc.M900656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berney C, Pawlowski J. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc Biol Sci. 2006;273:1867–1872. doi: 10.1098/rspb.2006.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiersma HI, et al. A role for coccidian cGMP-dependent protein kinase in motility and invasion. Intl. J. Parasit. 2004;34:369–380. doi: 10.1016/j.ijpara.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 43.Treeck M, et al. The calcium-dependent protein kinase 3 of Toxoplasma influences basal calcium levels and functions beyond egress as revealed by quantitative phosphoproteome analysis. PLoS Pathog. 2014;10:e1004197. doi: 10.1371/journal.ppat.1004197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collins CR, et al. Malaria parasite cGMP-dependent protein kinase regulates blood stage merozoite secretory organelle discharge and egress. PLoS Pathog. 2013;9:e1003344. doi: 10.1371/journal.ppat.1003344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brochet M, et al. Phosphoinositide metabolism links cGMP-dependent protein kinase G to essential Ca2+ signals at key decision points in the life cycle of malaria parasites. PLoS Biol. 2014;12:e1001806. doi: 10.1371/journal.pbio.1001806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holder AA, et al. Calcium dependent protein kinase 1 and calcium fluxes in the malaria parasite. Microbes Infect. 2012;14:825–830. doi: 10.1016/j.micinf.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Green JL, et al. The motor complex of Plasmodium falciparum: phosphorylation by a calcium-dependent protein kinase. J. Biol. Chem. 2008;283:30980–30989. doi: 10.1074/jbc.M803129200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ridzuan MA, et al. Subcellular Location, Phosphorylation and Assembly into the Motor Complex of GAP45 during Plasmodium falciparum Schizont Development. PLoS One. 2012;7:e33845. doi: 10.1371/journal.pone.0033845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azevedo MF, et al. Inhibition of Plasmodium falciparum CDPK1 by conditional expression of its J-domain demonstrates a key role in schizont development. Biochem J. 2013;452:433–441. doi: 10.1042/BJ20130124. [DOI] [PubMed] [Google Scholar]
- 50.Jebiwott S, et al. Plasmodium berghei calcium dependent protein kinase 1 is not required for host cell invasion. PLoS One. 2013;8:e79171. doi: 10.1371/journal.pone.0079171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Treeck M, et al. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites' boundaries. Cell Host Microbe. 2011;10:410–419. doi: 10.1016/j.chom.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB Journal. 1995;9:576–596. [PubMed] [Google Scholar]
- 53.Eswaran J, Knapp S. Insights into protein kinase regulation and inhibition by large scale structural comparison. Biochim Biophys Acta. 2010;1804:429–432. doi: 10.1016/j.bbapap.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 55.Taylor SS, et al. Assembly of allosteric macromolecular switches: lessons from PKA. Nat Rev Mol Cell Biol. 2012;13:646–658. doi: 10.1038/nrm3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor SS, et al. Evolution of the eukaryotic protein kinases as dynamic molecular switches. Philos Trans R Soc Lond B Biol Sci. 2012;367:2517–2528. doi: 10.1098/rstb.2012.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kornev AP, et al. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci U S A. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kornev AP, et al. A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci U S A. 2008;105:14377–14382. doi: 10.1073/pnas.0807988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shaw AS, et al. Kinases and pseudokinases: lessons from RAF. Mol Cell Biol. 2014;34:1538–1546. doi: 10.1128/MCB.00057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat Chem Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- 62.Smith CC, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485:260–263. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lamba V, Ghosh I. New inhibitors in targetting protein kinases: focusing upon true allosteric and bivalent inhibitors. Curr. Pharm. Design. 2012;18:2936–2945. doi: 10.2174/138161212800672813. [DOI] [PubMed] [Google Scholar]
- 64.Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 65.Bishop AC, et al. Design of allele-specific inhibitors to probe protein kinase signaling. Curr Biol. 1998;8:257–266. doi: 10.1016/s0960-9822(98)70198-8. [DOI] [PubMed] [Google Scholar]
- 66.Bishop AC, Shokat KM. Acquisition of inhibitor-sensitive protein kinases through protein design. Pharmacol Ther. 1999;82:337–346. doi: 10.1016/s0163-7258(98)00060-6. [DOI] [PubMed] [Google Scholar]
- 67.Liu Y, et al. Structural basis for selective inhibition of Src family kinases by PP1. Chem Biol. 1999;6:671–678. doi: 10.1016/s1074-5521(99)80118-5. [DOI] [PubMed] [Google Scholar]
- 68.Liu Y, et al. A molecular gate which controls unnatural ATP analogue recognition by the tyrosine kinase v-Src. Bioorg Med Chem. 1998;6:1219–1226. doi: 10.1016/s0968-0896(98)00099-6. [DOI] [PubMed] [Google Scholar]
- 69.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 70.Zhang C, et al. Structure-guided inhibitor design expands the scope of analog-sensitive kinase technology. ACS Chem Biol. 2013;8:1931–1938. doi: 10.1021/cb400376p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lopez MS, et al. The logic and design of analog-sensitive kinases and their small molecule inhibitors. Methods Enzymol. 2014;548:189–213. doi: 10.1016/B978-0-12-397918-6.00008-2. [DOI] [PubMed] [Google Scholar]
- 72.Allen JJ, et al. Bio-orthogonal affinity purification of direct kinase substrates. J Am Chem Soc. 2005;127:5288–5289. doi: 10.1021/ja050727t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hertz NT, et al. Chemical genetic approach for kinase-substrate mapping by covalent capture of thiophosphopeptides and analysis by mass spectrometry. Curr Protoc Chem Biol. 2010;2:15–36. doi: 10.1002/9780470559277.ch090201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ansell KH, et al. Biochemical and antiparasitic properties of inhibitors of the Plasmodium falciparum calcium-dependent protein kinase PfCDPK1. Antimicrob Agents Chemother. 2014;58:6032–6043. doi: 10.1128/AAC.02959-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chapman TM, et al. Substituted imidazopyridazines are potent and selective inhibitors of Plasmodium falciparum calcium-dependent protein kinase 1 (PfCDPK1) Bioorg Med Chem Lett. 2013;23:3064–3069. doi: 10.1016/j.bmcl.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Large JM, et al. Imidazopyridazines as potent inhibitors of Plasmodium falciparum calcium-dependent protein kinase 1 (PfCDPK1): Preparation and evaluation of pyrazole linked analogues. Bioorg Med Chem Lett. 2013;23:6019–6024. doi: 10.1016/j.bmcl.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lemercier G, et al. Identification and characterization of novel small molecules as potent inhibitors of the plasmodial calcium-dependent protein kinase 1. Biochemistry. 2009;48:6379–6389. doi: 10.1021/bi9005122. [DOI] [PubMed] [Google Scholar]
- 78.Murphy RC, et al. Discovery of potent and selective inhibitors of calcium-dependent protein kinase 1 (CDPK1) from C. parvum and T. gondii. ACS Med Chem Lett. 2010;1:331–335. doi: 10.1021/ml100096t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ojo KK, et al. Toxoplasma gondii calcium-dependent protein kinase 1 is a target for selective kinase inhibitors. Nat. Struct. Molec. Biol. 2010;17:602–607. doi: 10.1038/nsmb.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Z, et al. Potent and selective inhibitors of CDPK1 from and based on a 5-aminopyrazole-4-carboxamide scaffold. ACS Med Chem Lett. 2014;5:40–44. doi: 10.1021/ml400315s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson SM, et al. Development of Toxoplasma gondii calcium-dependent protein kinase 1 (TgCDPK1) inhibitors with potent anti-toxoplasma activity. J Med Chem. 2012;55:2416–2426. doi: 10.1021/jm201713h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Larson ET, et al. Multiple determinants for selective inhibition of apicomplexan calcium-dependent protein kinase CDPK1. J Med Chem. 2012;55:2803–2810. doi: 10.1021/jm201725v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lourido S, et al. Optimizing small molecule inhibitors of calcium-dependent protein kinase 1 to prevent infection by Toxoplasma gondii. J Med Chem. 2013;56:3068–3077. doi: 10.1021/jm4001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sugi T, et al. 1-NM-PP1 treatment of mice infected with Toxoplasma gondii. J Vet Med Sci. 2011;73:1377–1379. doi: 10.1292/jvms.11-0085. [DOI] [PubMed] [Google Scholar]
- 85.Doggett JS, et al. Bumped kinase inhibitor 1294 treats established Toxoplasma gondii infection. Antimicrob Agents Chemother. 2014;58:3547–3549. doi: 10.1128/AAC.01823-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sugi T, et al. Identification of mutations in TgMAPK1 of Toxoplasma gondii conferring resistance to 1NM-PP1. Int J Parasitol Drugs Drug Resist. 2013;3:93–101. doi: 10.1016/j.ijpddr.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Castellanos-Gonzalez A, et al. A novel calcium-dependent protein kinase inhibitor as a lead compound for treating cryptosporidiosis. J Infect Dis. 2013;208:1342–1348. doi: 10.1093/infdis/jit327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ojo KK, et al. Neospora caninum calcium-dependent protein kinase 1 is an effective drug target for neosporosis therapy. PLoS One. 2014;9:e92929. doi: 10.1371/journal.pone.0092929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ojo KK, et al. Transmission of malaria to mosquitoes blocked by bumped kinase inhibitors. J Clin Invest. 2012;122:2301–2305. doi: 10.1172/JCI61822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vidadala RS, et al. Development of potent and selective Plasmodium falciparum calcium-dependent protein kinase 4 (PfCDPK4) inhibitors that block the transmission of malaria to mosquitoes. Eur J Med Chem. 2014;74:562–573. doi: 10.1016/j.ejmech.2013.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ojo KK, et al. A specific inhibitor of PfCDPK4 blocks malaria transmission: chemical-genetic validation. J Infect Dis. 2014;209:275–284. doi: 10.1093/infdis/jit522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dar AC, et al. Small molecule recognition of c-Src via the Imatinib-binding conformation. Chem Biol. 2008;15:1015–1022. doi: 10.1016/j.chembiol.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kato N, et al. Gene expression signatures and small-molecule compounds link a protein kinase to Plasmodium falciparum motility. Nat. Chem. Biol. 2008;4:347–356. doi: 10.1038/nchembio.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Le Manach C, et al. Medicinal chemistry optimization of antiplasmodial imidazopyridazine hits from high throughput screening of a SoftFocus kinase library: part 1. J Med Chem. 2014;57:2789–2798. doi: 10.1021/jm500098s. [DOI] [PubMed] [Google Scholar]
- 95.Chapman TM, et al. Optimization of an imidazopyridazine series of inhibitors of Plasmodium falciparum calcium-dependent protein kinase 1 (PfCDPK1) J Med Chem. 2014;57:3570–3587. doi: 10.1021/jm500342d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tamura K, et al. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]