Abstract
The highly conserved molecular chaperones Hsp90 and Hsp70 are indispensible for folding and maturation of a significant fraction of the proteome, including many proteins involved in signal transduction and stress response. To examine the dynamics of chaperone-client interactions after DNA damage, we applied quantitative affinity-purification mass spectrometry (AP-MS) proteomics to characterize interactomes of the yeast Hsp70 isoform Ssa1 and Hsp90 isoform Hsp82 before and after exposure to methyl methanesulfonate. Of 256 proteins identified and quantified via 16O/18O labeling and LC-MS/MS, 142 are novel Hsp70/90 interactors. Nearly all interactions remained unchanged or decreased after DNA damage, but 5 proteins increased interactions with Ssa1 and/or Hsp82, including the ribonucleotide reductase (RNR) subunit Rnr4. Inhibiting Hsp70 or 90 chaperone activity destabilized Rnr4 in yeast and its vertebrate homolog hRMM2 in breast cancer cells. In turn, pre-treatment of cancer cells with chaperone inhibitors sensitized cells to the RNR inhibitor gemcitabine, suggesting a novel chemotherapy strategy. All MS data have been deposited in the ProteomeXchange with identifier PXD001284.
1. Introduction
Damage to genomic DNA must be quickly repaired to maintain cell viability and allow cell proliferation. As such, the response to DNA damage is a tightly regulated process involving modulation and co-regulation of diverse pathways, including cell cycle progression, metabolism and DNA repair. Studies of the global response of cells to a variety of DNA damaging agents have revealed dramatic changes in post-translational modification, sub-cellular localization, expression and degradation of key effector proteins that play a critical role in the DNA damage response (DDR). Indeed, one such study in budding yeast observed 14% of proteins changed localization or abundance in response to DNA damage agents [1, 2]. These and other studies have established a paradigm where DNA damage induces rapid accumulation and modification of DDR proteins that are critical for checkpoint arrest and DNA repair, such as p53. These observations provide a rationale for targeting the abundance and/or modification of DDR effector proteins as a means to sensitize cancer cells to radiotherapy or genotoxic drugs.
Potentially lethal DNA damage can be induced by a wide range of external agents including ionizing radiation, UV radiation and radiomimetic agents such as the DNA alkylating agent methyl methanesulphonate (MMS). Although not a current chemotherapy drug, MMS is commonly used as an alternative to X-rays to induce experimental DNA damage in both mammalian and yeast cells. Like X-irradiation, MMS induces damage throughout the genome that requires both single strand and double strand break repair [3]. MMS treated cells typically display a prolonged S phase, reflecting activation of intra-S phase checkpoints.
Perturbation of DNA metabolism can arise through lack of sufficient deoxyribonucleotides (dNTPs), typically leading to stalled and collapsed replication forks and cell cycle delay in S Phase. dNTP synthesis is blocked upon inhibition of the key enzyme in dNTP formation, ribonucleotide reductase (RNR) [4]. RNR constitutes a complex of pairs of large (R1) and small (R2) subunits. R1 (RRM1 in vertebrates, Rnr1/Rnr3 in yeast) forms the catalytic domain while R2 (p53R2/RRM2 in vertebrates, Rnr2/Rnr4 in yeast) serves a regulatory role. Although the subunits are expressed at varying levels according to cell cycle stage, all are essential for cell viability [5, 6].
RNR is a well-validated therapeutic target [7, 8]. Since RNR function is required for DNA replication, loss of RNR activity slows proliferation, with eventual arrest in S phase. The first small-molecule RNR inhibitor, hydroxyurea (hydroxycarbamide, HU), was approved in 1967. HU and other agents, including the nucleoside analog gemcitabine (Gemzar), remain important agents in cancer chemotherapy. These agents are commonly combined with radiotherapy and/or genotoxic chemotherapy, which potentiate RNR inhibitors via exposing the requirement for dNTPs in DNA repair [4, 7]. It would be highly desirable to identify agents that can enhance the therapeutic benefit of RNR inhibitors without incurring additional toxicity.
The molecular chaperones Hsp90 and Hsp70 are essential for viability, and particularly important for responses to stresses such as heat shock, osmotic stress, oxidative stress and nutrient deprivation [9, 10]. Hsp90 and Hsp70 perform diverse functions including refolding denatured proteins, stabilizing protein-protein interactions, and mediating protein transport and degradation [11–13]. Consistent with their roles in stress tolerance, molecular chaperones have previously been linked to the DDR [14]. Via their role in stabilizing oncoproteins, cancer cells may become “addicted” to chaperones for proliferation [15, 16]. Given these considerations, chaperones have long been proposed as attractive targets for cancer drugs. Indeed, clinical studies have validated anti-cancer activity for geldanamycin-related Hsp90 inhibitors.
The budding yeast S. cerevisiae genome encodes four cytosolic Hsp70s, Ssa1–4, that differ in expression pattern but are together essential for cell viability [17, 18]. Yeast expresses two Hsp90 isoforms, with Hsc82 constitutively expressed and Hsp82 induced by stresses such as heat shock [13]. When yeast are treated with MMS to induce DNA damage, Ssa1 binds directly to the DDR mediator protein Rad9, presumably to facilitate Rad9 oligomerization and Rad9-dependent signal transduction of the DNA damage response signal [14, 19]. Consequently, deletion of Ssa1 and a second Hsp70, Ssa2 results in cellular sensitivity to DNA damage. In mammalian cells, Hsp90 is phosphorylated immediately after DNA damage and is recruited to foci containing DNA repair enzymes [20]. A recent study in mammalian cells revealed that inhibition of Hsp90 by the geldanamycin analog 17-DMAG triggered degradation of several proteins associated with DNA damage response [21].
Although several global proteomic and genetic screens for interactors (or “clients”) of Hsp70/90 have revealed a large number of clients in both yeast and mammalian cells [10, 22–25], heat shock proteins appear selective in the proteins they bind and fold. Thus, the interactions may be related to specific structural features of those proteins, rather than biological function [26]. Despite the prominent role of Hsp70/90 in multiple stress response pathways, no interactome studies exist that attempt to understand how global chaperone interactions change during those specific stress conditions.
Comprehensive global proteomic analysis of the effect of stress on cells remains technically challenging, costly and time consuming. It is thus easier to examine enriched or fractionated proteomes. This includes PTM enrichment such as phosphoprotein or phosphopeptide purification [27, 28]. Fractionation by cellular compartment is another way to simplify the proteome [29, 30]. There are many technical challenges involved with such methods. As such, we chose to sample a more manageable subset of the stress proteome, namely interactors of molecular chaperones Hsp70 and Hsp90. These proteins are a diverse, yet specific set of proteins required for cell survival under a variety of conditions [13, 16, 31]. Isolation of novel chaperone interacting proteins may allow us to understand how chaperones function to fold, stabilize and even degrade proteins under stress conditions.
Many clients of Hsp70 and Hsp90 can be destabilized by inhibition of chaperone function, often through binding of small molecules [10, 32, 33]. If these clients are current or potential cancer drug targets, then inhibitors of chaperones may sensitize cancer cells to inhibitors of these clients.
In this study, we have used comparative mass spectrometry to analyze how yeast Hsp90 (Hsc82) and Hsp70 (Ssa1) interactomes change upon treatment with the DNA damaging agent methyl methanesulfonate. In doing so, we have uncovered a role for chaperones in the stability of the ribonucleotide reductase subunit Rnr4/hRRM2. We exploit this chaperone dependency by demonstrating that cancer cells may be sensitized to RNR inhibitors such as gemcitabine by pretreatment of cells with chaperone inhibitors.
2. Materials and Methods
2.1. Yeast culture, strains and drug treatment
Yeast cultures were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) or in SD (0.67% yeast nitrogen base, 2% glucose) supplemented with the appropriate nutrients to select for plasmids and gene replacements. A full table of yeast strains and plasmids used can be found in Supplemental Table S1.
17-N-allylamino-17-demethoxygeldanamycin (17-AAG), VER-155008 and gemcitabine hydrochloride were purchased from Tocris Bioscience. Yeast transformed with a plasmid expressing HA-tagged Rnr4 were grown to mid-log phase. At this point, cells were treated with either DMSO (control), 17-AAG at a concentration of 1 or 10 µM or VER-155008 at 0, 10, 100 µM for 4 h (∼ 2 yeast cell divisions). Protein was extracted, run on SDS-PAGE gels and Western blotted with antibodies to either HA (Covance) or tubulin (Millipore).
2.2. Cancer cell tissue culture and drug treatment
The MCF-7 human breast cancer cell line was obtained from ATCC and were cultured in high-glucose Dulbecco’s Modified Eagle Medium (Invitrogen) with 10% fetal bovine serum (FBS, Clontech), 100 units/ml penicillin, and 100 µg/ml streptomycin at 5% CO2 and 37° C.
For experiments delineating the effect of chaperones on mammalian RNR stability, MCF-7 cells were plated in growth media at 40% confluency 1 day prior to drug treatment. Cells were treated with either DMSO (control), 17-AAG at a concentration of 1 or 10 µM or VER-155008 at 10 or 100 µM for 48 h (∼ 2 cell divisions). Protein was extracted, run on SDS-PAGE gels and Western blotted with antibodies to either hRRM2 or tubulin (Santa Cruz). For drug synergy assays, MCF-7 cells were plated in growth media at 20% confluency 1 day prior to initiation of drug treatment. On Day 1 of treatment, cells were treated with DMSO (control), 17-AAG at 3 µM or VER-155008 at 5 µM. After 24 h, cells were either further treated with DMSO (control) or gemcitabine (10–40 nM). At 24 h, cell viability was assessed by Trypan Blue staining. The data shown are the mean and SEM of three independent biological replicates. Statistical significance was determined by t-test and data were deemed significantly different where P < 0.05.
2.3. Purification of the Ssa1 and Hsp82 interactomes from yeast
100 ml of SKY4364 [24] were grown to an OD600 of 0.5 in YPD media. Cells were split into two flasks, one untreated and one which was subjected to 0.02% MMS for 3 h. Cells were harvested and HIS6-tagged Ssa1 along with the associated interactome was isolated as follows: Protein was extracted via bead beating in 500 µl Binding/Wash Buffer (50 mM Na-phosphate pH 8.0, 300 mM NaCl, 0.01% Tween-20). 200 µg of protein extract was incubated with 50 µl His-Tag Dynabeads (Invitrogen) at 4° C for 15 min. Dynabeads were collected by magnet then washed 5 times with 500 µl Binding/Wash buffer. After final wash, buffer was aspirated and beads were incubated with 100 µl Elution buffer (300 mM imidazole, 50 mM Na-phosphate pH 8.0, 300 mM NaCl, 0.01% Tween-20) for 20 min, then beads were collected via magnet. The supernatant containing purified HIS6-Ssa1 was transferred to a fresh tube, 25 µl of 5× SDS-PAGE sample buffer was added and the sample was denatured for 5 min at 95° C. 10 µl of sample was analyzed by SDS-PAGE. To isolate HIS6-tagged Hsp82, SKY4635 expressing HIS6-Hsp82 as the sole Hsp90 isoform in the cell were grown and processed identically to the SKY4364 cells as above.
2.4. LC-MS/MS data acquisition
2.4.1. Trypsin digestion of samples from SDS-PAGE Gels
Gel lanes to be analyzed were excised from 4–12% MOPS buffer SDS-PAGE gels by sterile razor blade and divided into 8 sections with the following molecular weight ranges (kD): 300-150, 150-110, 110-80, 80-75, 75-60, 60-52, 52-38 and 38-24. These were then chopped into ∼1 mm3 pieces. Each section was washed in dH2O and destained using 100 mM NH4HCO3 pH 7.5 in 50% acetonitrile. A reduction step was performed by addition of 100 µl 50 mM NH4HCO3 pH 7.5 and 10 µl of 10 mM tris(2-carboxyethyl)phosphine HCl at 37°C for 30 min. The proteins were alkylated by adding 100 µl of 50 mM iodoacetamide and allowed to react in the dark at 20°C for 30 min. Gel sections were washed in water, then acetonitrile, and vacuum dried. Trypsin digestion was carried out overnight at 37° C with 1:50 enzyme-protein ratio of sequencing grade-modified trypsin (Promega) in 50 mM NH4HCO3 pH 7.5, and 20 mM CaCl2. Peptides were extracted with 5% formic acid and vacuum dried.
2.4.2. Isotopic labeling
Peptide digests were reconstituted with 60 µl of Tris-HCl Buffer Solution (10 mM of Tris-HCl, 150 mM NaCl, 20 mM CaCl2, pH 7.6), then split into two vials with 30 µl each (16O vial and 18O vial) and vacuum dried. In a separate vial, 30 µl of Mag-Trypsin beads (Clontech) was washed 5 times with 500 µl of Tris-HCl Buffer Solution, then vacuum dried. 30 µl of either 16O H2O or 97% 18O H2O (Cambridge Isotopes) was added to the respective 16O or 18O vials and vortexed for 20 min to reconstitute the peptide mixture, which was then added to the prepared Mag-Trypsin bead vial and allowed to exchange overnight at 37° C. After 18O exchange, the solution was removed and any free trypsin in solution was inactivated with 1 mM PMSF for 30 min at 4° C. For each sample the +/− MMS digests were combined 1:1 as follows: Forward (FWD) Sample Set: (−MMS) 16O : (+MMS) 18O and Reversed (REV) Sample Set: (+MMS) 16O : (−MMS) 18O, dried and stored at −80° C until analysis. Three biological replicate experiments were performed per sample.
2.4.3. HPLC for mass spectrometry
All samples were re-suspended in Burdick & Jackson HPLC-grade water containing 0.2% formic acid (Fluka), 0.1% TFA (Pierce), and 0.002% Zwittergent 3–16 (Calbiochem), a sulfobetaine detergent that contributes the following distinct peaks at the end of chromatograms: MH+ at 392, and in-source dimer [2M+H+] at 783, and some minor impurities of Zwittergent 3–12 seen as MH+ at 336. The peptide samples were loaded to a 0.25 µl C8 OptiPak trapping cartridge custom-packed with Michrom Magic (Optimize Technologies) C8, washed, then switched in-line with a 20 cm by 75 µm C18 packed spray tip nano column packed with Michrom Magic C18AQ, for a 2-step gradient. Mobile phase A was water/acetonitrile/formic acid (98/2/0.2) and mobile phase B was acetonitrile/isopropanol/water/formic acid (80/10/10/0.2). Using a flow rate of 350 nl/min, a 90 min, 2-step LC gradient was run from 5% B to 50% B in 60 min, followed by 50%–95% B over the next 10 min, hold 10 min at 95% B, back to starting conditions and re-equilibrated.
2.4.4. LC-MS/MS Analysis
The samples were analyzed via electrospray tandem mass spectrometry (LC-MS/MS) on a Thermo LTQ Orbitrap XL, using a 60,000 RP survey scan, m/z 375–1950, with lockmasses, followed by 10 LTQ CAD scans on doubly and triply charged-only precursors between 375 Da and 1500 Da. Ions selected for MS/MS were placed on an exclusion list for 60 sec.
2.5. LC-MS/MS data analysis, statistical analysis and visualization
Data were analyzed and filtered on MaxQuant version 1.2.2 searching the SPROT Yeast database using 20 ppm error tolerance, an FDR setting of 1% and a cutoff of at least 2 high confidence peptides to assign a quantitation ratio. Proteins were removed from the file if they were flagged by MaxQuant as “Contaminants”, “Reverse” or “Only identified by site”. Complete MaxQuant settings used are reported in Supplemental Document S1. Three biological replicates were performed, with each biological replicate split into technical replicates (18O forward (FWD) labeling and 18O reverse (REV) labeling). The abundance data from each biological replicate were normalized to the ratio of the bait protein in that run (e.g. normalized to the + MMS:- MMS His6-SSA1 ratio or His6-Hsp82 ratio). This produced a list of interactors and their respective quantitated changes upon DNA damage.
Statistical analysis was performed using the R statistical package (http://www.r-project.org/). Proteins with three out of six observations within each group (SSA1 and HSP82) were retained. Missing values were imputed using row mean imputation. Z-score normalization was performed on the log of all protein ratios. An ANOVA test was then performed to identify proteins that indicate significant variability (P-value < 0.05) between biological replicates within each group. These were removed from consideration. A list of proteins identified and corresponding ratios can be found in Supplemental Table S2. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [34] with the dataset identifier PXD001284.
Gene Ontology analysis was performed using GO Slim Mapper on the Saccharomyces Genome Database (http://www.yeastgenome.org/cgi-bin/GO/goSlimMapper.pl). Network analysis was performed in Cytoscape. Nodes were colored using the MultiColoredNodes plug-in for Cytoscape [35].
3. Results
3.1. Quantitative affinity-purification mass spectrometry analysis of yeast Hsp70 and Hsp90 interactomes during the DNA damage response
Characterizing the interactome of a target protein by affinity-purification mass spectrometry (AP-MS, [36–39]) offers a powerful approach to understanding its role in the cell. Further information comes from analysis of dynamic interactomes, as proteins may change their interactions in response to cellular events such as cell cycle progression and signal transduction. We employed quantitative AP-MS to assess the dynamics of the yeast Hsp70 (Ssa1) and Hsp90 (Hsp82) interactomes upon treatment with the DNA damaging agent methyl methanesulfonate (MMS). Thus, we pulled down His-epitope-tagged Ssa1 and Hsp82 along with any associated proteins before and after MMS treatment, and performed quantitative LC-MS/MS proteomics using 18O exchange-labeling ([40, 41], Fig. 1A). For identification of Ssa1-specific interactors, we used SKY4364, a yeast strain in which all four SSA (Hsp70) genes have been deleted and functionally complemented by expression of an N terminal His6-tagged Ssa1 [24, 42]. For identification of Hsp82-specific interactors, we used SKY4365, a yeast strain that expresses a functional N terminally HIS6-tagged Hsp82 as the sole Hsp90 in the cell [43]. This approach facilitated identification of the interactomes of Ssa1 and Hsp82 without competition from other Hsp70 or Hsp90 isoforms.
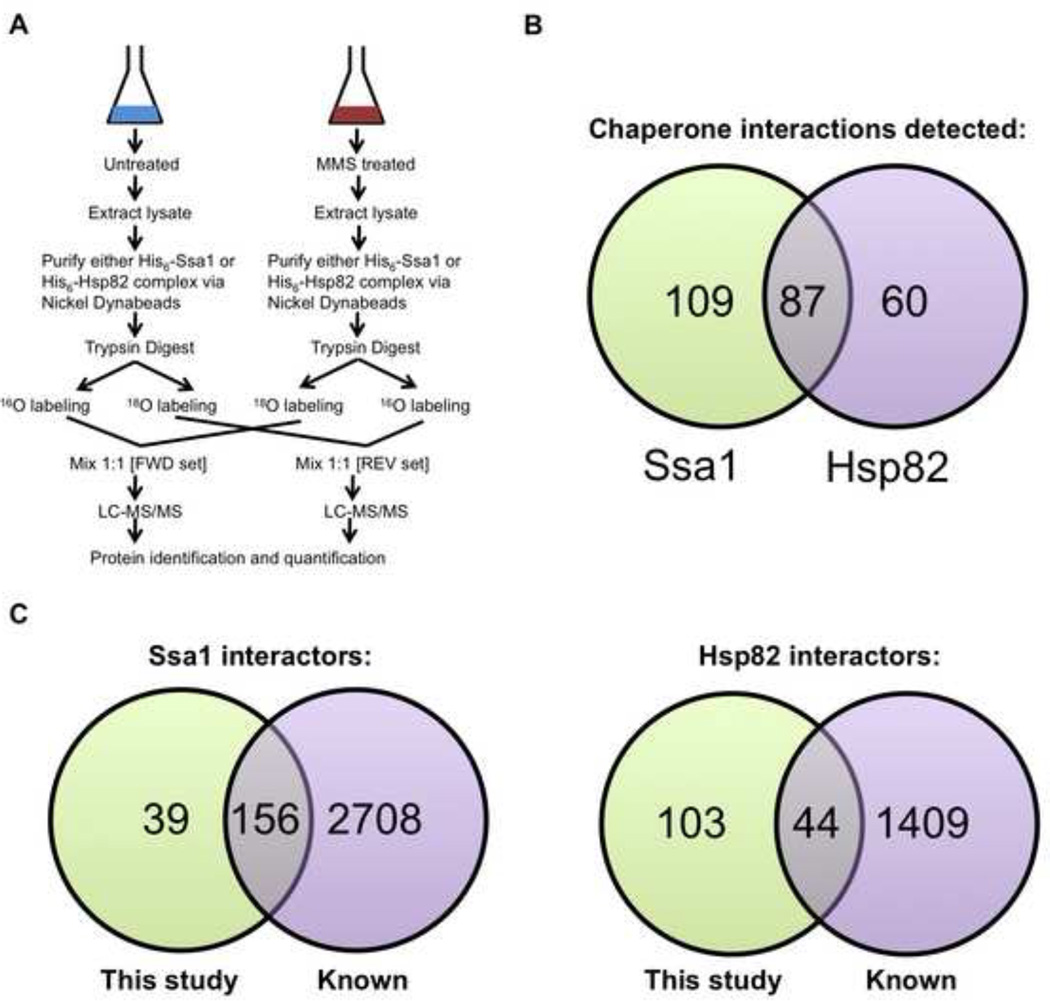
Figure 1. Quantitative affinity-purification mass spectrometry strategy for analysis of dynamic chaperone interactomes during DNA damage response.
(A) Growing yeast cells expressing His6-tagged Ssa1 or Hsp82 were left untreated or exposed to 0.02% methyl methanesulfonate (MMS) for 3 h. Chaperone interactomes were isolated by nickel-NTA magnetic bead affinity purification. Following PAGE and in-gel trypsin digestion and peptides were 18O or 16O labeled by trypsin-mediated exchange. Then, the heavy and light samples were combined and analyzed by Orbitrap LC-MS/MS and MassQuant informatic analysis, allowing identification of interacting proteins and determination of their relative enrichment after DNA damage. Each experiment was performed in biological triplicate from which each pair of samples was analyzed as technical replicates by forward and reverse 18O labeling.
(B) Venn diagram of candidate yeast Ssa1 and Hsp82 interactors remaining after applying statistical filters.
(C) Comparison of candidate yeast Ssa1 and Hsp82 interactors detected in our study vs. previously identified interactors in public databases.
Yeast cells were grown to mid-log phase and then either treated with 0.02% MMS for 3 h or left untreated. These conditions for MMS treatment were chosen because of their previous use in several proteomic studies examining the global response of cells to DNA damage [1, 2]. This provided us with a way to compare and contrast our data with those already obtained. HIS6-Ssa1 or -Hsp82 interactomes were isolated from yeast cell lysates and then subjected to SDS-PAGE and in-gel proteolysis. Peptides were extracted and isotopically labeled using 18O exchange with 97% 18O H2O or incubated in 16O H2O. For each sample the +/− MMS digests were combined 1:1 in both forward (FWD): (−MMS) 16O: (+MMS) 18O and reverse (REV): (+MMS) 16O : (−MMS) 18O. For each interactome, three affinity purifications were performed (biological replicates) and the peptides labeled with 16O and 18O and combined in both forward and reverse (technical replicates), resulting in six samples (Fig. 1A). The samples were analyzed by electrospray tandem mass spectrometry (LC-MS/MS) on a Thermo LTQ Orbitrap XL. Using Mascot analysis [44], we identified 663 proteins present in the Ssa1 complex and 434 in the Hsp82 complex.
After rigorous filtering and ANOVA testing, 196 proteins for Ssa1 and 147 proteins for Hsp82 remained (Supplemental Table S2). Of these interactions, 87 proteins were found to bind both Ssa1 and Hsp82 (Fig. 1B). After the stringent statistical filtering imposed on our data sets, the final list of quantitated chaperone interactors represented a fraction of known interactors. Comparing our results to published Ssa1 and Hsp82 interactomes [22, 23, 25, 26, 45, 46], we found that 80% of Ssa1 interactors seen in our screen had previously been reported as Ssa1 clients (Fig. 1C). Interestingly, only 30% of Hsp82 interactors we isolated had previously been identified as Hsp82 clients (Fig. 1D).
3.2. Gene Ontology (GO) analysis of Ssa1/Hsp82 interactomes
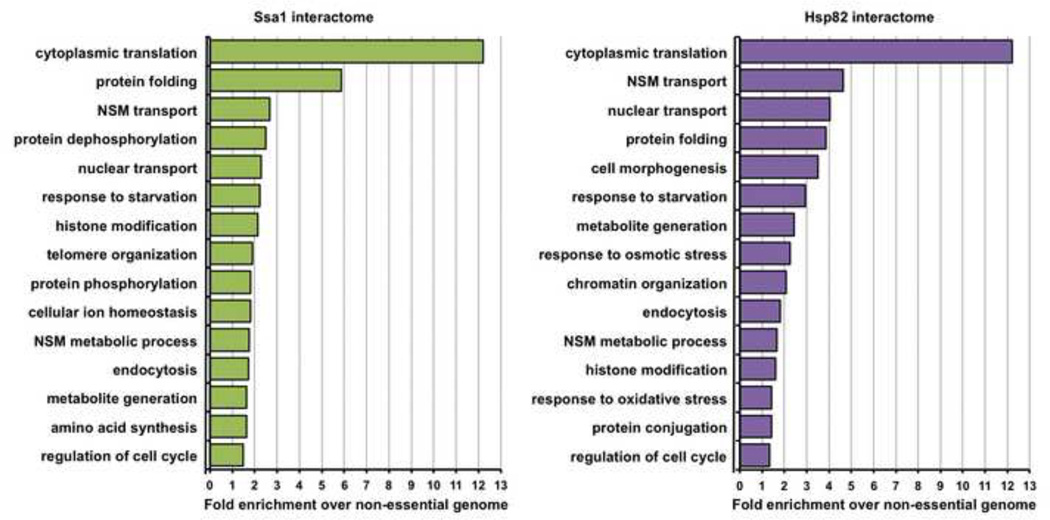
Gene Ontology (GO) analysis of the 256 candidate Ssa1/Hsp82 partners revealed significant enrichment of multiple cellular functions (Fig. 2). For both Ssa1 and Hsp82, the GO term cytoplasmic translation was most enriched, representing the important role these chaperones have in folding newly synthesized proteins [47, 48]. 29 out of 76 (38%) ribosomal 60S subunit proteins were identified as chaperone interactors, along with 25 out of 56 (52%) ribosomal 40S subunit proteins. In addition, a large number of ribosome biosynthesis, transcriptional elongation and initiation proteins were detected as both interactors of Hsp70 and Hsp90.
Figure 2. Gene Ontology (GO) term analysis of Ssa1/Hsp82 interactors.
Functional classification of the Ssa1/Hsp82 interactome. Interactors were categorized by cellular function using GO Slim analysis and relative enrichment compared to occurrence in the non-essential genome was calculated. The top 15 enriched cellular processes are shown for the Ssa1 and Hsp82 interactomes.
As expected, the term protein folding was also highly enriched, reflecting the wide range of chaperones and co-chaperones that bind to both Ssa1 and Hsp82 (Fig. 2). These chaperones are known to interact with each other in both yeast and mammalian cells. Indeed, in our study, Hsp82 was identified as in the interactome of Ssa1. Conversely, Ssa1 was identified in the interactome of Hsp82. Several Hsp70 isoforms were detected along with their respective co-chaperones, including the cytosolically-localized Ssa2 and Sse1, the ribosome-associated Ssb1 and the mitochondrial-associated Ssc1.
Consistent with the role of chaperones in cellular adaptation to nutrient availability [49], yeast proteins associated with glucose metabolism (including glyceraldehyde-3-phosphate dehydrogenases Tdh1, 2 and 3, UDP-glucose phosphorylases Udp1 and Ugp1) and amino acid metabolism (such as Leu4, Ilv6, Lys20 and Met17) were identified. We also observed chaperone interaction with both Nma1 and Pnc1, part of the nicotinamide salvage pathway, which is connected to both metabolism and the DNA damage response (see below and [50]).
Given our use of an alkylating agent such as MMS, it was not surprising that the chaperone interactomes contained proteins associated with the GO terms nucleobase-containing compound transport and modification. These included Mhr1, involved in homologous recombination in mitochondria, the Hos3 and Sin3 histone deacetylases (HDACs), Rvb2 and Abf1 chromatin remodeling factors and Xrn1 exonuclease. Proteins responsible for DNA base synthesis (critical for repair of damaged DNA) such as Rnr2, Rnr4 and Amd1 were isolated as novel chaperone interactors.
3.4. Analysis of the dynamic Ssa1/Hsp82 interactomes
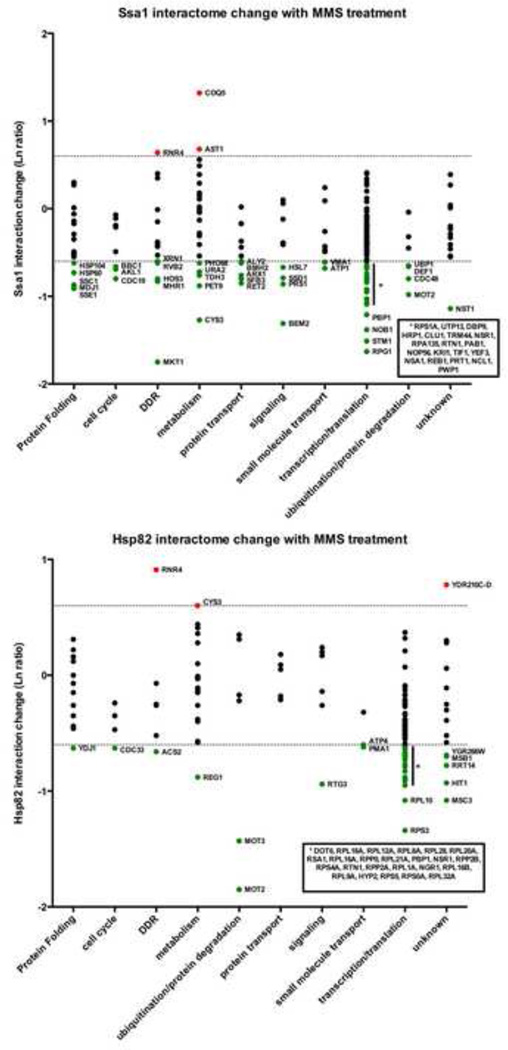
Interactome analysis for proteins with a large number of protein partners can prove challenging. To represent the dynamics of chaperone interactions, the average natural log (Ln) ratio change in interaction upon MMS treatment for each quantitated interacting protein was calculated. Any change in interaction greater than two-fold up or down (>0.69, <-0.69) was considered significant. We sorted our interactomes by non-redundant GO terms and graphed their average change in interaction with either Ssa1 or Hsp82 upon MMS treatment (Fig. 3). For Ssa1 clients, 135 of 196 (69%) interactions remained unchanged with 58 (29.5%) decreasing and 3 (1.5%) increasing, respectively, after MMS treatment. For Hsp82 clients, 105 of 147 (71%) interactions remained unchanged, with only 39 (26.5%) decreasing and 3 (2.0%) increasing after MMS treatment (Supplemental Table S2).
Figure 3. The dynamic Ssa1 and Hsp82 interactomes during the DNA damage response.
Interactors were organized into functional categories and plotted against interaction change (Ln ratio) with either A) Ssa1 or B) Hsp82 following MMS treatment. The dotted lines represent a two-fold interaction change up or down. Interactors are colored according to change in interaction as follows: red (significant increase), green (significant decrease) or black (no significant change).
We considered the possibility that certain pathways/classes of proteins may act in a correlated manner in terms of interaction change under stress. Analysis of proteins that increased or decreased the most during MMS treatment revealed little enrichment for any particular class of protein suggesting that chaperone interactions may be client specific, with unique patterns of association and dissociation. For Ssa1, the ribonucleotide reductase subunit Rnr4, the Coq5 coenzyme Q protein and the medium-chain dehydrogenase/reductases Ast1 were the only proteins to increase in interaction significantly upon MMS-exposure. For Hsp82, Rnr4, Cys3 (cysteine and glutathione synthesis) and an ORF of unknown function, Ydr210c were observed to bind Hsp82 at a higher enrichment after MMS.
Although previous studies have indicated that ribosomal genes are generally repressed during DNA damage, ribosomal protein abundance increases [1]. Given the high abundance of ribosomal proteins and the role that chaperones play in the folding of newly synthesized proteins, it was unsurprising that proteins that make up/regulate the ribosome formed a major part of the Hsp82/Ssa1 interactors isolated. Interestingly, a large proportion of ribosomal interactors of Ssa1 and Hsp82 dissociated upon MMS (28% and 35% respectively).
Molecular chaperones function both in complex with other chaperones and with regulatory proteins known as co-chaperones. An expected outcome of stressing cells with DNA damage might be the upregulation of chaperone function, and therefore an increased association of Ssa1 and Hsp82 with proteins that activate their ATPase activities and thus their cellular function. Many of the detected yeast co-chaperone proteins for Ssa1 including Ssb1, Kar2, Ydj1, Sis1 and Scj1 remained unaltered for binding under MMS treatment. Unexpectedly, a decreased interaction was observed for the non-cytoplasmic chaperone/co-chaperone proteins Ssc1, Mdj1 and Hsp60. In contrast, all of the chaperone/co-chaperone interactors of Hsp82 remained unchanged except Ydj1 (Fig. 3.).
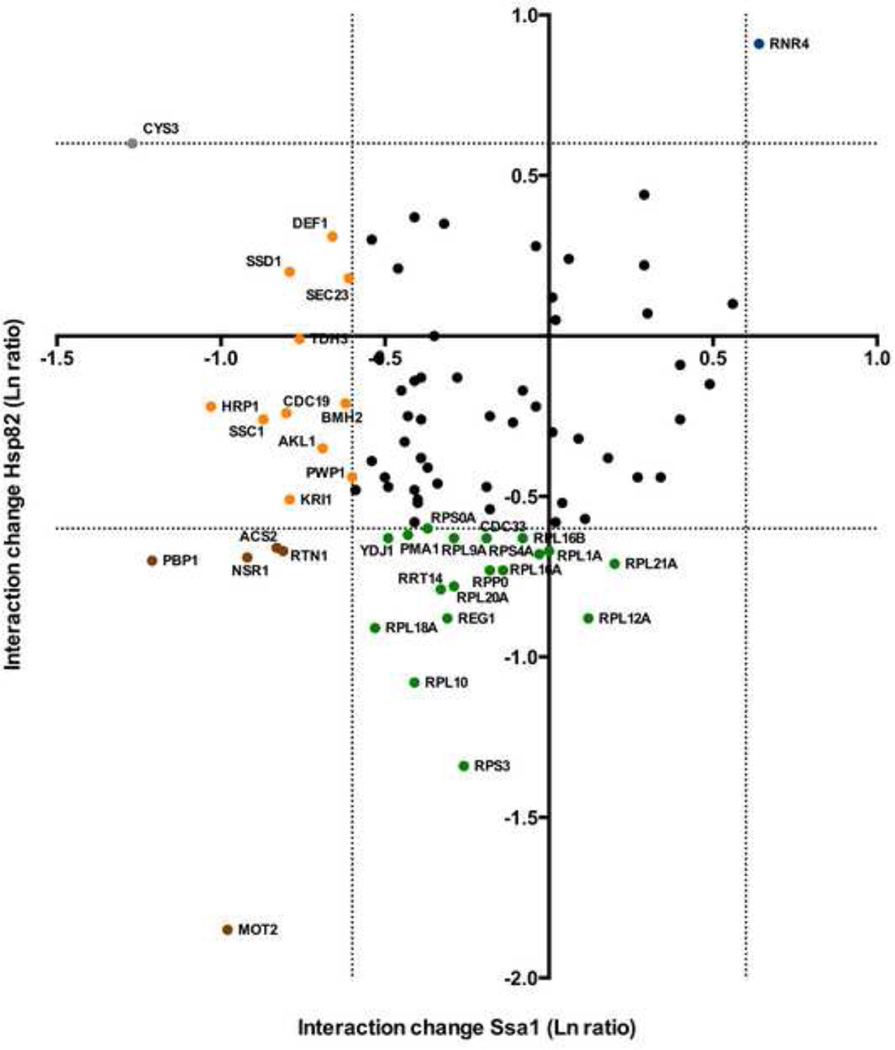
Hsp82 and Ssa1 work co-operatively in the cell to fold proteins. In certain conditions, interacting proteins may be transferred from one chaperone to another. This occurs often during the folding process, where Ssa1 will fold a protein 90% of the way and then transfer it to Hsp82 to achieve full activity. Conversely, proteins may be targeted for degradation by chaperones under stress conditions, potentially rebinding Ssa1. To see if we could observe any interactors transitioning from an Ssa1-binding to an Hsp82-binding state under MMS treatment, we plotted where possible for each interactor the change in Hsp82 interaction vs. Ssa1 interaction (Fig. 4). The majority of interacting proteins were unchanged for any interaction, but there were exceptions. Cys3 appeared to lose Ssa1 binding while gaining Hsp82 binding; Acs2, Rtn1, Nsr1 and Pbp1 dissociated from both chaperones under stress. Out of all interactors seen, only one protein, Rnr4 increased significantly in interactions with both Ssa1 and Hsp82 under MMS treatment.
Figure 4. A direct comparison of interaction changes seen for each client against both Ssa1 and Hsp82.
For each interactor observed to bind both Ssa1 and Hsp82 in this study, change in association for Ssa1 (Ln ratio, X-axis) vs. change in association for Hsp82 (Ln ratio, Y-axis) was plotted. The dotted lines represent an interaction change of up or down two-fold. Interactors are colored according to significant change in chaperone association upon DNA damage as follows: blue (increased with both chaperones), brown (decreased with both chaperones), yellow (decreased with Ssa1), green (decreased with Hsp82), grey (increased with Hsp82), or black (no significant change with either chaperone).
3.4 Comparison of client interaction vs. client abundance
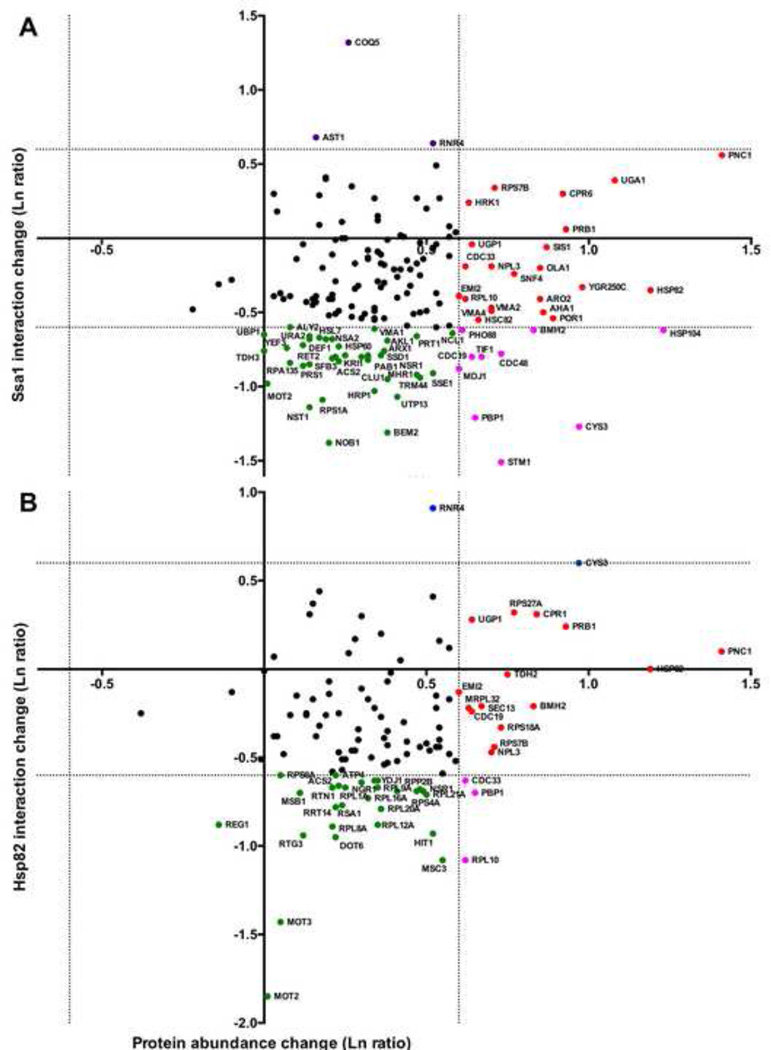
Several studies have analyzed the global effects of DNA damage on the yeast proteome or transcriptome [1, 2, 45, 51]. To address whether the interaction changes we observed for our interactomes could be attributed to client protein abundance changes alone or might reflect capacity to interact with chaperones, we plotted chaperone interaction change (from our study) vs. protein abundance change obtained from a prior study performed under similar conditions [2]. Interestingly for both chaperones, all of the clients detected in this study were proteins whose levels have been observed to either remain constant or increase during MMS treatment (Fig. 5). Of the interactors that significantly increased or decreased in chaperone interaction, the majority were proteins whose abundance do not substantially change during MMS treatment (Fig. 5A and 5B, green and purple-colored nodes).
Figure 5. Analysis of Ssa1/Hsp82 interactions in comparison to protein abundance changes during MMS treatment.
Where possible, interactors were plotted with the x-value corresponding to change in chaperone interaction (our data) and y-axis value the total protein abundance change (obtained from [2]). The dotted lines represent a change in either chaperone binding or protein levels of two-fold up or down. Interactors were colored according to significant change in chaperone association and/or protein abundance upon DNA damage as follows: green (decreased chaperone association), red (increased protein abundance), pink (decreased association, increased abundance), or black (no significant changes in association or abundance).
3.5. Chaperone function is required for stability of yeast ribonucleotide reductase subunits Rnr4 and Rnr2
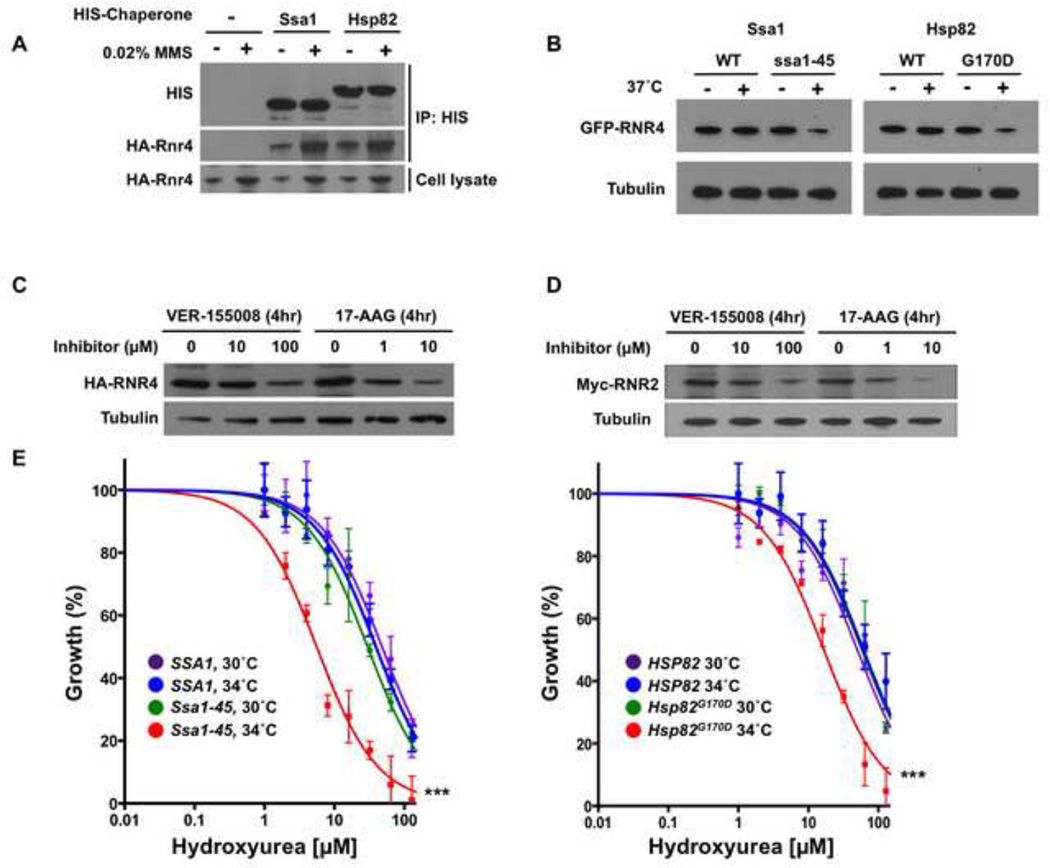
Many client proteins have been shown to depend on chaperones for their activity and/or stability [10]. We reasoned that any interactor that displayed increased association with both Ssa1 and Hsp82 upon DNA damage would be especially likely to be dependent on these chaperones for function. In this study, only one interactor, the ribonucleotide-diphosphate reductase (RNR) small subunit protein Rnr4, fulfilled this criterion. Rnr4 is particularly biologically interesting, due to its key role in cell cycle progression and DNA damage resistance [4, 5, 52]. To validate the chaperone-Rnr4 interactions detected by mass spectrometry, we performed IMAC bead pull-downs from lysates of cells expressing HA-tagged Rnr4 and untagged or HIS6-tagged Ssa1 or Hsp82. No HA-Rnr4 was detected in pull-downs from lysates lacking tagged chaperones (Fig. 6A). However, HA-Rnr4 was pulled down along with the tagged Ssa1 or Hsp82, and the apparent interaction was further enhanced by DNA damage.
Figure 6. Ribonucleotide reductase is a client of Hsp70 and Hsp90 in yeast cells.
A) Yeast expressing HA-tagged Rnr4 and either untagged chaperones, HIS6-Ssa1 or HIS6-Hsp82 were grown to early mid-log phase. Cells were lysed, pull-down was performed with His-Tag Dynabeads, and captured proteins were separated by SDS-PAGE, blotted and probed with anti-hexahistidine or anti-HA antibody.
B) Yeast expressing GFP-tagged Rnr4 along with wild-type chaperones or temperature-sensitive alleles Ssa1-45 or Hsp82G170D were incubated at 25° or 37° C for 90 min. GFP-Rnr4 levels were determined via Western blot of cell lysates with anti-GFP antibody. Equal loading was determined using anti-tubulin immunoreactivity.
C) Yeast cells expressing HA-tagged Rnr4 were grown to early mid-log phase and treated with DMSO, Hsp70 inhibitor VER-155008 or Hsp90 inhibitor 17-AAG for 4 h. Cells were lysed and HA-Rnr4 levels were determined via Western blot with anti-HA epitope antibody and anti-tubulin loading control.
D) Yeast cells expressing Myc-tagged Rnr2 were grown to early mid-log phase and treated with DMSO, Hsp70 inhibitor VER-155008 or Hsp90 inhibitor 17-AAG for 4 h. Cells were lysed and Myc-Rnr2 levels were determined via Western blot with anti-Myc epitope antibody and anti-tubulin loading control.
E) Yeast expressing wild-type chaperones or temperature-sensitive alleles Ssa1-45 or Hsp82G170D were diluted to OD600 of 0.01 and grown for 8 h in rich media at either 25° or 34° C in the presence of hydroxyurea (HU) at the indicated concentrations. OD600 was determined, normalized to 0 µM HU (100%), the data for each condition fit to a nonlinear curve (Prism software) and plotted. Data shown are mean ± SEM of three replicates (*** P<0.001) compared to WT strain at 34° C, t-test)
To decipher the role of chaperones in Rnr4 function, we examined Rnr4 protein levels in conditional yeast mutants under conditions of limiting chaperone activity. Cells expressing Ssa1–45 or Hsp82G170D as the sole Ssa1 or Hsp82, respectively, display temperature sensitive (ts) growth [53, 54]. At the permissive temperature of 25°C, cells proliferate normally, but display increasing growth defects at temperatures above 34°C. Cells expressing GFP-Rnr4 along with Ssa1–45, Hsp82G170D or WT controls and were grown at 25°C until early mid log phase and split into two flasks, one of which was shifted to 37°C. Cells were lysed after 90 mins and GFP-Rnr4 levels examined by Western blot. Incubation at 37°C did not affect GFP-Rnr4 levels in cells expressing wild-type Ssa1 or Hsp82, but decreased GFP-Rnr4 abundance in both strains bearing temperature-sensitive alleles (Fig. 6B).
As a complementary test, we inhibited Ssa1 or Hsp82 function in cells expressing HA-tagged Rnr4 with either the Hsp70 inhibitor VER-155008 or Hsp90 inhibitor 17-allylamino-17-demethoxygeldanamycin (17-AAG, tanespimycin). After 4 h at 25°C, cells were lysed and HA-Rnr4 levels examined by Western blot. Addition of either inhibitor decreased abundance of GFP-Rnr4 (Fig. 6C). This result is consistent with Rnr4 acting as a bona fide chaperone client in yeast, requiring Hsp70 and/or Hsp90 activity for stability.
The RNR holoenzyme is composed of 4 subunits, Rnr1-4 [5, 6]. In yeast, the two homologous R2 subunits, Rnr4 and Rnr2, heterodimerize. Insofar as Rnr4 is required for Rnr2 folding and function, we considered whether Hsp70 and Hsp90 may similarly regulate Rnr2. Mass spectrometry had suggested Rnr2 as an interactor for both chaperones but revealed no change after exposure to MMS (Supplemental Table S2). Following the strategy for Rnr4, cells expressing epitope-tagged Rnr2 were treated with the Hsp70 and Hsp90 inhibitors VER-155008 and 17-AAG after which Rnr2 levels were measured. As with Rnr4, Rnr2 abundance decreased after addition of either inhibitor (Fig. 6C).
Like many inhibitors of ribonucleotide reductase, hydroxyurea (HU) [7, 8] inhibits RNR by scavenging the tyrosyl free radical from the R2 subunit [55]. Impaired RNR expression and/or function sensitize yeast cells to HU [56, 57]. The growth of cells expressing Ssa1-45, Hsp82G170D or wild-type controls were compared after incubation for 8 h at 25°C or 34°C (semi-permissive temperature) in increasing concentrations of HU. Both the Ssa1-45 and Hsp82G170D strains displayed supersensitivity to HU at 34°C, consistent with a critical role for chaperones in RNR function (Fig. 6E).
3.6. Identification of human RNR subunit protein hRRM2 as an Hsp70 and Hsp90 client in breast cancer cells
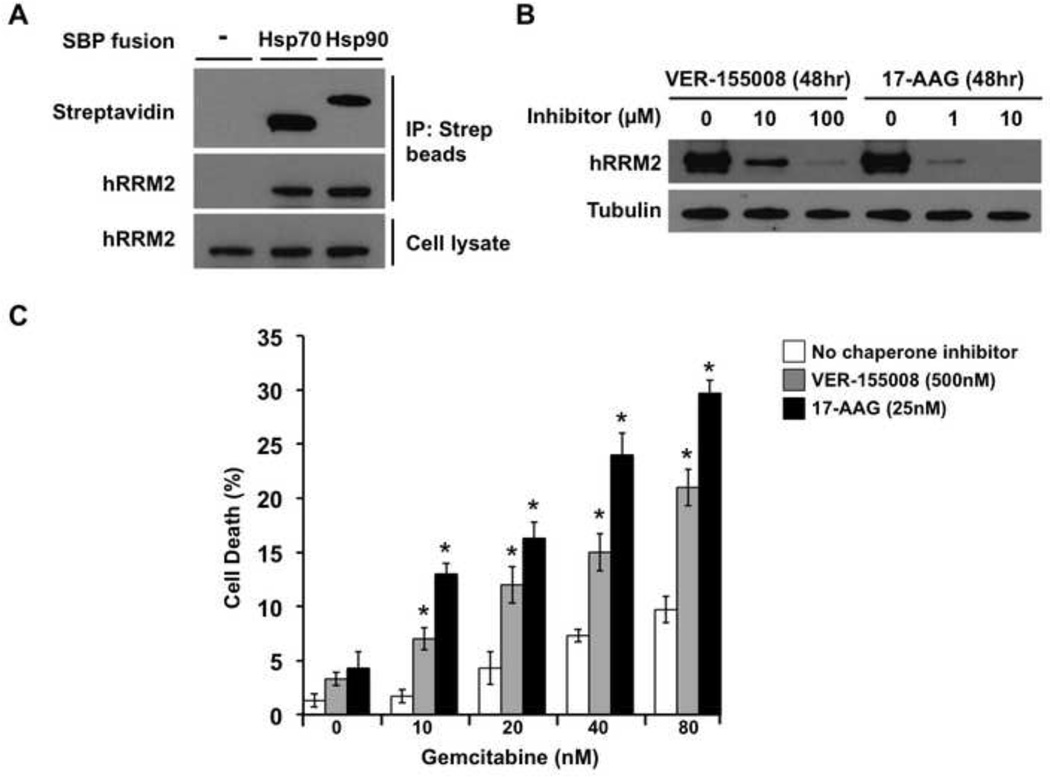
Ribonucleotide reductase is an important chemotherapeutic target in cancer. We considered the possibility that hRRM2, the human homolog of yeast Rnr4, might similarly interact with the Hsp70 and Hsp90 molecular chaperones in cancer cells. To examine this, we transfected MCF-7 breast cancer cells with Streptavidin Binding Protein (SBP) epitope-tagged Hsp70 or Hsp90, pulled-down these chaperones using streptavidin-magnetic beads and probed for associated hRRM2 (Fig. 7A). Interaction of the RNR subunit was seen with both Hsp70 and Hsp90, consistent with a conserved role for the chaperones. Indeed, when we treated mid-confluent cells with VER-155008 or 17-AAG for 48 h, hRRM2 protein levels decreased (Fig. 7B).
Figure 7. Ribonucleotide reductase is a client of Hsp70 and Hsp90 in human breast cancer cells.
A) MCF-7 human breast cancer cells were transiently transfected with no DNA (control) or plasmids expressing SBP-tagged Hsp70 or Hsp90. After 48 h recovery in media, cells were lysed and pull-down was performed with streptavidin-conjugated magnetic beads. Captured proteins were separated by SDS-PAGE, transferred and probed with streptavidin-HRP or anti-hRRM2 antibody and secondary antibody HRP conjugate.
B) MCF-7 cells grown to 50% confluence were treated either with DMSO, Hsp70 inhibitor VER-155008, or Hsp90 inhibitor 17-AAG for 48 h. Cells were lysed and hRRM2 levels were determined by Western blot with anti-hRRM2 antibody. Equal loading was determined using anti-tubulin immunoreactivity.
C) MCF-7 cells were incubated without chaperone inhibitors or with subtoxic concentrations of VER-155008 to inhibit Hsp70 or 17-AAG to inhibit Hsp90. After 24 h, gemcitabine was added at the indicated concentrations. After another 24 h, cell death was determined by Trypan Blue dye exclusion assay. Data shown are mean ± SEM of three replicates (* P < 0.05 compared to no chaperone inhibitor, t-test).
3.7. Sensitization of MCF-7 cells to gemcitabine using chaperone inhibitors
We reasoned that depletion of hRRM2 upon treatment with chaperone inhibitors might sensitize MCF-7 cells to RNR inhibition, leading to increased depletion of deoxyribonucleotides. As a model RNR inhibitor, we tested the nucleoside analog gemcitabine (4-amino-1-(2-deoxy-2,2-difluoro-β-D-erythro-pentofuranosyl)pyrimidin-2(1H)-on), an agent currently used in treatment of lung, pancreatic, bladder and breast cancer. MCF-7 cells were treated with VER-155008 or 17-AAG for 24 h and then with gemcitabine or DMSO for a further 24 h, whereupon cell viability was measured using Trypan Blue staining. Cell death was significantly greater in the cancer cells treated with chaperone inhibitors and gemcitabine than that induced by gemcitabine alone (Fig. 7C, P < 0.05, t-test). This compound effect is consistent with chemical synthetic lethality wherein inhibition of Hsp70 or Hsp90 results in depletion of RNR subunit abundance, thereby enhancing the toxicity of the RNR inhibitor gemcitabine.
4. Discussion
4.1. Functions of chaperone-client interactions during MMS treatment
Cells respond to stress by activating specific signaling pathways, many of which are dependent on chaperones for activity. A proportion of clients associate with chaperones to maintain stability and activity, whereas others must dissociate from chaperones to achieve full activity or correct cellular localization. In our study, we observed that the majority of proteins that bind to Ssa1/Hsp82 either remained constant or dissociated from them during MMS stress despite many being up regulated at the protein abundance level. As any protein that increases during exposure to DNA damage is likely to be required for cell survival and recovery, this then indicates that Ssa1 and Hsp82 are important mediators of the cell’s DNA damage response.
In addition, a proportion of the chaperone-client interactions decreased during MMS (29.5% of Ssa1 interactions, 26.5% of Hsp82 interactions). This implies that although chaperone interaction may be important for these proteins under normal cellular conditions, it may prevent full client activity under times of stress. Several examples of this have been uncovered. In mammalian cells, upon ligand binding the progesterone receptor dissociates from Hsp90, translocates to the nucleus and regulates target gene expression [58]. Another example is phosphatidylinositol 4-kinase type IIβ (PI4KIIβ). Hsp90 stabilizes (PI4KIIβ), keeping it active until pathway activation induces chaperone dissociation and recruitment to membranes [59]. Finally the Erk5 MAP kinase binds to Hsp90, an association required for stability. For the protein to achieve full activity, it becomes phosphorylated by upstream kinases, dissociates with Hsp90 and translocates to the nucleus [60, 61]. Mutations in Hsp90 (such as E33A or T22A) that stabilize client-chaperone interactions result in impairment of client function [43, 62]. For Hsp70, client interactions have several functions. Firstly, Hsp70 will bind and fold denatured proteins, to help them regain lost activity. When client protein deterioration is too severe, Hsp70 can target proteins for degradation by the ubiquitin proteasomal system. Thus loss of Hsp70-client interaction can either promote client stability or destruction depending on the specific client and cell conditions [31, 63]. In a previous study, we also saw disruption of many Ssa1-client interactions during G1 arrest using mating pheromone [24]. It may be that controlled dissociation of chaperone and a subset of clients can silence proliferative pathways, providing a cell the time required to adapt to environmental changes. Ssa1 and Hsp82 interactions during the cell cycle are regulated by direct chaperone phosphorylation. This may also be a method of interactome regulation for response to DNA damage. Although we did not detect differential phosphorylations on either chaperone, a more targeted approach in the future may reveal otherwise.
4.2. Chaperone interactors connected directly to the DNA damage response
Treatment of cells with MMS causes alkylation of DNA resulting in replication fork blockade through inhibition of DNA polymerase [64]. It is interesting to note that although we isolated proteins involved in the response to DNA damage, core signaling proteins in the DDR signaling cascade such as Rad9, Mec1 and Tel1 were not detected. Rad9 has previously been isolated as an interactor of Ssa1 in yeast [14], but its low abundance (∼ 400 molecules per cell) makes it difficult to detect in global interactome screens.
We did detect interactions with proteins and protein complexes connected to the cellular response to DNA damage. One example is Mot2 (Not4), which decreased in interaction with Hsp82 and Ssa1 during MMS treatment. This protein exists as part of the Ccr4-Not1 complex in yeast, a complex that has been implicated in the DDR. A recent study of regulated protein-protein interactions in yeast determined the Ccr4-Not1 complex to be particularly dynamic under MMS treatment [51]. This is particularly interesting as the Ccr4-Not1 complex plays a role in the regulation of RNR complex in several studies [65–67]. The Ccr4-Not1 complex controls the recruitment of transcription factors to the RNR gene promoters and is also necessary for correct localization of RNR subunits [67].
Yeast cope with DNA damage using multiple parallel systems, one of which is transcription-coupled repair (TCR). When RNA polymerase II (RNAPII) becomes stalled at sites of DNA damage, Def1 (seen here as interacting with both chaperones) is recruited in a complex with Rad26. Def1 promotes the destruction of RNAPII when the lesion cannot be rapidly removed by Rad26-promoted DNA repair [68]. Histone proteins and HDACs play an important role in transcriptional activation during DNA damage. HDACs Hos3 and Sin3, responsible for chromosomal integrity, Rvb2 and Abf1 chromatin remodeling factors and the Xrn1 exonuclease were all seen in our client list.
We also observed chaperone interaction with two key proteins involved in the nicotinamide salvage pathway, Nma1 and Pnc1. This is particularly interesting, since DNA repair has long been shown to be an NAD-dependent process [50]. In addition, it has been recently shown that overexpression of NAD salvage proteins promotes the clearance of misfolded proteins, particularly those regulated by Hsp90 and Hsp70 [69]. Perhaps Nma1 and Pnc1 are both clients of these chaperones and also modify chaperone activity directly, particularly in response to DNA damage.
During the course of this study, we also isolated uncharacterized ORFs as Hsp90/70 interacting proteins, several of which were altered in chaperone binding upon MMS stress. Little is known about Nst1 (seen here as an interactor of Ssa1) except that it mediates salt resistance in yeast [70]. Nst1 does however physically interact with Mot3 (mentioned above), and thus may also potentially regulate RNR function. Hsp82 bound to five uncharacterized ORFs; Ygr266w, Msb1, Rrt14, Hit1, and Msb3. Msb3 interacts with Bem2 (also detected in this study) and relocalizes from the bud neck to cytoplasm upon replication stress.
4.3. Interactions between proteins that regulate the cellular response to oxidative stress and Ssa1/Hsp82 are modulated during MMS treatment
In addition to its documented effect as a DNA damaging agent, MMS treatment also increases reactive oxygen species [2, 71]. As such, many of the clients that were altered for binding are linked to the cellular response to oxidative stress. We see a clear dissociation of Ssa1 with Hsp104, Hsp60, Ssc1, Mdj1 and Sse1, proteins all involved in the cellular response to oxidative stress. When yeast cells are challenged with oxidative stress, many mitochondrial proteins become oxidized/unfolded [72]. Protein folding inside mitochondria is promoted through a successive action of the Hsp70 complex (Scc1 with co-chaperone Mdj1) and the Hsp60 complex [73]. In addition to Ssc1-Mdj1 protein folding, Ssc1 can form a complex with the co-chaperone Mge1 to regulate resistance to oxidative stress [74]. Mge1-Mge1 dimers decrease in response to oxidative stress, promoting Ssc1-Mge1 interaction, thereby up-regulating chaperone activity to refold damaged proteins [74]. When yeast cells are challenged with oxidative stress, several mitochondrial proteins become oxidized including Hsp60 [72]. In turn, Hsp60 function is directly proportional to the ability of cells to cope with oxidative damage [72]. How modulation of chaperone-co-chaperone interactions regulates cellular response to oxidative stress remains to be elucidated. Sse1 and Sse2 are highly homologous to canonical Hsp70s such as Ssa1, but contain insertions and C-terminal extensions that increase their molecular mass leading to their categorization as members of the “Hsp110” family [75, 76]. Sse1 acts as a nuclear exchange factor (NEF) for Ssa1, exchanging ADP for fresh ATP to drive protein folding. In this study, the Sse1-Ssa1 interaction decreased significantly during DNA damage. Interestingly, we have previously observed that the Sse1-Ssa1 interaction is dynamic in response to phosphorylation of Ssa1 and stress to the cell [24]. It may be that MMS treatment promotes an as yet undetected post-translational modification on Ssa1 that regulates global interactor binding.
Aside from chaperone-co-chaperone interactions, several other clients associated with the response to oxidative stress displayed highly regulated interactions upon MMS treatment. The cystathionine-gamma-lyase increases in abundance at both transcript and protein level upon exposure to MMS [2, 77]. In our study, Cys3 displayed greater enrichment in the Hsp82 (but not Ssa1) interactome after MMS treatment, potentially a response to depletion of reduced thiols after reaction with MMS. This may be relevant in light of findings that Ssa1 is a direct target of thiol-reactive molecules [78]. The mitochondrial DNA repair and replication factor Mhr1 interacted only with Ssa1 and dissociated upon MMS treatment. Upon oxidative damage to the mtDNA, the base excision-repair enzyme Ntg1 introduces a DSB at the mtDNA replication origin, triggering activation of Mhr1, which initiates mtDNA replication. As a result, oxidative stress indirectly regulates mtDNA copy number [79]. It may be that dissociation from Hsp70 is required for Mhr1 to achieve full activity.
The COQ5 gene encodes a C-methyltransferase involved in the biosynthesis of ubiquinone or coenzyme Q [80]. Ubiquinone functions as the electron carrier in the mitochondrial respiratory chain for energy production. Importantly, ubiquinone also participates in other cellular processes, such as control of cellular redox status and detoxification of harmful reactive oxygen species [81, 82]. It is interesting then that Coq5 displayed the largest positive change in chaperone binding under MMS treatment. As with Rnr4, it may suggest that Coq5 stability is mediated by chaperones during MMS and may be necessary for the cell to tolerate high ROS levels.
It should be noted that although Hsp82 has been detected in association with proteins localized to the mitochondria matrix, such as the F1F0 ATPase subunits Atp1, 2 and 3 [83–85], little evidence exists to support Ssa1 being present inside the mitochondria. Ssa1 does bind mitochondrial proteins, but has been ascribed to a role in mitochondrial protein import rather than localization to mitochondria per se [86, 87]. Capturing transport intermediates may explain the Ssa1/Mhr1 interaction. On the other hand, some mitochondrial proteins appear to relocalize to the cytoplasm under MMS treatment [2], offering a way for them to interact with cytplasmically localized Ssa1 and Hsp82. Nonetheless, we cannot rule out that a subset of the Hsp70/Hsp90 interacting proteins identified here are simply artifacts of cell lysis and/or tagged-protein purification, despite our best efforts to optimize biochemical methods and use of rigorous statistical criteria for proteomic data analysis.
4.5. Ribonucleotide reductase subunit stability is regulated by molecular chaperones
The yeast RNR subunit Rnr4 plays an important role in DNA replication during S phase and the response to DNA damage. RNR has been studied for several decades and a variety of inhibitors of the enzyme are commercially available [7, 8]. Unlike all of the other interactors detected in this study, Rnr4 substantially increased in interaction with both chaperones after DNA damage. In turn, inhibition of either Hsp70 or Hsp90 (either through small molecules or chaperone mutation) resulted in a substantial decrease in Rnr4 protein levels. Like regulation of gene expression, control of protein stability provides a powerful mechanism that can adapt protein abundance to cellular needs [88]. In our study, we detected loss of interaction between Ssa1/Hsp82 and Mot2/3. As these proteins indirectly regulate RNR gene transcription, it is tempting to hypothesize that chaperone inhibition may regulate RNR levels through the Ccr-Not complex. In our yeast system however, Rnr4 levels decreased upon chaperone inhibition even when expressed from the constitutive Gal4 promoter (Figure 6B). This suggests the effect most likely occurs through increased degradation rather than decreased transcription. The RNR subunit Rnr2 was also identified as an Ssa1 and Hsp82 interactor, although its association remained unchanged upon treatment with MMS. However, like Rnr4, we observed Rnr2 abundance also depended on chaperone activity. This may reflect a requirement for Rnr4 in folding of Rnr2 [89]. Thus, via their dynamic interaction with Rnr4, chaperones may be able to regulate both Rnr4 and Rnr2 and thereby control RNR activity. It is interesting to note that Rnr2 and Rnr4 do not heterodimerize easily, and have low intrinsic activity when produced and purified from bacteria [89].
Suggesting broad conservation of this mechanism of RNR regulation, treating breast cancer cells with chaperone inhibitors decreased hRMM2 abundance and sensitized cells to the clinically useful RNR inhibitor gemcitabine. Synergy between these two classes of inhibitors has not been demonstrated previously. Given the emergence of Hsp90-targeted agents in the clinic [16], combining ribonucleotide reductase inhibitors such as gemcitabine with chaperone inhibitors such as 17-AAG might have value in targeting proliferative diseases.
5. Conclusions
Many different techniques have been employed to assess protein-protein interactions [39]. Affinity-purification mass spectrometry offers a thorough way to interrogate these interactions on a global scale [36–39]. Observing interactome dynamics during response to perturbations such as DNA damage adds an additional level of information. Given that chaperones have critical roles in protein function, following dynamic interactomes of Hsp70/90 may reveal a function for clients in DNA damage response. In particular, this may lead to DNA damage response proteins particularly prone to destabilization via chaperone inhibition. Using yeast as a model, interactome analysis of the Hsp70 and Hsp90 proteins during DNA damage revealed interactions with shared client proteins, such as ribonucleotide reductase subunits Rnr4 and Rnr2, both of whose stability appears to depend on chaperone activity. Following this logic, we found that chaperone inhibition enhanced the effects of an RNR inhibitor on breast cancer cells in culture, suggesting a new approach to targeting cancer cell proliferation in vivo.
Supplementary Material
A table of yeast strain and plasmids used in the study are described, along with their original source, genotypes and selectable markers.
Chaperone interactors identified in our mass spectrometry studies are listed. For each interactor, Column B gives the systematic yeast ORF ID, Column C gives the commonly used ORF name. Columns D and E give the Ln ratio change of chaperone interaction from untreated to MMS treated cells. Column F is the Ln ratio change in total protein abundance for each interactor obtained from [2].
Full settings for MaxQuant used in this study.
Biological significance.
This study provides the dynamic interactome of the yeast Hsp70 and Hsp90 under DNA damage which suggest key roles for the chaperones in a variety of signaling cascades. Importantly, the cancer drug target ribonucleotide reductase was shown to be a client of Hsp70 and Hsp90 in both yeast and breast cancer cells. As such, this study highlights the potential of a novel cancer therapeutic strategy that exploits the synergy of chaperone and ribonucleotide reductase inhibitors.
Acknowledgements
The authors thank M. Mollapour for helpful comments. We are grateful to A. Mogk, M. Huang, P. Piper and P. Muller for providing materials used in this study. We acknowledge the PRIDE team for the deposition of our data to the ProteomeXchange consortium. This project was supported by NCI R01 CA164492 (SJK), the Ludwig Center for Metastasis Research and NCATS UL1 TR000430.
Abbreviations
- RNR
Ribonucleotide reductase
- GO
Gene ontology
- DDR
DNA damage response
- DSB
double strand break
- IP
immunoprecipitation
- IR
Ionizing radiation
- MMS
Methyl methanesulphonate
- ROS
Reactive oxygen species
- dNTP
deoxyribonucleotide
- STRING
Search Tool for the Retrieval of Interacting Genes/Proteins
- HSP
Heat Shock Protein
REFERENCES
- 1.Mazumder A, et al. Genome-wide single-cell-level screen for protein abundance and localization changes in response to DNA damage in S. cerevisiae. Nucleic Acids Res. 2013;41(20):9310–9324. doi: 10.1093/nar/gkt715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tkach JM, et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol. 2012;14(9):966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem Res Toxicol. 2006;19(12):1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanvisens N, de Llanos R, Puig S. Function and regulation of yeast ribonucleotide reductase: cell cycle, genotoxic stress, and iron bioavailability. Biomedical journal. 2013;36(2):51–58. doi: 10.4103/2319-4170.110398. [DOI] [PubMed] [Google Scholar]
- 5.Kolberg M, et al. Structure, function, and mechanism of ribonucleotide reductases. Biochim Biophys Acta. 2004;1699(1–2):1–34. doi: 10.1016/j.bbapap.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 7.Cerqueira NM, Fernandes PA, Ramos MJ. Ribonucleotide reductase: a critical enzyme for cancer chemotherapy and antiviral agents. Recent patents on anti-cancer drug discovery. 2007;2(1):11–29. doi: 10.2174/157489207779561408. [DOI] [PubMed] [Google Scholar]
- 8.Cerqueira NM, et al. Overview of ribonucleotide reductase inhibitors: an appealing target in anti-tumour therapy. Curr Med Chem. 2005;12(11):1283–1294. doi: 10.2174/0929867054020981. [DOI] [PubMed] [Google Scholar]
- 9.Newnam GP, et al. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol. 1999;19(2):1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Z, Moghaddas Gholami A, Kuster B. Systematic identification of the HSP90 candidate regulated proteome. Mol Cell Proteomics. 2012;11(6):M111. doi: 10.1074/mcp.M111.016675. 016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62(6):670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meimaridou E, Gooljar SB, Chapple JP. From hatching to dispatching: the multiple cellular roles of the Hsp70 molecular chaperone machinery. J Mol Endocrinol. 2009;42(1):1–9. doi: 10.1677/JME-08-0116. [DOI] [PubMed] [Google Scholar]
- 13.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11(7):515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert CS, et al. The budding yeast Rad9 checkpoint complex: chaperone proteins are required for its function. EMBO Rep. 2003;4(10):953–958. doi: 10.1038/sj.embor.embor935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goloudina AR, Demidov ON, Garrido C. Inhibition of HSP70: a challenging anti-cancer strategy. Cancer Lett. 2012;325(2):117–124. doi: 10.1016/j.canlet.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Trepel J, et al. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10(8):537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray JV, et al. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. The EMBO journal. 1997;16(16):4924–4937. doi: 10.1093/emboj/16.16.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasin N, et al. Global transcript and phenotypic analysis of yeast cells expressing Ssa1, Ssa2, Ssa3 or Ssa4 as sole source of cytosolic Hsp70-Ssa chaperone activity. BMC Genomics. 2014;15:194. doi: 10.1186/1471-2164-15-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van den Bosch M, Lowndes NF. Remodelling the Rad9 checkpoint complex: preparing Rad53 for action. Cell Cycle. 2004;3(2):119–122. [PubMed] [Google Scholar]
- 20.Quanz M, et al. Heat shock protein 90alpha (Hsp90alpha) is phosphorylated in response to DNA damage and accumulates in repair foci. J Biol Chem. 2012;287(12):8803–8815. doi: 10.1074/jbc.M111.320887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma K, et al. Quantitative proteomics reveals that Hsp90 inhibition preferentially targets kinases and the DNA damage response. Mol Cell Proteomics. 2012;11(3):M111. doi: 10.1074/mcp.M111.014654. 014654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong Y, et al. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol Syst Biol. 2009;5:275. doi: 10.1038/msb.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Millson SH, et al. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p) Eukaryot Cell. 2005;4(5):849–860. doi: 10.1128/EC.4.5.849-860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Truman AW, et al. CDK-dependent Hsp70 Phosphorylation controls G1 cyclin abundance and cell-cycle progression. Cell. 2012;151(6):1308–1318. doi: 10.1016/j.cell.2012.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao R, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120(5):715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 26.Taipale M, et al. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150(5):987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loroch S, et al. Phosphoproteomics--more than meets the eye. Electrophoresis. 2013;34(11):1483–1492. doi: 10.1002/elps.201200710. [DOI] [PubMed] [Google Scholar]
- 28.Leitner A, Sturm M, Lindner W. Tools for analyzing the phosphoproteome and other phosphorylated biomolecules: a review. Analytica chimica acta. 2011;703(1):19–30. doi: 10.1016/j.aca.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Drissi R, Dubois ML, Boisvert FM. Proteomics methods for subcellular proteome analysis. The FEBS journal. 2013;280(22):5626–5634. doi: 10.1111/febs.12502. [DOI] [PubMed] [Google Scholar]
- 30.Lee YH, Tan HT, Chung MC. Subcellular fractionation methods and strategies for proteomics. Proteomics. 2010;10(22):3935–3956. doi: 10.1002/pmic.201000289. [DOI] [PubMed] [Google Scholar]
- 31.Wegele H, Muller L, Buchner J. Hsp70 and Hsp90--a relay team for protein folding. Rev Physiol Biochem Pharmacol. 2004;151:1–44. doi: 10.1007/s10254-003-0021-1. [DOI] [PubMed] [Google Scholar]
- 32.Balaburski GM, et al. A modified HSP70 inhibitor shows broad activity as an anticancer agent. Molecular cancer research : MCR. 2013;11(3):219–229. doi: 10.1158/1541-7786.MCR-12-0547-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prodromou C, et al. Structural basis of the radicicol resistance displayed by a fungal hsp90. ACS Chem Biol. 2009;4(4):289–297. doi: 10.1021/cb9000316. [DOI] [PubMed] [Google Scholar]
- 34.Vizcaino JA, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;2013;41(Database issue):D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warsow G, et al. ExprEssence--revealing the essence of differential experimental data in the context of an interaction/regulation net-work. BMC Syst Biol. 2010;4:164. doi: 10.1186/1752-0509-4-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 37.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415(6868):141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 38.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 39.Kocher T, Superti-Furga G. Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat Methods. 2007;4(10):807–815. doi: 10.1038/nmeth1093. [DOI] [PubMed] [Google Scholar]
- 40.Yao X, et al. Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal Chem. 2001;73(13):2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 41.Ye X, et al. 18O stable isotope labeling in MS-based proteomics. Brief Funct Genomic Proteomic. 2009;8(2):136–144. doi: 10.1093/bfgp/eln055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaiswal H, et al. The chaperone network connected to human ribosome-associated complex. Molecular and cellular biology. 2011;31(6):1160–1173. doi: 10.1128/MCB.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panaretou B, et al. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17(16):4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perkins DN, et al. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 45.Chen SH, et al. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J Biol Chem. 2010;285(17):12803–12812. doi: 10.1074/jbc.M110.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Millson SH, et al. Investigating the protein-protein interactions of the yeast Hsp90 chaperone system by two-hybrid analysis: potential uses and limitations of this approach. Cell Stress Chaperones. 2004;9(4):359–368. doi: 10.1379/CSC-29R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albanese V, Reissmann S, Frydman J. A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J Cell Biol. 2010;189(1):69–81. doi: 10.1083/jcb.201001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Craig EA, Eisenman HC, Hundley HA. Ribosome-tethered molecular chaperones: the first line of defense against protein misfolding? Curr Opin Microbiol. 2003;6(2):157–162. doi: 10.1016/s1369-5274(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 49.Verghese J, et al. Biology of the heat shock response and protein chaperones: budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol Mol Biol Rev. 2012;76(2):115–158. doi: 10.1128/MMBR.05018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pittelli M, et al. Pharmacological effects of exogenous NAD on mitochondrial bioenergetics, DNA repair, and apoptosis. Mol Pharmacol. 2011;80(6):1136–1146. doi: 10.1124/mol.111.073916. [DOI] [PubMed] [Google Scholar]
- 51.Rochette S, et al. Modulation of the yeast protein interactome in response to DNA damage. J Proteomics. 2014;100:25–36. doi: 10.1016/j.jprot.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 52.Elledge SJ, et al. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays. 1993;15(5):333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- 53.Nathan DF, Lindquist S. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15(7):3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winkler J, et al. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol. 2012;198(3):387–404. doi: 10.1083/jcb.201201074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yarbro JW. Mechanism of action of hydroxyurea. Semin Oncol. 1992;19(3 Suppl 9):1–10. [PubMed] [Google Scholar]
- 56.Strauss M, et al. RNR4 mutant alleles pso3-1 and rnr4Delta block induced mutation in Saccharomyces cerevisiae. Curr Genet. 2007;51(4):221–231. doi: 10.1007/s00294-007-0120-7. [DOI] [PubMed] [Google Scholar]
- 57.Wang PJ, et al. Rnr4p, a novel ribonucleotide reductase small-subunit protein. Mol Cell Biol. 1997;17(10):6114–6121. doi: 10.1128/mcb.17.10.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Picard D. Chaperoning steroid hormone action. Trends Endocrinol Metab. 2006;17(6):229–235. doi: 10.1016/j.tem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 59.Jung G, et al. Stabilization of phosphatidylinositol 4-kinase type IIbeta by interaction with Hsp90. J Biol Chem. 2011;286(14):12775–12784. doi: 10.1074/jbc.M110.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Erazo T, et al. Canonical and kinase activity-independent mechanisms for extracellular signal-regulated kinase 5 (ERK5) nuclear translocation require dissociation of Hsp90 from the ERK5-Cdc37 complex. Mol Cell Biol. 2013;33(8):1671–1686. doi: 10.1128/MCB.01246-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Truman AW, et al. Expressed in the yeast Saccharomyces cerevisiae, human ERK5 is a client of the Hsp90 chaperone that complements loss of the Slt2p (Mpk1p) cell integrity stress-activated protein kinase. Eukaryot Cell. 2006;5(11):1914–1924. doi: 10.1128/EC.00263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mollapour M, et al. Threonine 22 phosphorylation attenuates Hsp90 interaction with cochaperones and affects its chaperone activity. Mol Cell. 2011;41(6):672–681. doi: 10.1016/j.molcel.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends in biochemical sciences. 2002;27(7):368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 64.Groth P, et al. Methylated DNA causes a physical block to replication forks independently of damage signalling, O(6)-methylguanine or DNA single-strand breaks and results in DNA damage. J Mol Biol. 2010;402(1):70–82. doi: 10.1016/j.jmb.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 65.Collart MA, Panasenko OO. The Ccr4--not complex. Gene. 2012;492(1):42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 66.Mulder KW, Winkler GS, Timmers HT. DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4-Not complex. Nucleic Acids Res. 2005;33(19):6384–6392. doi: 10.1093/nar/gki938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takahashi S, et al. Caf1 regulates translocation of ribonucleotide reductase by releasing nucleoplasmic Spd1-Suc22 assembly. Nucleic Acids Res. 2007;35(4):1187–1197. doi: 10.1093/nar/gkm015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woudstra EC, et al. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature. 2002;415(6874):929–933. doi: 10.1038/415929a. [DOI] [PubMed] [Google Scholar]
- 69.Ocampo A, Liu J, Barrientos A. NAD+ salvage pathway proteins suppress proteotoxicity in yeast models of neurodegeneration by promoting the clearance of misfolded/oligomerized proteins. Hum Mol Genet. 2013;22(9):1699–1708. doi: 10.1093/hmg/ddt016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goossens A, Forment J, Serrano R. Involvement of Nst1p/YNL091w and Msl1p, a U2B” splicing factor, in Saccharomyces cerevisiae salt tolerance. Yeast. 2002;19(3):193–202. doi: 10.1002/yea.815. [DOI] [PubMed] [Google Scholar]
- 71.Kitanovic A, Wolfl S. Fructose-1,6-bisphosphatase mediates cellular responses to DNA damage and aging in Saccharomyces cerevisiae. Mutat Res. 2006;594(1–2):135–147. doi: 10.1016/j.mrfmmm.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 72.Cabiscol E, et al. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J Biol Chem. 2000;275(35):27393–27398. doi: 10.1074/jbc.M003140200. [DOI] [PubMed] [Google Scholar]
- 73.Petit MA, et al. Sequential folding of UmuC by the Hsp70 and Hsp60 chaperone complexes of Escherichia coli. J Biol Chem. 1994;269(38):23824–23829. [PubMed] [Google Scholar]
- 74.Marada A, et al. Mge1, a nucleotide exchange factor of Hsp70, acts as an oxidative sensor to regulate mitochondrial Hsp70 function. Mol Biol Cell. 2013;24(6):692–703. doi: 10.1091/mbc.E12-10-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Easton DP, Kaneko Y, Subjeck JR. The hsp110 and Grp1 70 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5(4):276–290. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shaner L, et al. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J Biol Chem. 2005;280(50):41262–41269. doi: 10.1074/jbc.M503614200. [DOI] [PubMed] [Google Scholar]
- 77.Caba E, et al. Differentiating mechanisms of toxicity using global gene expression analysis in Saccharomyces cerevisiae. Mutat Res. 2005;575(1–2):34–46. doi: 10.1016/j.mrfmmm.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 78.Wang RY, et al. A temperature sensitive mutant of heat shock protein 70 reveals an essential role during the early steps of tombusvirus replication. Virology. 2009;394(1):28–38. doi: 10.1016/j.virol.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hori A, et al. Reactive oxygen species regulate DNA copy number in isolated yeast mitochondria by triggering recombination-mediated replication. Nucleic Acids Res. 2009;37(3):749–761. doi: 10.1093/nar/gkn993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dibrov E, Robinson KM, Lemire BD. The COQ5 gene encodes a yeast mitochondrial protein necessary for ubiquinone biosynthesis and the assembly of the respiratory chain. J Biol Chem. 1997;272(14):9175–9181. doi: 10.1074/jbc.272.14.9175. [DOI] [PubMed] [Google Scholar]
- 81.Kawamukai M. Biosynthesis, bioproduction and novel roles of ubiquinone. J Biosci Bioeng. 2002;94(6):511–517. doi: 10.1016/s1389-1723(02)80188-8. [DOI] [PubMed] [Google Scholar]
- 82.Turunen M, Olsson J, Dallner G. Metabolism and function of coenzyme Q. Biochim Biophys Acta. 2004;1660(1–2):171–199. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 83.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 84.McClellan AJ, et al. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131(1):121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 85.Papathanassiu AE, et al. F1F0-ATP synthase functions as a co-chaperone of Hsp90-substrate protein complexes. Biochem Biophys Res Commun. 2006;345(1):419–429. doi: 10.1016/j.bbrc.2006.04.104. [DOI] [PubMed] [Google Scholar]
- 86.Voos W. A new connection: chaperones meet a mitochondrial receptor. Mol Cell. 2003;11(1):1–3. doi: 10.1016/s1097-2765(03)00002-9. [DOI] [PubMed] [Google Scholar]
- 87.Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112(1):41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- 88.Belle A, et al. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci U S A. 2006;103(35):13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chabes A, et al. Yeast ribonucleotide reductase has a heterodimeric iron-radical-containing subunit. Proc Natl Acad Sci U S A. 2000;97(6):2474–2479. doi: 10.1073/pnas.97.6.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A table of yeast strain and plasmids used in the study are described, along with their original source, genotypes and selectable markers.
Chaperone interactors identified in our mass spectrometry studies are listed. For each interactor, Column B gives the systematic yeast ORF ID, Column C gives the commonly used ORF name. Columns D and E give the Ln ratio change of chaperone interaction from untreated to MMS treated cells. Column F is the Ln ratio change in total protein abundance for each interactor obtained from [2].
Full settings for MaxQuant used in this study.