SUMMARY
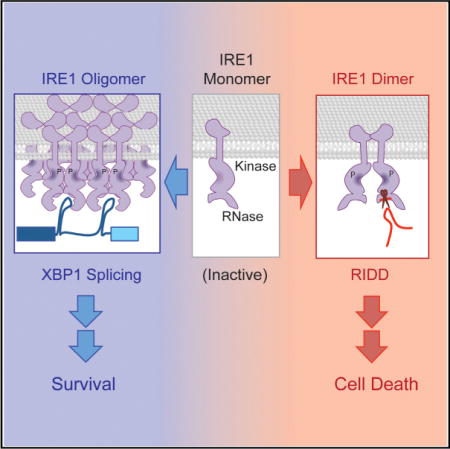
An evolutionarily conserved unfolded protein response (UPR) component, IRE1, cleaves XBP1/HAC1 introns in order to generate spliced mRNAs that are translated into potent transcription factors. IRE1 also cleaves endoplasmic-reticulum-associated RNAs leading to their decay, an activity termed regulated IRE1-dependent decay (RIDD); however, the mechanism by which IRE1 differentiates intron cleavage from RIDD is not well understood. Using in vitro experiments, we found that IRE1 has two different modes of action: XBP1/HAC1 is cleaved by IRE1 subunits acting cooperatively within IRE1 oligomers, whereas a single subunit of IRE1 performs RIDD without cooperativity. Furthermore, these distinct activities can be separated by complementation of catalytically inactive IRE1 RNase and mutations at oligomerization interfaces. Using an IRE1 RNase inhibitor, STF-083010, selective inhibition of XBP1 splicing indicates that XBP1 promotes cell survival, whereas RIDD leads to cell death, revealing modulation of IRE1 activities as a drug-development strategy.
Graphical abstract
INTRODUCTION AND RESULTS
IRE1 is a transmembrane receptor kinase located on the surface of the endoplasmic reticulum (ER) that initiates the unfolded protein response (UPR) pathway and is required for the ER to function properly. Upon activation, IRE1 becomes an active endoribonuclease (RNase) that cleaves an intron of an mRNA coding for a bZIP transcription factor, HAC1 in yeast or XBP1 in mammalian cells (Calfon et al., 2002; Gonzalez et al., 1999; Kawahara et al., 1997; Sidrauski and Walter, 1997). Cleaved exons are then ligated by tRNA ligase in the case of HAC1 (Siduraski et al., 1996) and RtcB for XBP1 (Lu et al., 2014) generating the spliced form of HAC1 or XBP1 mRNA. The IRE1-dependent splicing step is critical for mounting the UPR, as only the spliced form of HAC1 or XBP1 mRNA produces a potent transcription factor that induces UPR gene expression needed for re-establishment of ER functions.
Recently, UPR-activated IRE1 in several organisms— including S. pombe, mammalian, Drosophila, and plant cells— has been reported to also cleave a subset of mRNA that is associated with the ER membrane (Han et al., 2009; Hollien et al., 2009; Hollien and Weissman, 2006; Kimmig et al., 2012; Mishiba et al., 2013) In these cases, however, IRE1-mediated cleavage is followed by degradation, a process that has been termed regulated IRE1-dependent decay (RIDD) (Hollien et al., 2009). At present, a detailed, mechanistic understanding of IRE1 engaged in either RIDD or XBP1 mRNA intron cleavage is lacking. IRE1 that cleaves the XBP1 intron must be coordinated with a ligase to generate the spliced form of XBP1. In contrast, IRE1 engaged in RIDD must be coupled with mRNA degradation enzymes in order to prevent either translating or ligating cleavage products. Curiously, RIDD has never been reported to occur in the budding yeast, S. cerevisiae.
To investigate the mechanistic relationship between XBP1/HAC1 splicing and RIDD, we first asked if IRE1 from the budding yeast S. cerevisiae can perform RIDD cleavage events. After treating cells with tunicamycin (Tm), a well-characterized inducer of UPR, levels of two mRNAs, DAP2 (DPAPB) and MFα1 (α-Factor) coding for secretory pathway proteins (Figure S1A), decreased in agreement with genome-wide transcription analyses (Gasch et al., 2000; Travers et al., 2000). The decrease in mRNA levels required IRE1 but not HAC1 (Figure S1A) and was well correlated with the kinetics of HAC1 splicing (Figure S1B). Using an in vitro RNase assay, recombinant yeast IRE1 (yIre1) can cleave HAC1 RNA (Figure S1C) and in vitro transcribed radiolabeled DAP2 (Figure 1A) and MFα1 (Figure S1D) RNA upon incubation with ADP. The RNA cleavage fragments generated by yIre1 cleaving mammalian mRNAs that are known substrates for RIDD such as BLOS1 or INSULIN were essentially identical to those generated by human recombinant IRE1 (hIre1) (compare Figures 1A and 1B). Notably, recombinant yeast and human Ire1 did not cleave the mRNA of ACTIN, a nonsecretory protein, demonstrating that Ire1 is not a random nonspecific RNase (Figure S1C). Furthermore, mutating the invariant guanosine (Gonzalez et al., 1999; Han et al., 2009; Kawahara et al., 1997) located at the RIDD cleavage site of the INSULIN mRNA abolished cleavage at that site and instead generated aberrantly cleaved fragments (Figure S1E), reminiscent of other RNA processing events (Yang, 2011). These results demonstrate that RIDD activity is conserved in yIre1.
Figure 1. In Vitro Cleavage of RIDD Substrate RNA Is Distinct from XBP1/HAC1 Cleavage.
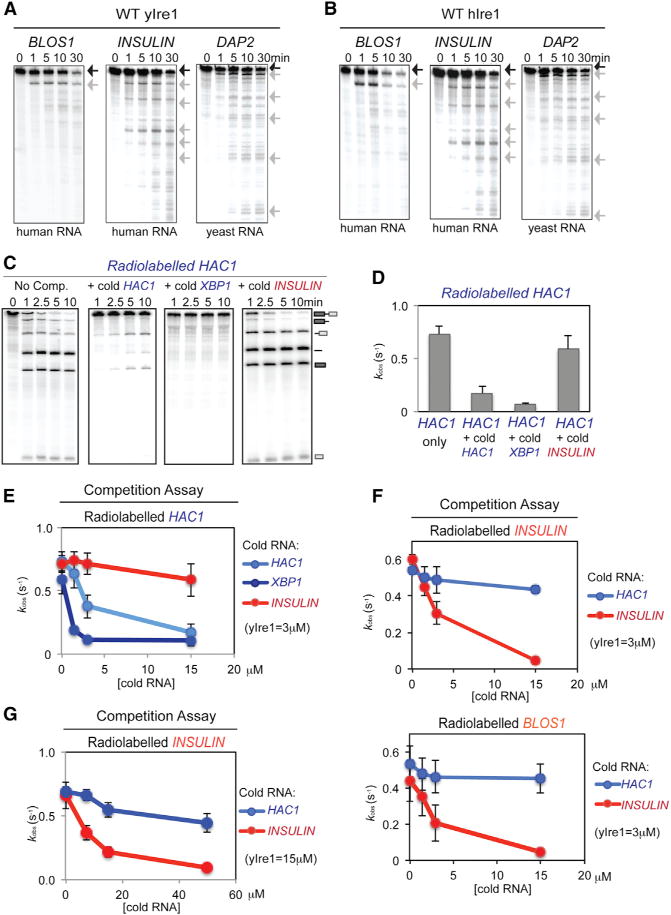
(A and B) In vitro cleavage of RIDD. In vitro transcribed radiolabeled mammalian (BLOS1, INSULIN) and yeast (DAP2) RNA substrates (0.5 nM) were incubated with ADP (2 mM) and 1 μM recombinant WT yIre1 (A) or WT hIre1 (B) for up to 30 min. Black arrow, full-length RNA; gray arrow, cleaved RNA fragments.
(C and D) Using single-turnover conditions (Figure S1F), HAC1 RNA cleavage reactions were competed by excess unlabeled (cold) HAC1 or XBP1 RNA (15 μM) but not with RIDD RNA (15 μM). Reactions were performed with 2 mM ADP for up to 10 min. (C) kobs calculated from these reactions are shown in (D). Error bars of all the experiments in this figure represent at least three independent repeats.
(E) HAC1 RNA cleavage reactions with different concentrations of various competitor RNAs (representative reactions shown Figures S1G and S1H).
(F) RIDD reactions are competed by RIDD substrate RNA but not by HAC1 RNA (representative reactions shown in Figures S1K and S1L).
(G) RIDD RNA cleavage competition reactions performed under single turnover conditions (Figure S2A) (representative reactions shown in Figure S2B).
XBP1/HAC1 RNA Cleavage Is Not Competed by RIDD Substrates
To determine the molecular relationship between IRE1-mediated cleavage of HAC1 and RIDD substrate RNAs, we performed in vitro yIre1 RNase reactions with radiolabeled RNA substrates in the presence of unlabeled (cold) RNA competitors. If IRE1 uses the same active site for binding and cleavage of both HAC1 and RIDD substrates, then both substrate RNAs should compete for the same active site. However, if the active sites are different, then HAC1 and RIDD RNA substrates should not compete equally well under single turnover conditions (Figure S1F). Cleavage of radiolabeled HAC1 RNA decreased upon addition of increasing amounts of cold HAC1 RNA, resulting in the accumulation of uncleaved radiolabeled HAC1 RNA (Figures 1C–1E and S1G). The addition of cold XBP1 RNA also resulted in decreased cleavage of radiolabeled HAC1 RNA (Figures 1C–1E, and S1H). Conversely, cold HAC1 also inhibited cleavage of radiolabeled XBP1 (Figures S1I and S1J). In contrast, addition of cold INSULIN RNA at a high concentration (15 μM) did not notably affect the cleavage of radiolabeled HAC1 RNA (Figures 1C–1E and S1G). The converse was also true: cleavage of radiolabeled INSULIN RNA was inhibited by excess amounts of cold INSULIN RNA but not HAC1 RNA (Figures 1F and S1K). Furthermore, the cleavage of another RIDD substrate, BLOS1 RNA, was also inhibited by excess amounts of cold INSULIN RNA (Figures 1F and S1L). During these competition experiments, we noted that overall RIDD cleavage (either with INSULIN or BLOS1 RNA) was rather less effective than HAC1 or XBP1 RNA cleavage, suggesting that 3 μM yIre1 might not truly represent a single turnover condition for RIDD cleavage. In fact, further experiments revealed that RIDD reaction was saturated at ~10 μM (Figure S2A). Thus, we performed a similar set of RIDD competition at yIre1 concentrations (15 μM) and found essentially the same results: cold INSULIN but not HAC RNA competed with INSULIN cleavage (Figures 1G and S2B). Taken altogether, these results suggest that Ire1 cleaves HAC1 or RIDD RNA substrates using two distinctive binding and/or catalytic sites.
To determine whether these two forms can interchange, we performed competition assays under steady-state conditions using a decreased amount of protein (0.1 μM). The addition of 0.15, 0.3, and 1.5 μM cold HAC1 RNA to radiolabeled HAC1 RNA resulted in dose-dependent inhibition (Figure S2C). In Lineweaver-Burk plot, we found that inhibition of radiolabeled HAC1 cleavage by cold HAC1 RNA showed competitive inhibition (Figures 2A and S2E). In contrast, we found that cold INSULIN inhibited the cleavage of radiolabeled HAC1 RNA (Figure S2D) in the mode of noncompetitive inhibition (Figures 2B and S2E), providing additional support that Ire1 has two distinct RNA substrate binding and/or catalytic sites for cleaving HAC1 or RIDD RNA substrates.
Figure 2. RIDD Requires the Nucleotide Binding in the IRE1 Kinase Domain and Exhibits Noncompetitive Inhibition with the HAC1 RNA Cleavage Reaction.
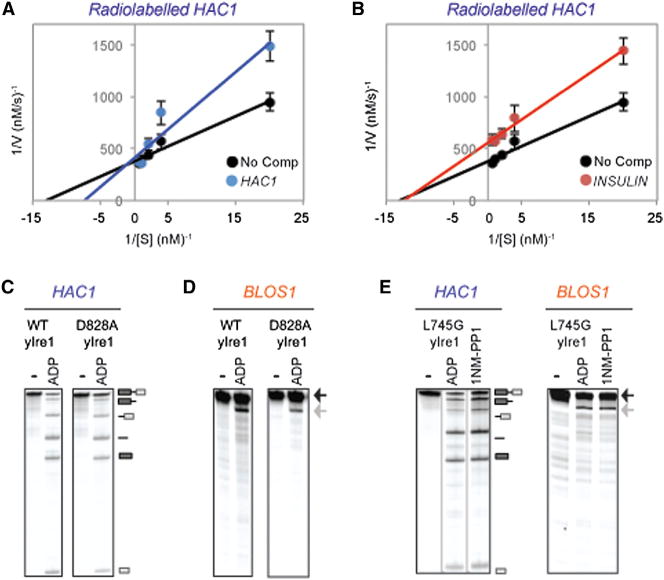
(A and B) Lineweaver-Burk plots of steady-state yIre1 RNase reactions cleaving radiolabeled HAC1 containing no competitor (black), cold HAC1 (blue) (A), or INSULIN RNA (red) (B). Error bars represent three independent repeats (representative reactions shown in Figure S2E).
(C and D) RNA cleavage reactions of HAC1 (C) or BLOS1 (D) RNA were performed with WT yIre1 or D828A yIre1 (1 μM) for 15 min. RNA cleavage fragments are indicated schematically.
(E) L745G yIre1 (1 μM) cleaves BLOS1 and HAC1 RNA in the presence of ADP or the nonhydrolyzable ATP homolog 1NM-PP1 (20 μM).
IRE1 Kinase Domain Requirements for RIDD
Results of competition experiments were unexpected and raised a question of what conferred the mechanistic differences of Ire1 cleaving XBP1/HAC1 or RIDD RNAs. Nucleotide binding is known to play a key role in activating Ire1 RNase for XBP1/HAC1 RNA cleavage (Lee et al., 2008). D828 in yIre1 is important for coordinating magnesium (Mg2+) in the active site, and a D828A yIre1 mutant is still able to bind ATP but is unable to transfer phosphates (Chawla et al., 2011). In agreement with previous work, WT and D828A yIre1 were able to cleave HAC1 RNA (Figure 2C) (Chawla et al., 2011). Similarly, we found that WT and D828A yIre1 were also able to cleave BLOS1 RNA (Figure 2D). The importance of nucleotide binding for both HAC1 and RIDD RNA cleavage was further confirmed by the use of the L745G mutant yIre1, which has an altered nucleotide binding pocket capable of binding the nonhydrolyzable ATP homolog 1NM-PP1 (Papa et al., 2003). Upon binding to 1NM-PP1, L745G yIre1 was capable of cleaving both HAC1 and BLOS1 RNA (Figure 2E), indicating that binding in the nucleotide pocket is sufficient for RIDD activation. Taken altogether, the importance of the nucleotide binding in the Ire1 kinase domain is the same for cleavage of both HAC1 and RIDD RNA and thus is unlikely to be the key determinant that differentiates HAC1 and RIDD substrate cleavage.
Differences in IRE1 Cooperativity Distinguish between XBP1/HAC1 and RIDD Substrate RNA Cleavage
The kinase domain of Ire1 is necessary for oligomer formation, an important step leading to XBP1/HAC1 splicing, and the oligomer of Ire1 can be monitored by foci formation using IRE1-GFP (Aragón et al., 2009; Ishiwata-Kimata et al., 2013; Korennykh et al., 2009; Li et al., 2010). At 1 hr after treating HEK293 cells with thapsigargin (Tg), a well-characterized UPR-activating drug that disrupts calcium levels in the ER, the IRE1-GFP foci were detected (Figure S3A). These foci became larger at 4 hr and then had dispersed by 8 hr (Figure S3A). Samples collected at the same time showed XBP1 splicing detectable within 1 hr of treatment and continuing for 4 hr before starting to decline (Figure S3B) as reported previously (Li et al., 2010). In contrast, significant levels of RIDD activity, which was calculated as a percentage of substrates cleaved (BLOS1 and SCARA3), did not appear until 2 hr after UPR induction and continued to increase throughout the time course (Figure S3B). At the 8 hr time point, RIDD activity was at its highest, and no IRE1-GFP foci were present (Figures S3A and S3B). Our finding in vitro that both XBP1 and RIDD substrate RNA cleavage occurred within a minute of incubation revealed that IRE1 was capable of cleaving either substrate with similar kinetics (Figures 1A, 1B, and S1C). The differences observed here in vivo might come from differential availabilities of these RNA substrates. However, both XBP1 and BLOS1 mRNA were associated with the ER membrane even prior to ER stress induction (Figure S3C). This suggests that a regulatory step(s) beyond RNA localization is unlikely to be responsible for generating the differences in activation kinetics of XBP1 splicing and RIDD in vivo. Although factors other than the RNA cleavage by IRE1, including the ligation step for XBP1 splicing or degradation of the cleaved fragments for RIDD, affect the appearance of the spliced form of XBP1 or overall levels of RIDD substrate mRNA in cells, these findings hinted that formation of IRE1 higher ordered structures, which correlate with XBP1 mRNA splicing, might not be necessary for RIDD activity, and warranted for further examination.
To investigate contributions of higher order structure of Ire1 in the XBP1 or RIDD RNA cleavage, we tested cooperativity of Ire1 and determined the Hill coefficient to be 2.13 ± 0.38 for yIre1-cleaving HAC1 RNA (Figures 3A and S3D) and similarly the Hill coefficient for hIre1 cleaving XBP1 RNA to be 3.07 ± 0.65 (Figures 3A and S3E), indicating that Ire1 RNase for HAC1 or XBP1 consists of Ire1 complex with the presence of cooperativity (Korennykh et al., 2009). In contrast, the Hill coefficients for the RIDD reactions were 1.15 ± 0.19 for yIre1 and 1.10 ± 0.29 for hIre1, indicating essentially no cooperativity (Figures 3A, S3D, and S3E). In addition, we found that the Hill coefficient value for a HAC1 substrate with only one cleavage due to a mutation at the 3′ splice site (Figure S3F) was similar to that for the WT HAC1 RNA, revealing that the number of cleavage sites does not matter for cooperativity.
Figure 3. Cleavage of RIDD Substrate RNA Does Not Require Cooperative IRE1 Oligomerization.
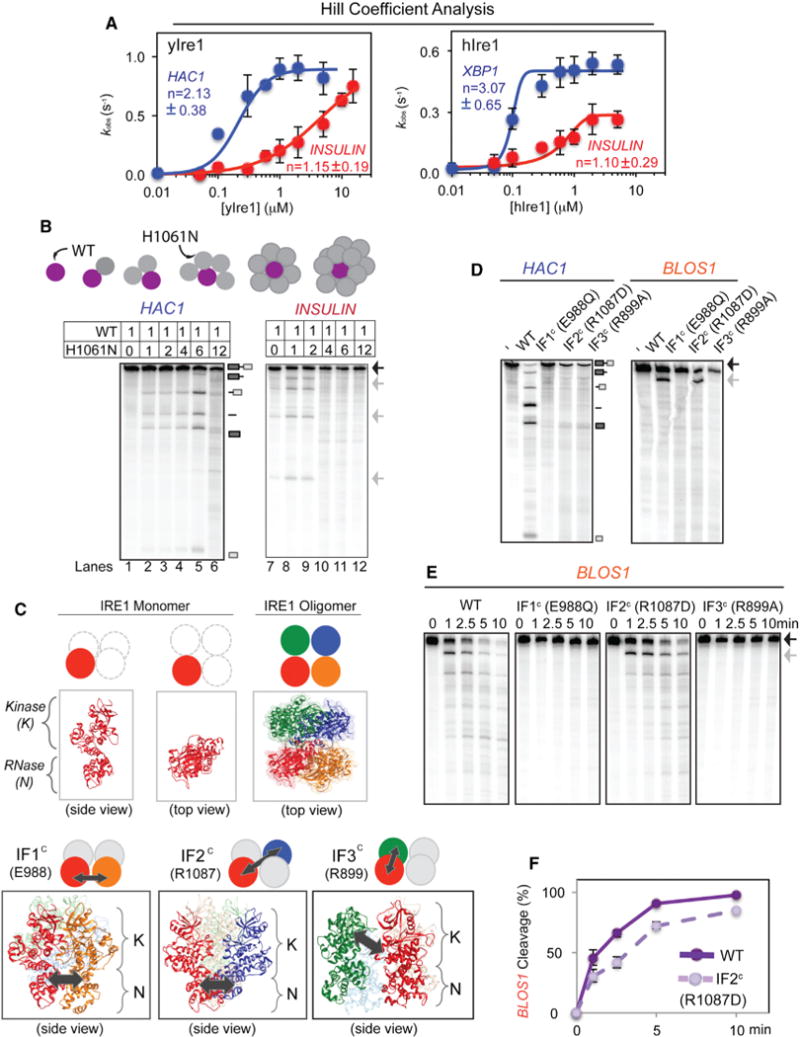
(A) Cooperative activation modes were observed for yIre1 and hIre1 cleaving HAC1 and XBP1 RNA, respectively, but not for cleaving INSULIN RNA. Cooperative Hill coefficients for yIre1/HAC1 (2.13 ± 0.38) and hIre1/XBP1 (3.07 ± 0.65) were found, while noncooperative values were found for yIre1/INSULIN (1.15 ± 0.19) and hIre1/INSULIN (1.10 ± 0.29). We interpret a noncooperative Hill coefficient of “1” as Ire1 acting as a dimer since unassociated, monomeric Ire1 is inactive (Lee et al., 2008), and the minimal active unit for Ire1 is a dimer (Figure S4A). All error bars in this figure were calculated from at least three independent experiments (representative reactions shown in Figures S3D and S3E).
(B) Transcomplementation assay between WT yIre1 and H1061N yIre1. WT yIre1 was kept at a concentration below the oligomerization threshold (0.083 μM) (Figure S3H) and inactive. Increasing amounts of catalytically inactive H1061N yIre1 at molar ratios from 1:0–1:12 ([WT]:[H1061N]) were added. A schematic of the experiment is shown in the top panel.
(C) Three interfaces in yIre1 oligomer (IF1c E988, IF2c R1087, and IF3c R899) previously identified from X-ray crystallographic structure studies (Korennykh et al., 2009). One monomer of IRE1 (red) can contact three different monomers (orange, blue, green) within a tetramer, leading to the three interfaces, IF1c, IF2c, and IF3c, respectively (bottom panel). Locations of IF1c (E988), IF2c (R1087), and IF3c (R899) interactions are shown (arrows).
(D) Nuclease reactions were performed with either WT yIre1 or interface mutants IF1c E988Q, IF2c R1087D, and IF3c R899A yIre1 (1 μM) for 10 min. (E and F) Time course of BLOS1 RNA cleavage reaction with WT yIre1 or yIre1 interface mutants (E) and the quantitation (F) of BLOS1 RNA cleavage activities for WT and IF2c yIre1.
In addition to Hill coefficients, we also performed transcomplementation assays using WT and H1061N (a catalytically inactive mutant) yIre1. Previous studies found that H1061 participates in the proton relay mechanism necessary for cleaving HAC1 RNA (Korennykh et al., 2011). A mutation at this site prevents cleavage of HAC1 RNA without disrupting yIre1 oligomer formation (Figures S3G and S3H) (Korennykh et al., 2011). Furthermore, we found that H1061N yIre1 was also inactive for RIDD substrate cleavage (Figure S3G). In the transcomplementation assay, we kept the concentration of WT yIre1 constant at 0.083 μM where yIre1 remained as a monomer upon performing the previously reported oligomerization assay (Figure S3H) (Korennykh et al., 2011). Furthermore, WT yIre1 showed very little cleavage activity for either HAC1 or RIDD substrates (Figure 3B, lanes 1 and 7). As increasing amounts of H1061N yIre1 were added, the heterocomplexes became active for HAC1 cleavage (Figure 3B, lanes 2–5), consistent as previously reported (Korennykh et al., 2011). However, once the concentration of H1061N yIre1 far exceeded that of WT yIre1, HAC1 RNA cleavage became inactive (Figure 3B, lane 6). In contrast, RIDD RNA cleavage was restored when H1061N yIre1 was added at an equal or 2-fold higher concentration of WT yIre1 (Figure 3B, lanes 7–9), but higher levels of H1061N yIre1 inhibited RIDD cleavage (Figure 3B, lanes 10–12).
Oligomerization State of IRE1 Can Distinguish between HAC1 and RIDD Substrate RNA Cleavage
These observations also provided a prediction that disruption of the interfaces within the yeast Ire1 oligomers has little impact on RIDD cleavage. To test this idea, we prepared three mutant forms of yeast Ire1 carrying a mutation at one of the three interfaces: IF1c Glu 988, IF2c Arg 1087, and IF3c Arg 899, generating E988Q, R1087D, and R899A yIre1, respectively (Korennykh et al., 2009) (Figure 3C). All three yIre1 interface mutants did not generate oligomers based on the oligomerization assay (Figure S3H) (Korennykh et al., 2009). In agreement with previous reports (Korennykh et al., 2009; Lee et al., 2008), we found that alterations in any of these residues inactivated HAC1 RNA cleavage (Figure 3D). In contrast, IF2c-R1087D yIre1 remained active for RIDD at the rate similar to WT yIre1 (Figures 3E and 3F), whereas E988Q and R899A yIre1 did not show RIDD cleavage (Figures 3D and 3E). R1087 is localized between the two RNase interface in the oligomer (Figure 3C), and the ability of R1087D yIre1 to retain the cleavage of RIDD RNA, but not that of HAC1 RNA, provided a further support for the different nature of RIDD RNase reactions.
Curiously, we found that both E988Q yIre1 and R1087D yIre1 were active kinases able to undergo autophosphorylation (Figure S3I) (Chawla et al., 2011; Han et al., 2009; Lee et al., 2008). E988Q yIre1 is an inactive RNase for both HAC1 RNA and RIDD, suggesting that activation of the kinase domain alone is not sufficient to generate an active Ire1 RNase. This is a notable result as the current model proposes that Ire1 initially forms “face-to-face” dimers where the nucleotide binding pockets are facing each other within Ire1 dimer (Figure S4A), and structural studies reveal that the two RNase domains are not in close proximity (Figure S4A) (Ali et al., 2011). Presumably, formation of dimers in a “back-to-back” form where the nucleotide binding pockets are facing away from each other brings two RNase domains together to create a catalytically active RNase (Lee et al., 2008). The back-to-back dimer is present in the crystalized oligomer that corresponds to the IF1c interface (compare Figure S4A [back-to-back dimer] with Figure 3C [IF1c]) (Korennykh et al., 2009). Recently, quercetin (Q) has been shown to induce the formation of yIre1 back-to-back dimers by binding to the Q site, an interface between the kinase and RNase domain (Figure S4A) (Wiseman et al., 2010). Q binding can occur independent of Ire1 kinase domain and activates Ire1 RNase for cleavage of HAC1 RNA (Figures 4A and S4B) (Wiseman et al., 2010). Similarly, we found that the binding of Q to Ire1 was sufficient to promote cleavage of RIDD substrate RNA (Figures 4A and S4B). Given that RIDD does not require oligomer formation (Figures 3A and 3D–3F), these results suggest that the Ire1 back-to-back dimer itself is sufficient for RIDD cleavage.
Figure 4. IRE1 RIDD Contributes to Cell Death.
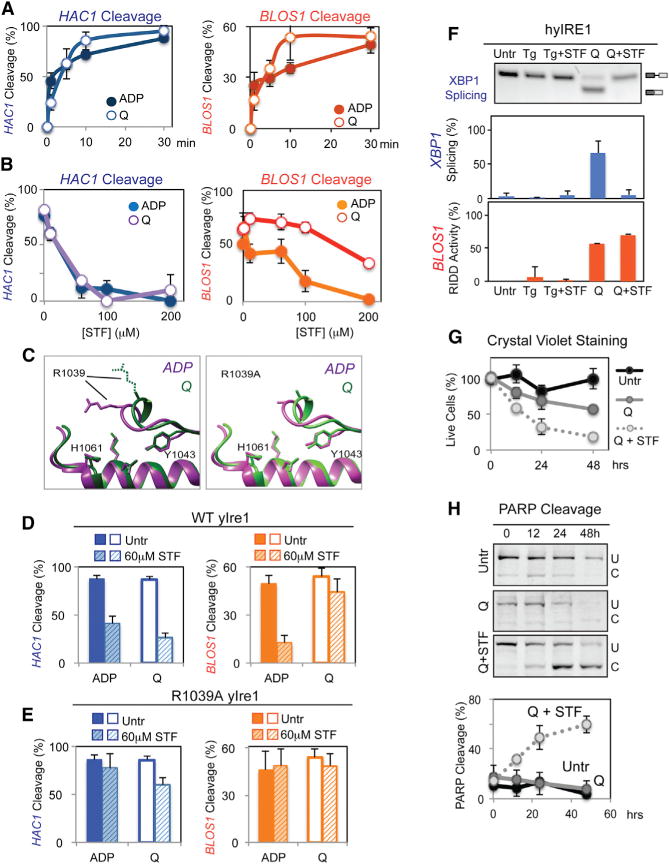
(A) Cleavage of HAC1 (left) or BLOS1 RNA (right) with WT yIre1 (1 μM) activated with ADP (2 μM) (closed circle) or Q (100 μM) (open circle). Error bars in this figure were calculated from at least three independent experiments.
(B) Nuclease reactions were performed with varying concentrations of STF-083010 (STF), an inhibitor, on WT yIre1 (1 μM) activated with either ADP (2 μM) (closed circles) or Q (100 μM) (open circles).
(C) The core RNase catalytic center between oligomerized Ire1, the ADP activated form “ADP” (purple) (Korennykh et al., 2009) and Q activated “Q” (green) yIre1 (Wiseman et al., 2010) is similar at the molecular level, except for the spatial orientation of the R1039 residue. The right panel shows structure predictions of R1039A yIre1 activated by ADP (purple) or Q (green).
(D and E) HAC1 or RIDD cleavage by (D) WT yIre1 (1 μM) or (E) R1039A yIRE1 was activated by Q (100 μM) or ADP (2 μM) with and without STF (60 μM) for 30 min.
(F) STF inhibited XBP1 mRNA splicing but not RIDD for Q-activated hybrid IRE1 (hyIRE1) in vivo with or without STF (60 μM) for 2 hr.
(G and H) Preferential inhibition of XBP1 splicing but not RIDD in hyIRE1 induced (G) cell death and (H) apoptosis.
To further understand the mechanistic differences in Ire1 for HAC1 RNA and RIDD cleavage, we examined the effect of the Ire1 RNase specific inhibitor STF-083010 (STF) on yIre1 activated by ADP or Q. We found that HAC1 cleavage by ADP- or Q-activated yIre1 was effectively inhibited by STF, both with an IC50 around 30 μM (Figure 4B) (Papandreou et al., 2011). STF also potently inhibited cleavage of RIDD substrates when yIre1 was activated by ADP (Figures 4B [orange closed circles] and 4D). In contrast, however, STF was not able to inhibit Q-induced RIDD as effectively as ADP activated RIDD even at higher concentration (Figure 4B [compare open with closed orange circles] and 4D). Structural comparisons of ADP-bound Ire1 (Figure 4C, in purple) (Korennykh et al., 2009) and Q-bound Ire1 (in green) (Wiseman et al., 2010) revealed that the core catalytic residues, including H1061 and Y1043, remain unchanged but that the orientation of R1039 side chain became significantly different (Figure 4C, left). R1039 has been identified as a HAC1 RNA binding residue (Korennykh et al., 2011). In fact, we found that altering R1039 to Ala in yIre1 diminished RNase activity for HAC1 RNA as anticipated, but R1039A yIre1 remained active for the cleavage of BLOS1 RNA similarly to WT yIre1 (Figure S4C), revealing that RIDD cleavage reaction does not engage R1039 residue as it does for HAC1 RNA cleavage. Furthermore, R1039A yIre1 became insensitive to STF for the cleavage of both HAC1 and BLOS1 RNA (Figures 4E and S4D). Taken together, these observations suggest that R1039 residue of yIre1 is involved in HAC1 RNA cleavage but does not play a significant role in the RIDD.
Differential Activation of IRE1 Determines Cell Fate Decisions
Preferential inhibition of Q-activated Ire1 cleavage of HAC1 RNA but not RIDD substrates would provide a useful tool to dissect functional consequences of these two activities of Ire1. Q was not effectively taken up by yeast cells (Wiseman et al., 2010), which precluded the use of yeast for our analyses. Elegantly, a previous report demonstrated that a chimeric hybrid IRE1 (hyIRE1) where a human luminal domain was fused with the yeast cytosolic domain could be activated by Q when expressed in ire1 knockout mouse embryonic fibroblasts (MEFs) (Wiseman et al., 2010). Using hyIRE1, we confirmed that hyIRE1 was not activated when Tg was used to induce ER stress, as reported previously (Figure 4F; Tg and Tg + STF). In contrast, incubation of hyIRE1 cells with Q activated hyIRE1 RNase for both XBP1 RNA splicing (Q, blue bar) and RIDD cleavage (Figure 4F; Q, orange bar). Furthermore, STF treatment inhibited XBP1 splicing (Figure 4F; Q + STF, blue bar), whereas cleavage of BLOS1 RIDD RNA occurred normally even in the presence of STF (Figure 4F; Q + STF, orange bar), recapitulating our in vitro results (Figures 4B and 4D).
Thus, we tested the functional consequence of XBP1 mRNA splicing inhibition, but leaving RIDD intact by examining the extent of cell death. Upon treatment of hyIRE1 cells with Q and STF, numbers of cells stained with Crystal violet were reduced (Figure 4G), and PARP cleavage was elevated (Figure 4H) when compared with those treated with Q alone, revealing that RIDD activation without XBP1 splicing induces more predominant cell death. Similarly, we also tested the effect of STF on Q-activated mammalian cells carrying the endogenous WT IRE1 (Figures S4F–S4K), as we found that Q was able to activate hIre1for both XBP1 RNA and BLOS1 RNA cleavage in vitro (Figure S4E). Preferential inhibition of XBP1 splicing but not RIDD brought by Q and STF resulted in more diminish numbers of crystal violet stained cells and increased PARP cleavage, in comparison to both XBP1 splicing and RIDD activation in Q-treated cells (Figures S4I and S4J). Finally, experiments with ire1 knockout MEFs conferred that the increase in apoptotic events observed in WT MEFs was an IRE1-dependent event (Figures S4K–S4M). Altogether, these findings, consistent with previous studies (Han et al., 2009; Upton et al., 2012), indicate that activation of RIDD without XBP1 RNA promotes cell death.
DISCUSSION
Our data suggest that activated IRE1 RNase has different mechanisms for cleaving XBP1/HAC1 RNA or RIDD substrates. Specifically, a catalytically active IRE1 unit engaged in HAC1 or XBP1 mRNA splicing is generated within the IRE1 oligomer, while IRE1 engaged in RIDD resides within an IRE1 monomer/dimer. An active catalytic core for XBP1/HAC1 mRNA cleavage is unlikely to consist of all subunits within the IRE1 oligomer, but rather, oligomerization will lead to formation of the catalytically active pocket by establishing a specific orientation of catalytic residues. The structural conformation of the RNA binding and/or RNase catalytic residues within the catalytic core unit must differ such that HAC1 or XBP1 RNA cannot compete the cleavage reaction of RIDD substrates and vice versa under single turnover conditions (Figures 1C–1G). Consistent with this idea, the Hill coefficient for IRE1 engaged in HAC1 or XBP1 RNA cleavage showed cooperativity among IRE1 subunits, while IRE1 engaged in RIDD cleavage displayed no significant cooperativity and may not form higher order structures as it does not require the IF2c (R1087) interface (Figures 3D–3F). It is possible, however, that IRE1 engaged in RIDD could also exist within an oligomer without any cooperative impact from other IRE1 subunits in the complex.
Structural and biochemical studies have revealed that the H1061 residue of yeast IRE1 plays a critical role in catalysis of HAC1 cleavage (Korennykh et al., 2011). Similarly, we found that H1061 was also important for RIDD substrate cleavage (Figures 3B and S3G). Previous studies have also demonstrated that R1039 residue is involved in binding to HAC1 RNA (Korennykh et al., 2011). Mutation of R1039 to Alanine did not affect ability of yIre1 to cleave RIDD substrate, while it decreased that of HAC1 RNA (Figure S4C). Curiously, comparisons of Ire1 structure bound to Q highlight a difference in the spatial orientation of R1039 residue (Figure 4C). The ability of an IRE1 RNase inhibitor STF-083010 (STF) to inhibit HAC1 RNA cleavage occurred regardless of the side-chain orientation of R1039, while the cleavage of BLOS1 RNA by Q-induced yIre1, but not by ADP-induced yIre1, was no longer inhibited by STF-083010 (Figures 4B and 4D). Together with the noncompetitive inhibition between RIDD and HAC1 substrates under steady-state competition experiments, these results point that binding sites for XBP1/HAC1 RNA and RIDD substrate may differ, while both reactions share a catalytic site, including H1061 residue. Future work will require understanding of how RIDD substrates bind to IRE1 at the molecular level.
Notably, our model for IRE1 activation is different from what has previously been proposed (Han et al., 2009) where higher ordered structures were assigned to the RIDD active form of IRE1. Experiments reported by Han et al. (2009) utilized murine I642G IRE1, a homolog of L745G yeast IRE1, that binds to a modified nucleotide, 1NM-PP1. Upon expressing I642G IRE1 in INS-1 cells, the addition of 1NM-PP1 (without ER stress induction) activated XBP1 mRNA splicing but not RIDD. In INS-1 cells expressing WT IRE1, instead of I642G IRE1, both XBP1 mRNA splicing and RIDD occurred. Since I642G IRE1 does not autophosphorylate, it was concluded that I642G hIRE1 was unable to perform RIDD due to the lack of both phosphorylation and its accompanying oligomerization. However, we found that L745G yIre1 itself, a yeast homolog of I642G IRE1, was active for RIDD in vitro upon binding to 1NM-PP1 (Figure 2E), revealing that a lack of RIDD may not be an intrinsic property of L745G (or I642G) IRE1. Further studies have also reported that I642G IRE1 is active for RIDD upon addition of 1NM-PP1 (Upton et al., 2012) and with ER stress when introduced into ire1−/− cells (Hollien et al., 2009). These experiments suggest that, in addition to the occupancy of the kinase nucleotide-binding pocket, the ER stress-induced conformational change of the cytosolic portion of IRE1 containing both kinase and RNase domains holds a key to activation of I642G IRE1. The importance of the conformational change(s) through the ER luminal, transmembrane, and linker domains for activation of IRE1 RNase has previously been described (Credle et al., 2005; Korennykh et al., 2009; Volmer et al., 2013; Zhou et al., 2006). Such conformational changes may also trigger oligomerization and ultimately full activation of IRE1. In vitro, L745G yIre1 binding to 1NM-PP1 caused a mobility shift to heavier fractions on a density gradient sedimentation assay similar to ADP-bound WT yIre1 (Papa et al., 2003), revealing that L745G yIre1 is capable of forming a higher order structure similar to WT yIre1 without the ER luminal domain.
Additionally, in these previous experiments (Han et al., 2009), I642G IRE1 was introduced into INS-1 cells that express the endogenous WT IRE1. Here, we demonstrated that even a catalytically inactive H1061N yIre1 could reconstitute WT yIRE1 present below its active concentration. Importantly, we demonstrate that at higher molar ratios of WT to mutant IRE1, HAC1 cleavage but not RIDD can be reconstituted (Figure 3B), suggesting that overexpressing mutant IRE1 may favor XBP1 splicing over RIDD. Finally, our results revealed that the HAC1 RNA cleavage activity of yIre1 is intrinsically more active than RIDD (Figures S1F and S2A), and thus, comparisons of activities between different forms of IRE1 should be performed carefully. Future work will require more understanding of I642G/L745G mutant IRE1.
This report shows that RIDD can occur in yeast, S. cerevisiae. We have previously reported that the only RNA cleaved by IRE1 in yeast is HAC1 RNA (Niwa et al., 2005). However, the yeast recombinant IRE1 used in the previous study differed slightly from the one used in this study, where the linker domain was shortened in the yIre1 used for this study. We and others have noted that yIre1 with the full-length linker domain is less active for HAC1 RNA cleavage (Korennykh et al., 2009). In addition, yIre1 with the full-length linker domain does not show significant RIDD cleavage activity. The molecular reasons for this observation are not clear, but it is in agreement with recent studies highlighting the importance of the linker region (Volmer et al., 2013).
Since IRE1 exhibits dual RNase activities, this calls into question the role of RIDD in vivo. Previously, contributions of RIDD versus XBP1 mRNA splicing in vivo were assessed by comparing ire1 knockout and xbp1 knockout cells with a rationale that subtracting the XBP1 contribution in xbp1 knockout cells would allow assessment of RIDD functions (Hur et al., 2012). However, since both unspliced and spliced XBP1 mRNA is not present in xbp1 knockout cells, results from this approach also include contributions from an absent unspliced XBP1 protein and changes in transcription due to a lack of XBP1, not simply from RIDD activation alone. Furthermore, while no additional IRE1 splicing substrate mRNA beyond XBP1 has been identified, splicing of such RNA would take place normally in xbp1 knockout cells. Initially, activation of the IRE1 branch of the UPR was thought to be protective by virtue of XBP1 splicing (Lin et al., 2007). However, our finding that RIDD promotes cell death highlights the importance of re-evaluating the functional significance of IRE1 activation. In S. pombe, where either HAC1 or XBP1 is absent, making RIDD the sole function for IRE1, a recent report revealed that IRE1 cleaves KAR2/BiP, a homolog of GRP78, within its 3′ UTR as a RIDD substrate (Kimmig et al., 2012), resulting in increased translation, rather than degradation. In addition, the kinetics or intensity of RIDD versus XBP1/HAC1 splicing may differ depending on the cell or tissue type and the nature of the ER stress and ultimately may change the overall outcomes. Thus, careful evaluation of both the mechanistic and functional consequences between RIDD activation versus XBP1/HAC1 splicing will provide a greater understanding of how ER stress affects cell physiology and also provide rationales for development of drugs and more effective treatment strategies for ER stress-related diseases.
EXPERIMENTAL PROCEDURES
In vitro transcription of IRE1 substrates, protein purification, and in vitro nuclease assay; determination of Hill coefficient; and determination of XBP1 splicing and RIDD in vivo are described in the Supplemental Experimental Procedures.
Competition Assays and Michaelis-Menten Kinetics
For single-turnover conditions, reactions were set up by incubating 3 μM of yIre1 (Figures 1E and 1F) or 15 μM yIre1 (Figure 1G) with buffer followed by addition of unlabeled (cold) RNA at the indicated concentrations. To this reaction mixture, 0.5 nM (Figures 1E and 1F) or 0.25 nM (Figure 1G) of radiolabeled RNA substrate were added and incubated at 30°C for 5 min. Reactions were then started upon addition of 2 mM ADP and stopped at the indicated times, as described in in vitro nuclease assay (Supplemental Experimental Procedures). For steady-state competition, reactions were performed similarly, except that 0.1 μM yIre1 was used and the concentration of radiolabeled RNA substrate was varied from 0.05–1.50 nM, as indicated and competed with 1.5 μM cold RNA. Samples were analyzed on a 6% urea gel, and molar values for uncleaved and cleaved products were calculated as described above. Velocity (nM/s) was calculated with the slope of the linear regression line of the graph plotting cleaved substrate (nM) over time (s). To generate the Lineweaver-Burk plots, the reciprocal of substrate concentration was plotted against the reciprocal of the velocity. Linear regression trend lines were then generated and graphed in Figures 2A and 2B.
trans-Complementation Assay
For transcomplementation assays, in vitro nuclease reactions were set up with nuclease reaction buffer, 2mM ADP, and 0.083 μM of WT yIre1. At this concentration of WT yIre1, efficient cleavage of either HAC1 or RIDD RNA did not take place (Figure 3B, lanes 1 and 7). H1061N yIre1 was added to the reaction at molar ratios of 1:1, 1:2, 1:4, 1:6, or 1:12 ([WT]:[H1061N]). Reactions were started upon addition of 0.5 nM radiolabeled substrate and proceeded for 10 min at 30°C.
Supplementary Material
Highlights.
Cleavage of XBP1/HAC1 intron by IRE1 is a distinct and separable activity from RIDD
Oligomerization of active IRE1 is required for XBP1/HAC1 cleavage but not RIDD
Both activities use the same catalytic residues but different substrate binding sites
Selective activation of RIDD promotes cell death, whereas XBP1/HAC1 splicing supports survival
Acknowledgments
We are grateful to Drs. Douglass Forbes, Peter E. Geiduschek, and Andrew Shiau for many discussions throughout the course of this study and Drs. Forbes and Geiduschek for comments on the manuscript. This work was supported by NIH RO1GM087415 and the ACS 118765-RSG-10-027-01-CSM to M.N., PO1 CA-67166 to A.K., and NIH T32, UCSD/LIAI allergy postdoctoral training grant for A.T.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.09.016.
References
- Ali MM, Bagratuni T, Davenport EL, Nowak PR, Silva-Santisteban MC, Hardcastle A, McAndrews C, Rowlands MG, Morgan GJ, Aherne W, et al. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 2011;30:894–905. doi: 10.1038/emboj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, Walter P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–740. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Chawla A, Chakrabarti S, Ghosh G, Niwa M. Attenuation of yeast UPR is essential for survival and is mediated by IRE1 kinase. J Cell Biol. 2011;193:41–50. doi: 10.1083/jcb.201008071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez TN, Sidrauski C, Dörfler S, Walter P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 1999;18:3119–3132. doi: 10.1093/emboj/18.11.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur KY, So JS, Ruda V, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V, Iwawaki T, Glimcher LH, Lee AH. IRE1a activation protects mice against acetaminophen-induced hepatotoxicity. J Exp Med. 2012;209:307–318. doi: 10.1084/jem.20111298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwata-Kimata Y, Promlek T, Kohno K, Kimata Y. BiP-bound and nonclustered mode of Ire1 evokes a weak but sustained unfolded protein response. Genes to cells: devoted to molecular & cellular mechanisms. 2013;18:288–301. doi: 10.1111/gtc.12035. [DOI] [PubMed] [Google Scholar]
- Kawahara T, Yanagi H, Yura T, Mori K. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol Biol Cell. 1997;8:1845–1862. doi: 10.1091/mbc.8.10.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmig P, Diaz M, Zheng J, Williams CC, Lang A, Aragón T, Li H, Walter P. The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. eLife. 2012;1:e00048. doi: 10.7554/eLife.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, Stroud RM, Walter P. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh AV, Korostelev AA, Egea PF, Finer-Moore J, Stroud RM, Zhang C, Shokat KM, Walter P. Structural and functional basis for RNA cleavage by Ire1. BMC Biol. 2011;9:47. doi: 10.1186/1741-7007-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Dey M, Neculai D, Cao C, Dever TE, Sicheri F. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Korennykh AV, Behrman SL, Walter P. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc Natl Acad Sci USA. 2010;107:16113–16118. doi: 10.1073/pnas.1010580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Liang FX, Wang X. A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol Cell. 2014;55:758–770. doi: 10.1016/j.molcel.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiba K, Nagashima Y, Suzuki E, Hayashi N, Ogata Y, Shimada Y, Koizumi N. Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc Natl Acad Sci USA. 2013;110:5713–5718. doi: 10.1073/pnas.1219047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Patil CK, DeRisi J, Walter P. Genome-scale approaches for discovering novel nonconventional splicing substrates of the Ire1 nuclease. Genome Biol. 2005;6:R3. doi: 10.1186/gb-2004-6-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa FR, Zhang C, Shokat K, Walter P. Bypassing a kinase activity with an ATP-competitive drug. Science. 2003;302:1533–1537. doi: 10.1126/science.1090031. [DOI] [PubMed] [Google Scholar]
- Papandreou I, Denko NC, Olson M, Van Melckebeke H, Lust S, Tam A, Solow-Cordero DE, Bouley DM, Offner F, Niwa M, Koong AC. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L, Truitt M, McManus MT, Ruggero D, Goga A, et al. IRE1a cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338:818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volmer R, van der Ploeg K, Ron D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc Natl Acad Sci USA. 2013;110:4628–4633. doi: 10.1073/pnas.1217611110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman RL, Zhang Y, Lee KP, Harding HP, Haynes CM, Price J, Sicheri F, Ron D. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol Cell. 2010;38:291–304. doi: 10.1016/j.molcel.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu CY, Back SH, Clark RL, Peisach D, Xu Z, Kaufman RJ. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci USA. 2006;103:14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


