Abstract
The M protein of coronavirus plays a central role in virus assembly, turning cellular membranes into workshops where virus and host factors come together to make new virus particles. We investigated how M structure and organization is related to virus shape and size using cryo-electron microscopy, tomography and statistical analysis. We present evidence that suggests M can adopt two conformations and that membrane curvature is regulated by one M conformer. Elongated M protein is associated with rigidity, clusters of spikes and a relatively narrow range of membrane curvature. In contrast, compact M protein is associated with flexibility and low spike density. Analysis of several types of virus-like particles and virions revealed that S protein, N protein and genomic RNA each help to regulate virion size and variation, presumably through interactions with M. These findings provide insight into how M protein functions to promote virus assembly.
Keywords: Cryo-electron microscopy, Cryo-electron tomography, Pleomorphic virus structure, Coronavirus, Viral matrix protein
1. Introduction
Every type of virus architecture has its own structural constraints. In other words, there is a limit to the variation in shape, size or protein configuration that can be realized by a particular set of structural proteins. When this tolerance is exceeded, the result becomes uncertain: it is possible that the assembly process may fail, produce misshapen but otherwise infectious particles, or yield non-infectious particles. Pleomorphic enveloped viruses represent the most extreme cases of natural variation during assembly. Pleomorphic virions can vary considerably in size, as in the case of arenaviruses (Neuman et al., 2005) or shape, as in the case of influenza A virus (Harris et al., 2006). Although this variation has been documented, little is known about how the assembly components shape the overall particle architecture. Our understanding of virus assembly has practical implications: a new generation of HIV-1 assembly inhibitors acts at the level of particle architecture by reducing the fidelity of the assembly process (Tang et al., 2003) or by blocking connections between Gag proteins (Sticht et al., 2005).
In this study we have chosen to analyze the relationship between composition and architecture for three pleomorphic coronaviruses: Mouse hepatitis virus (MHV), Severe acute respiratory syndrome coronavirus (SARS-CoV) and Feline coronavirus (FCoV). Recent electron microscopy studies have confirmed that coronavirus particles vary considerably in size, and so can safely be described as pleomorphic. However, there is disagreement over the extent of variation in virion shape (Barcena et al., 2009, Beniac et al., 2006, Neuman et al., 2006, Risco et al., 1996), although a range of morphologies is represented in each study. These three coronaviruses make an interesting dataset because each is built from a conserved set of components, but amino acid identity between the homologous structural proteins is typically less than 30%.
Four structural proteins are important for coronavirus infectivity: the integral membrane protein M adapts a region of membrane for virus assembly and captures other structural proteins at the budding site, the N protein chaperones and protects the viral RNA genome, spikes consisting of three copies of the S glycoprotein promote receptor-binding and membrane fusion, and the small membrane protein E is present in sub-stoichiometric amounts and acts as an enhancer of budding (Hogue and Machamer, 2007). In this study, we will focus on the role of M in assembly and in determining particle morphology.
M proteins from MHV (Klumperman et al., 1994, Rottier and Rose, 1987), FCoV (Klumperman et al., 1994), SARS-CoV (Nal et al., 2005), Infectious bronchitis virus (Machamer and Rose, 1987), Transmissible gastroenteritis virus, (TGEV) (Klumperman et al., 1994) and Bovine coronavirus (Nguyen and Hogue, 1997) are targeted to the vicinity of the Golgi apparatus. Reverse genetic studies and VLP assembly studies suggest that M protein promotes assembly by interacting with viral ribonucleoprotein (RNP) and S glycoproteins at the budding site (de Haan et al., 1999, Escors et al., 2001a, Escors et al., 2001b, Kuo and Masters, 2002, Narayanan et al., 2000, Nguyen and Hogue, 1997, Opstelten et al., 1995, Sturman et al., 1980), and by forming a network of M–M interactions that is capable of excluding some host membrane proteins from the viral envelope (de Haan et al., 2000, Neuman et al., 2008b). M proteins interact through both the transmembrane domain and endodomain (de Haan et al., 2000). M can also interact with RNA that carries the genomic packaging signal (Narayanan et al., 2003). Coronavirus assembly is then completed at the membrane of a pre-Golgi compartment, as shown most recently in a tomography study of intracellular structures involved in virus replication and assembly (Knoops et al., 2008). Packets of virions are then shuttled out of the cell along the secretory pathway (reviewed in (Hogue and Machamer, 2007)). The minimum requirement for MHV virus-like particle (VLP) production is co-expression of M and E proteins (Vennema et al., 1996), although in some expression systems, the additional co-expression of N increases the efficiency of VLP production (Boscarino et al., 2008).
Recent studies have begun to reveal the structure of the coronavirus pre-fusion spike (Li et al., 2005), N protein (Chen et al., 2007, Fan et al., 2005, Huang et al., 2004, Jayaram et al., 2006, Saikatendu et al., 2007, Schutze et al., 2006), the hemagglutinin-esterase protein, which is found on some group 2 coronaviruses (Zeng et al., 2008), and the E protein (Pervushin et al., 2009). Also, transmembrane features have been identified as M on SARS-CoV, MHV, FCoV and TGEV particles using cryo-electron microscopy (Neuman et al., 2006) and cryo-electron tomography (Barcena et al., 2009), but the structure of M remains poorly characterized. The lack of detailed structural and functional information is largely due to its small size, close association with the viral envelope and a tendency to form insoluble aggregates when perturbed (Lee et al., 2005).
In this study we have attempted to provide a better understanding of the structure and function of M protein. First, we have used cryo-EM and tomography to probe the structure of M in the envelope of MHV, SARS-CoV and FCoV virions. Second, we have analyzed the structure of the MHV M protein in VLPs that lack S and RNP. This is because identification of M in electron micrographs of virions is complicated by the presence of transmembrane regions of spikes and RNP, and, more importantly, intermolecular interactions could potentially affect coronavirus morphology, including M–M, M–E, M–N, M–S, M–RNA and N–RNA interactions (reviewed in (Hogue and Machamer, 2007)), palmitin-mediated interactions involving S and E (Boscarino et al., 2008, Lopez et al., 2008, Thorp et al., 2006), and envelope stretching caused by the packaged helical ribonucleoprotein (Barcena et al., 2009). Together, these experiments reveal new facets of the structure and function of M, and demonstrate how pleomorphicity can be harnessed to reveal the function of membrane protein networks.
2. Materials and methods
2.1. Virus preparation
Growth, purification and imaging of SARS-CoV from Vero-E6 cells, FCoV from AK-D cells and MHV-OBLV60 (Gallagher et al., 1991) from DBT cells has been described previously (Neuman et al., 2006). A small plaque mutant derived from MHV-A59, called MHV-sp1, was isolated by plaque purification from the medium from persistently infected 17 clone 1 (17cl-1) cells that had survived infection with MHV-A59 and had been passaged more than 40 times (Sawicki, 1987). The MHV-sp1 S glycoprotein is not cleaved at the S1-S2 boundary and it is resistant to cleavage by trypsin, unlike the S-glycoprotein of MHV-A59. MHV-sp1 was purified by centrifugation. First, the virus from 450 ml of infected cell supernatant (∼5 × 1011 pfu) was pelleted by centrifugation for 3 h at 24,000 rpm at 4 °C using an SW28 rotor. The pellet was allowed to dissolve on ice overnight in buffered saline (0.15 M NaCl, 20 mM HEPES, pH 6.8). The 6 ml of the suspended virus was layer on top of linear gradient of 40% (w/w) potassium tartrate, 20 mM HEPES, pH 7.4 (bottom) to 15% (w/w) glycerol, 20 mM HEPES, pH 7.4 (top) and subjected to isopycnic centrifugation in a SW28 rotor (3 h at 24,000 rpm at 4 °C). The resulting milky band of virus was diluted with buffered saline and pelleted by centrifugation for 3 h at 24,000 rpm at 4 °C using an SW28 rotor. The pellet was allowed to dissolve on ice overnight in 1 ml of buffered saline.
2.2. VLP and SUV100 preparation
VLPs were produced by transfecting HEK-293T cells with pCAGGS expression vectors encoding M, E and N from MHV-A59 (Lokugamage et al., 2008). Briefly, flasks of near-confluent cells comprising approximately 1.5 m2 of total culture area were transfected with plasmid DNA using Lipofectamine and “Plus” reagent (Invitrogen) according to an appropriately scaled version of the manufacturer’s protocol. Virus-like particles were precipitated from culture medium 48 h after transfection using 10% polyethylene glycol and 2.2% w/v NaCl, then further purified by 10–30% sucrose density gradient ultracentrifugation as described previously (Neuman et al., 2008a).
Production and cryo-EM of unilamellar phospholipid vesicles (SUV100) created from a blend of 80 parts 1,2-dioleoyl-sn-glycero-3-phosphocholine, 10 parts 1,2-dioleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)], 2 parts 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[biotinyl(polyethylene glycol)2000], and 1 part 3,3′-dioctadecyl-oxacarbocyanine perchlorate has been described previously (Kunding et al., 2008).
2.3. Cryo-EM and tomography
Cryo-EM was done using standard low-dose imaging conditions. Images of MHV-OBLV60, VLPs and FCoV were recorded using Leginon (Suloway et al., 2005). Cryo-electron tomography of MHV-sp1 was done using a JEOL JEM-2200FS microscope with energy filter operated at 200 kV and 25,000 times magnification. Specimens were tilted along one axis through 140°, from +70° to −70°, and images were recorded to a 4 × 4 k CCD at a nominal resolution of 5.3 Å per pixel. Tomographic reconstruction was performed using the imod software suite (Kremer et al., 1996) by fiducial alignment of 10 nm gold particles in tilted images. Two-fold binning was performed at the time of imaging to produce a model with a calibrated resolution of 10.6 Å per pixel. Details of the equipment and conditions used in conventional cryo-EM are provided in Table 1 .
Table 1.
Microscopy conditions and particle count.
| Microscope | Imaging | kV | Defocus (μm) | Å/pixel | Images | Particles | Vesicles | |
|---|---|---|---|---|---|---|---|---|
| SUV100 | CM-120 | 1 k CCD | 120 | a | 7.5 | 36 | - | 479 |
| SARS-CoV | CM-120 | Scanned film | 120 | 2.3–5.1b | 1.8 | 16 | 609 | 10 |
| FCoV | Tecnai F20 | 4 k CCD | 120 | 2.3–4.3b | 2.3 | 140 | 466 | 25 |
| EM | Tecnai F20 | 4 k CCD | 120 | 1.8–2.8 | 2.3 | 46 | 107 | 85 |
| EMN | Tecnai F20 | 4 k CCD | 120 | 1.7–3.0 | 1.4–2.3 | 139 | 1094 | 15 |
| MHV-OBLV60 | Tecnai F20 | 4 k CCD | 120 | 2.0–3.3 | 2.3 | 74 | 338 | 16 |
| MHV-sp1 | JEM-2200FS | 4 k CCD | 200 | 3.0 | 10.6 | 3c | 206 | 0 |
Not reported.
Conventional cryo-EM images taken at a defocus of −3.3 μm or closer to focus were used for two-dimensional image analysis.
Denotes complete tomograms.
2.4. Image analysis
Before analysis, image contrast was inverted so that protein density appeared white rather than black. We corrected for the effects of phase reversal in the contrast transfer function using the EMAN module ctfit (Ludtke et al., 1999). The images used for two-dimensional reconstructions displayed Thon rings, which indicate the presence of image data, to 8–16 Å resolution. Micrographs were filtered in Fourier space to truncate high-frequency image data beyond the last visible Thon ring using the EMAN module proc2d. We then selected small images showing M using the EMAN module boxer.
Image clustering and refinement of class averages was performed using the startnrclasses and classalign2 modules. Full contrast transfer function correction was implemented during construction of class averages using classalign2. The clearest, most coherent images of M densities were obtained when correction was applied through 17 Å resolution.
For radial density analysis to assess the location of S, M and the RNP inside the virion, quadrants of particles were selected to minimize the distortion caused by small variations in curvature. Quadrants were selected for inclusion in the final average based on clarity and contrast in the envelope region. For radial density analysis of MCOMPACT and MLONG, radial density maps were constructed by selecting and aligning numerous small wedges centered at the particle edge, which were then averaged. Radial density analysis was performed using the SPIDER image analysis suite (Frank et al., 1996), and was normalized to the brightest and darkest datapoints, which were assigned relative brightness values of 100% and 0%, respectively. The signal from the envelope region was used as a fiducial mark for normalization of image intensity and alignment of radial density profiles.
2.5. Measurement and categorization
Two observers recorded two sets each of orthogonal measurements of the longest and shortest visible diameter of each virion or VLP, so that contributions of observer bias and measurement variation could be factored into our analysis. Average diameter (d AVG) was taken as a measure of particle size, and a ratio of the longest to the shortest visible diameter (d MAX/d MIN), was taken as a measure of particle shape (Supplementary Fig.S1). Diameters were measured to the outer edge of the membrane, leaving out spikes. Particles were excluded if part of the membrane was outside the captured image area, or if the membrane overlapped with the carbon support layer or another particle. Multilamellar, overlapping or tubular SUV100 particles, and exosomal vesicles that appeared to have inner contents or membrane-embedded proteins were also excluded.
To assess the precision of our measurements, the distance between lipid bilayer headgroup densities was measured repeatedly by two observers. The distance between the brightest parts of the lipid bilayer on vesicles matched expected results to within 1 nm (Nagle and Tristram-Nagle, 2000). Measurements of the longest and shortest diameter of vesicles were less precise, perhaps due to combined errors in measurement and identification of the maximum and minimum particle diameter (see Supplementary Fig.S2).
In order to assess whether virus size and shape varied from one preparation to another, we compared cryo-EM images of four preparations of MHV-OBLV60 and one preparation of MHV-sp1. MHV-OBLV60 virions were fixed with 1% phosphate-buffered formalin (pH 7.0) before imaging; MHV-sp1 virions were not fixed because of differences in local biosafety regulations between EM facilities. Particle diameter was similar for all five MHV preparations (Supplementary Fig.S3). The shape of particles in two OBLV60 preparations was significantly more elongated than the other three, so these particles were excluded from shape analysis.
Before marking particles as spike-depleted or containing an envelope thickness anomaly, we revised our criteria empirically by repeatedly examining images of SARS-CoV particles and tracking the agreement between observers. The criteria were revised until inter-observer agreement was consistently greater than 80%. The final criteria for categorization were: “Does closely-packed spike decoration extend around at least half of the particle edge?” and, “Is an abnormally thin region of no less than one-twelfth the virion circumference visible in the M density layer?” Statistics describing inter-observer agreement are presented in Supplementary Fig.S2.
2.6. Cloning and expression of M107–221
Complementary DNA was generated by reverse transcriptase PCR, and a fragment encoding amino acids 107-221 of the SARS-CoV-Tor2 M-protein was cloned into pET46Ek/LIC (Novagen, USA). The vector encodes an N-terminal poly-histidine tag and a flanking enterokinase cleavage site for tag removal. Expression of M107–221 was achieved in BL21 (DE3) E. coli (Invitrogen, USA) and induced at an OD600 of 0.6–0.8 with 1 mM IPTG followed by overnight growth at 18 °C. The majority of the expressed protein was present as insoluble inclusion bodies which were subsequently refolded.
2.7. Purification and refolding
Bacteria were lysed at 4 °C using an EmulsiFlex® C-3 cell disruptor (Avestin, Canada) at 15 kpsi in lysis buffer (20 mM Tris–HCl, 100 mM sodium chloride, 10 mM dithiothreitol, 1% Triton X-100 at pH 7.0). The insoluble fraction containing the inclusion bodies was isolated by low-speed centrifugation. Pellets containing the inclusion bodies were resuspended in lysis buffer and then pelleted by centrifugation three times to remove any soluble material that was present. The pellets of insoluble material were then washed three times, as above, with wash buffer (20 mM Tris–HCl, 100 mM sodium chloride, 10 mM dithiothreitol at pH 7.0) to remove residual detergent. The resultant white pellet was dissolved in guanidine buffer (6 M guanidine hydrochloride, 50 mM Tris–HCl, 10 mM dithiothreitol, 1 mM EDTA at pH 8.0) to a final concentration of 15 mg/ml as determined by Bradford protein assay (BioRad, USA).
Solubilized M107–221 inclusion bodies were refolded using the rapid dilution method. Solubilized inclusion body protein was rapidly added dropwise to refolding buffer (50 mM HEPES, 200 mM sodium chloride, 1 M NDSB-201, 10 mM beta-mercaptoethanol at pH 7.0, 4 °C) to give a final protein concentration of 0.1 mg/ml. The protein was allowed to refold for 1–3 h before being applied to a 5 ml nickel affinity column (GE Healthcare, USA). The column was washed with five column volumes of binding buffer (50 mM HEPES, 200 mM sodium chloride, 10 mM beta-mercaptoethanol at pH 7.0) and refolded protein was eluted in binding buffer supplemented with 250 mM imidazole and 1 mM EDTA. Further purification was achieved by passing 2 ml samples of the eluted protein over a pre-equilibrated Superdex™ 75 16/60 size exclusion column (20 mM HEPES, 200 mM sodium chloride, 5 mM dithiothreitol at pH 7.0). The purified monomeric protein was concentrated to 1.8 mg/ml using 0.5 ml Ultrafree Biomax 5 kDa concentrators (Millipore, USA). The protein was >95% pure as assessed by SDS–PAGE. Purified protein was stored in 20 mM HEPES, 200 mM sodium chloride, 5 mM dithiothreitol at pH 7.0.
2.8. Protein analysis
The stoichiometry of expressed M endodomain was assessed using perfluoro-octanoic acid polyacrylamide gel electrophoresis (PFO-PAGE; (Ramjeesingh et al., 1999)). To do this, 20, 10, 5 or 2 μl of purified protein was made up to 20 μl total by adding 20 mM HEPES, 200 mM sodium chloride, 5 mM dithiothreitol, pH 7.0. Samples were incubated at either 4 or 37 °C for 1 h then separated by electrophoresis on precast 4–20% acrylamide gels in Tris–glycine buffer containing 0.5% (wt/vol) PFO. Protein was detected by SYPRO-ruby staining (Invitrogen).
3. Results
3.1. Structural analysis of coronavirus particles and VLPs
The main problem with using cryo-EM to investigate protein structure is limited resolution, which usually does not allow for the structure to be interpreted by an atomic model (Stewart and Grigorieff, 2004). As a consequence, it can be difficult to identify small features such as M in cryo-EM images.
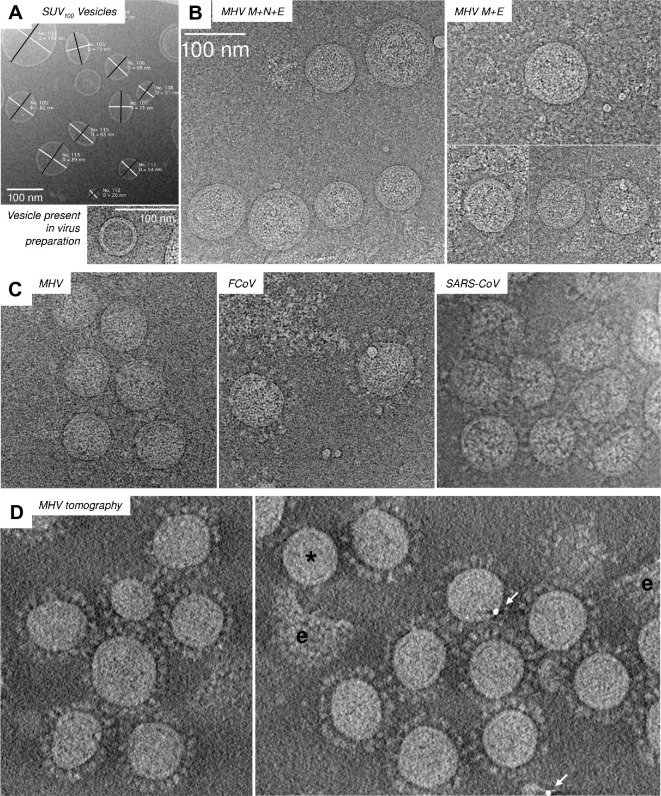
Previous studies have used what is known about protein size, topology and function to infer the structure of spikes and RNP (Barcena et al., 2009, Beniac et al., 2006, Cavanagh, 1983, Davies and Macnaughton, 1979, Neuman et al., 2006, Risco et al., 1996). However, none of these studies produced a clear view of M, which is obscured in micrographs by the viral membrane, the C-termini of S proteins and the RNP. To better understand the structure and organization of M, we examined by cryo-EM (Fig.1 A–C) and cryo-electron tomography (Fig.1D), coronavirus particles, vesicles, VLPs containing only M and E, and VLPs containing M, E and N proteins. In purified virus preparations we also found a small number of apparently protein-free empty vesicles, which presumably were released from infected cells, and which we refer to in this study as exosomes. Viruses and VLPs had thicker envelope regions than vesicles or exosomes due to the presence of M protein, as previously reported (Barcena et al., 2009, Neuman et al., 2006). The purification process used for MHV-sp1 also produced some free, spike-decorated envelopes. Free envelopes appeared thicker than ordinary phospholipid bilayers, suggesting that M was still present, but did not contain any trace of the RNP core (Fig.1D).
Fig. 1.
Coronavirus particles and vesicles in vitreous ice. Cryo-electron micrographs of small unilamellar vesicles (A), MHV-like particles (B), three coronaviruses (C) and cryo-electron tomography of MHV (D) are shown. The longest and shortest visible diameter of each particle was measured, as shown in panel A. Viral particles were distinguished from empty exosomes by the thickness of the envelope. A spikeless particle (∗) and free viral envelopes (e) are marked. Gold particles which were used as fiducial markers in construction of the tomogram are marked with arrows.
3.2. Extent of M inside the virion
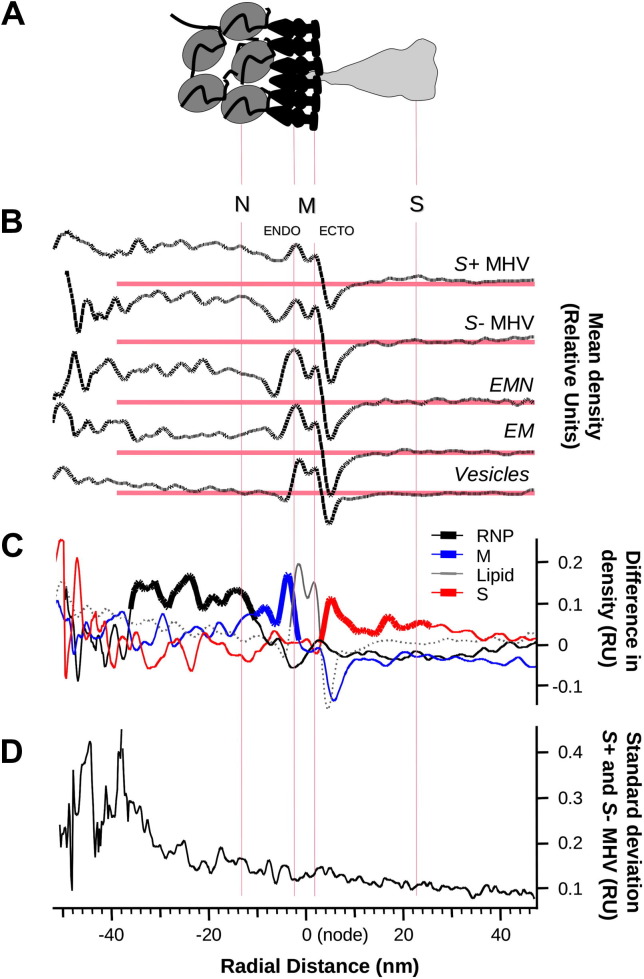
To determine the boundaries of packaged M (Fig.2 A), we examined radial density maps from groups of 18–22 empty exosomes, EM and EMN VLPs, low-spike and normal virus particles, as shown in Fig.2B. To identify individual components, we subtracted one set of appropriately scaled and aligned radial density data from another, as shown in Fig.2C.
Fig. 2.
Identification of M and ribonucleoprotein in viral particles. (A) A model viral envelope explains features of an MHV particle by comparing the (B) averages of ∼20 radial density distributions from MHV virus-like particles, apparently spike-depleted virions, normal virions and empty exosomal vesicles. (C) Individual features are mapped onto the radial density pattern of empty vesicles as follows: M protein (blue) is revealed as the difference between EM VLPs and vesicles, S (red) is the difference between normal and spikeless virions and ribonucleoprotein is the difference between EM VLPs and spikeless virions. (D) Differences in the consistency of core and envelope organization are revealed from the standard deviation of 40 MHV radial density plots.
From these data we determined that the endodomain of M extended inward ∼8 nm from the highest-density point of the outer membrane leaflet, in agreement with published measurements of MHV M protein from cryo-EM and tomography (Barcena et al., 2009, Neuman et al., 2006). The ectodomain of M did not give rise to a detectable radial density signal, suggesting that it may either be disordered or tightly membrane-associated. An area of consistent, high density which extended from 10 to 30 nm inside the particle was attributed to RNP, consistent with recent measurements by Barcena et al. (2009). Fig.2D shows that the variability in radial density was low from the virus exterior through the RNP feature, but innermost parts of the virion were highly variable. Taken together, these data show the extent of M and M-linked RNP inside the particle.
3.3. Appearance of packaged M protein
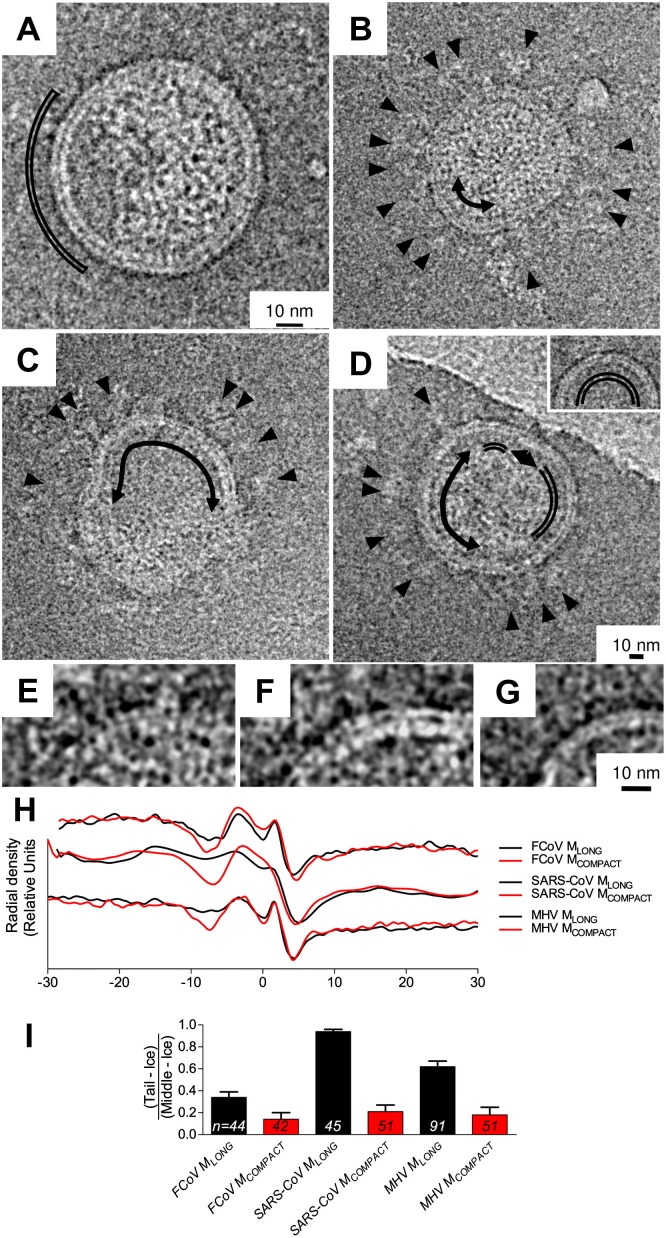
Viral particles were examined closely to identify individual M protein shapes. On most particles, M at the virion edge resembled tightly packed lines crossing the membrane and contacting the RNP (Fig.3 ). Small M-free areas which may represent budding scars were occasionally visible at the edge of virions and VLPs (Fig.3A). The observation that M remained tightly packed even on particles with excess M-free membrane provides structural validation of the existence of M–M interactions. The observation that M densities appear to make contact with the RNP core likewise can be taken as evidence of M–N or M–RNA interactions.
Fig. 3.
Evidence for two forms of M protein in virions. Distinct rod-like MLONG features can be seen in at the edge of cryo-EM images of MHV EMN VLPs (A) and feline coronavirus (B–F). Regions where M appears indistinct, compact and does not appear to contact the ribonucleoprotein at the particle edge are marked with curved arrows. Regions of membrane that resemble protein-free lipid bilayers (D, inset and G) are marked with a double-line. Enlargements of MLONG (E), MCOMPACT (F) and a vesicle (G) are shown below. Radial density maps (H) and M tail to body ratios (I) are shown to illustrate the difference in appearance between MLONG and MCOMPACT from three coronaviruses.
On a few particles, some of the M-like protein had a compact, blurred appearance and did not appear to make contact with the RNP. We have called the common form MLONG and the short, blurred form MCOMPACT. Viral spikes can be seen on both MLONG and MCOMPACT, but not on M-free membranes (Fig.3B–D). This was taken as further evidence of M–S interactions and confirmation that MCOMPACT is likely a form of M, perhaps an alternate conformer. Enlarged images showing the features we have termed MLONG, MCOMPACT and M-free membrane are shown in Fig.3E–G. Radial density maps showing the difference in appearance between MLONG and MCOMPACT are presented in Fig.3H.
In order to express M conformation numerically, we performed radial density analysis of small sections at the particle edge where MLONG or MCOMPACT was visible. The M endodomain density consisted of the tail region, extending 6 to 8 nm into the particle where the differences between M conformations were most apparent, and the denser body region, extending 3.5 to 5.5 nm into the particle. After subtracting the density of the ice outside the particle to correct for differences in background and scaling image brightness to a common mean, the ratio of M tail to middle was calculated for each virion or VLP. We found that the density in the “tail” region of MCOMPACT was similar to the density of the background ice, and the peak M endodomain was higher than for MLONG. To express M conformation more simply, we subtracted the background from the tail and body densities and calculated tail to body ratios for MLONG and MCOMPACT (Fig.3I). We found that the difference in tail to body ratios was statistically significant (T-test, P = 0.014 for FCoV, P = 2.0 × 10−17 for SARS-CoV and P = 3.0 × 10−6 for MHV). This suggests that the two forms of M represent different conformations of the same peptide chain, and the distribution of density at the virion edge can be used as an approximate readout for M conformation.
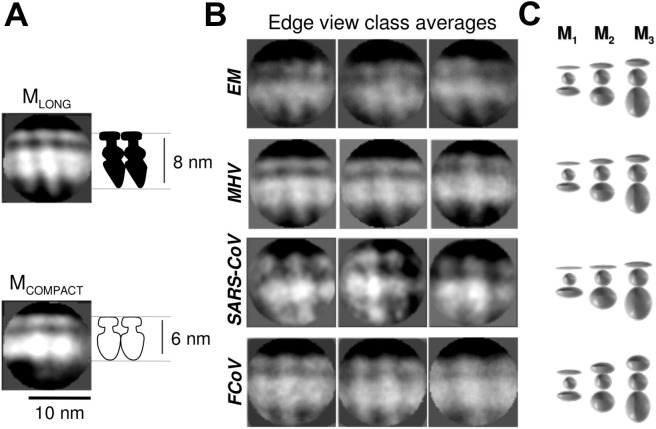
We next examined M proteins in more detail by classifying, aligning and averaging similar-looking regions from particle edges to produce class averages. MLONG resembles a dagger, with the ectodomain “pommel” resting on the outer membrane leaflet and the tip of the endodomain “blade” extending ∼8 nm and contacting the RNP (Fig.4 A-B). The endodomains of adjacent MLONG particles appeared to contact each other, suggesting that MLONG–MLONG interactions are mediated by the endodomain of M. MLONG on EM VLPs appeared similar to M on virions, thus confirming that MLONG is formed by M in the absence of the other high-copy virion proteins S and N (Fig.4B). Some class averages of MLONG also showed a distinct tilt relative to the membrane. The endodomain of MLONG was resolved into two globular components with crisp borders between adjacent molecules, while the endodomain of MCOMPACT appeared as an indistinct ellipsoid and extended only ∼6 nm (Fig.4A). The two globular components of the endodomain were not distinctly observed in class averages from EM VLPs (Fig.4B), suggesting that either the conformation is only formed in the presence of N or that N forms part of the two-lobed MLONG. MLONG and MCOMPACT were also visible in virtual slices through tomographic reconstructions of MHV. The difference in apparent length was most evident on tomographic slices through free envelopes, which showed thick convex MLONG membranes near the center giving way to thinner variably curved MCOMPACT membranes near the edges (Fig.5 A and B).
Fig. 4.
Edge views of packaged M. Reference-free class averages were made from groups of 300–2000 images clipped from viral particle edges. Full contrast transfer function correction through 17 Å resolution was implemented during class average reconstruction. Edge view class averages show MLONG (A, top) and MCOMPACT (A, bottom) from MHV EMN VLPs. Examples of class averages showing MLONG from EM VLPs and virions are shown in panel B. Panel C shows two-dimensional projections of M ectodomain, transmembrane domain and endodomain as ellipsoids with the width of MLONG from class averages and the expected volume of one, two or three copies of M protein (right).
Fig. 5.
Association of MLONG with convex, spike-decorated membranes. Panels A and B depict free viral envelopes that were found in MHV tomograms. Projections through thick (50 nm) and thin (1 nm) subtomograms are shown for comparison. Thicker regions of the membrane ascribed to MLONG are marked black, and thinner regions ascribed to MCOMPACT are marked white. Visible spikes are marked with circles to demonstrate membrane topology and intact virions (V) are also indicated.
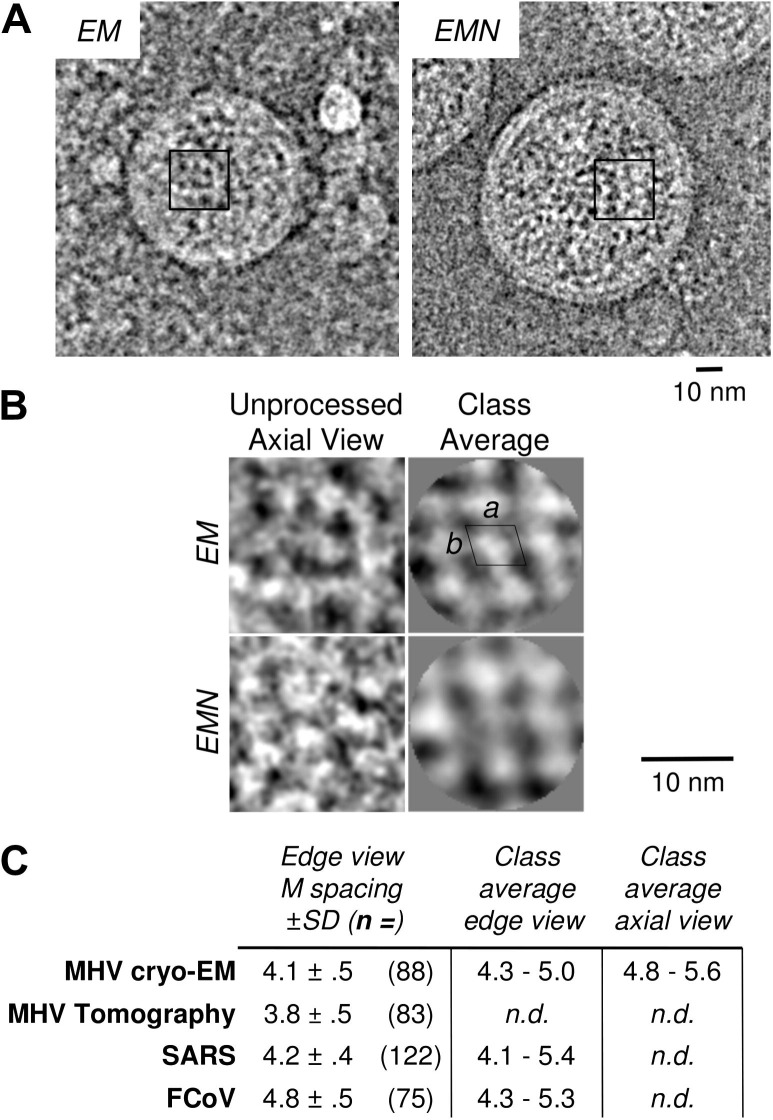
MLONG was spaced 4 to 5 nm apart in unprocessed cryo-EM images and edge view class averages (Fig.6 ). In axial views of M taken from the centers of EM and EMN VLPs, M sometimes appeared to form small ordered regions (Fig.6A) which could be classified, aligned and averaged to produce class averages (Fig.6B). M spacing in axial views was generally consistent with spacing in edge views (Fig.6C). While axial class averages suggest that M packing generally approximates a rhombus with sides of 4.0 and 4.5 nm and an interior angle of about 75°, the amount of heterogeneity in the unprocessed images is more consistent with a loosely-ordered network of M, as opposed to a rigid two-dimensional protein lattice.
Fig. 6.
Organization of M. Small areas which appear to show axial views of small clusters of organized M proteins are marked in images of VLPs (A), and enlarged below (B, left). Reference-free class averages were made from groups of 1500 (EM) or 3000 (EMN) images clipped from VLP centers. Full contrast transfer function correction through 17 Å resolution was implemented during class average reconstruction. Unit cell dimensions are approximately 4 nm (a) by 4.5 nm (b), with an interior angle of 75°. (C) Average spacing of M dimers as measured by various methods.
3.4. Oligomerization of M
Edge view class averages of MLONG from MHV, FCoV and SARS-CoV virions appeared about twice as large as expected for a single copy of the M protein, based on the partial specific volume of folded protein (Harpaz et al., 1994). This can be seen by comparing class averages showing M (Fig.4B) with projections of ellipsoids with the same spacing as M, and the volume of one, two or three copies of M, (Fig.4C). This suggested that each MLONG and MCOMPACT is large enough to contain at least two copies of M protein.
We next attempted to investigate the stoichiometry of purified M protein biochemically. Attempts to express SARS-CoV M protein in E. coli and baculovirus expression systems produced only insoluble aggregates. The largest M construct which we were able to express and purify consisted of residues 107 to 221 from SARS-CoV M, which covers the area of the endodomain from just after the third predicted transmembrane region to the C-terminus. M107–221 was expressed as an aggregate initially, but became soluble upon refolding.
Since a fusion protein incorporating the endodomain of MHV M was shown to co-immunoprecipitate with full-length M protein (de Haan et al., 2000), and class averages suggested that adjacent MLONG dimers make contact via two parts of the endodomain, we decided to investigate the oligomerization of M107–221 as a surrogate for full-length M. Soluble endodomain incubated in saline at 37 °C was present as monomers and aggregates as well as units of two, four and six, while endodomain incubated at 4 °C remained mostly monomeric (Supplementary Fig.S4). These observations support the interpretation that M functions as a homodimer.
3.5. Determinants of particle shape
In solution, the shape of a vesicle is determined by the interplay of opposing forces (Zhong-can and Helfrich, 1987). The ground state for vesicles in solution is spherical. Forces imparted by fluid motion or sample freezing in preparation for cryo-EM can temporarily distort vesicle shape. However, as a particle becomes less spherical the effects of hoop stress will increase, leading either to structural failure of the vesicle wall or driving the vesicle back to a more toward spherical, lower-stress shape.
Since enveloped virus particles and are essentially protein-decorated vesicles, we reasoned that their shape should be similar to the shape of vesicles unless modified by the effects of viral protein interactions. To test whether viral proteins significantly affected the shape of viral particles, we compared the shape of virions, VLPs and small unilamellar vesicles of the same size (SUV100) in cryo-EM images.
In our images, vesicles ranged from round to ellipsoidal, with the average particle having a long axis 1.08 times the length of the shortest axis (Supplementary Fig.S1). The shape of MHV and EM VLPs did not differ significantly from the shape of vesicles. However, EMN VLPs were significantly rounder than the combined population of SUV100 and endosomal vesicles (ANOVA, P = 0.005). Although EM particles were rounder on average than EMN particles or vesicles, the difference was not significant due to the small sample size and heterogeneity of the EM particles. This indicates that the protein component of EMN VLPs provides significant resistance to the deformative forces that affected vesicles under cryo-EM conditions.
We next sought an explanation for how E, M and N proteins can make spherical vesicles more rigid. N proteins can dimerize (Chen et al., 2007) and pack helically in the presence (Barcena et al., 2009) and absence (Saikatendu et al., 2007) of RNA, but neither of these interactions alone would be expected to produce a spherical arrangement. E protein is small and not abundant enough in viral particles to be a convincing explanation for overall shape. In contrast, M is abundant, interacts with other M proteins and with membranes, which are approximately spherical. We therefore hypothesized that particle shape is controlled primarily through M.
A striking difference between MLONG and MCOMPACT is that MLONG appears to contact the RNP while MCOMPACT does not. In cryo-EM images, the internal RNP appeared to be pulled away from the particle edge where either MCOMPACT or M-free membrane is present, as seen in Fig.3. Thus, MCOMPACT and M-free membranes can both be viewed as local disruptions of the M–RNP interaction. To test the hypothesis that M-mediated interactions control particle shape, we compared the shape of particles in which MLONG appeared to form a complete ring at the particle edge to the shape of particles in which the ring of MLONG was interrupted by either a M-free membrane or MCOMPACT.
Interruption of MLONG at the particle edge was associated with significant particle elongation in SARS-CoV (ANOVA, P = 5 × 10−12), FCoV (P = 0.01) and EM (P = 0.03) particles. A similar but non-significant trend was observed for MHV (Fig.7 A). While a relative increase in M anomalies was correlated with particle elongation, we noted that: The relative abundance of particles with interrupted M was quite different in the three coronaviruses, while the frequency of M anomalies was similar in MHV, EM and EMN particles. This result shows that an uninterrupted layer of MLONG is a marker for spherical morphology, but suggests that there may be inherent differences in the interaction affinities and membrane-bending properties of different coronavirus M proteins.
Fig. 7.
Relationship between M conformation and particle shape. (A) The ratio of the longest to the shortest diameter of coronavirus particles from MHV, SARS-CoV and FCoV micrographs and MHV tomograms is plotted against the percentage of particles which had at least one anomalously thin region of membrane. Each datapoint presents the average for 20 particles of similar shape. (B) Boxes were centered on the membrane at each end (stars) and side (circles) of seventy SARS-CoV particles to examine the relationship between M-form and membrane curvature. (C) The ratio of MLONG to MCOMPACT is plotted against average particle shape. Each datapoint represents two sides or ends from fourteen particles of similar shape. (A, C) Curves were fitted by logistic regression.
If only MLONG is associated with coronavirus-like membrane curvature, it is implicit that the ratio of MLONG to MCOMPACT should be related to local membrane curvature. To test this, we selected four parts of each particle edge, which were centered on either side of the longest and shortest particle diameter as shown in Fig.7B. Two independent observers counted the number of M dimers present and marked them as MLONG or MCOMPACT. The ratio of MLONG to MCOMPACT was about the same at all of the virion ends, but the proportion of M marked as MLONG on the flatter sides decreased as particles became more elongated (Fig.7C). From these results we conclude that clusters of MLONG mark round membranes.
We noted that some samples of purified MHV appeared quite ellipsoidal while others were mostly round, while virion size was similar in all of the MHV preparations (Supplementary Fig.S3). Purification over sucrose density gradients would at least temporarily expose viral particles to osmotic stress. Likewise, centrifugation would necessarily expose virus particles some mechanical stress. We suspected that in some cases the purification process might change virion shape.
3.6. Conversion between MLONG and MCOMPACT
To investigate whether virion shape could be changed experimentally after purification, one sample of purified virus was resuspended in HEPES-buffered 0.9% saline buffer, pH 7. Half the sample was kept in the resuspension buffer, while the other half was acidified to pH 5 for 5 min in order to simulate pH changes that might occur during entry via the endosomal route, and then re-buffered to pH 7. The shape and size of native MHV (Prep 1 in Supplementary Fig.S3) and acid-pulsed MHV (Prep 3) were examined by cryo-EM. Overall particle shape did not change significantly as a result of acidification (ANOVA, P = 0.93). However, the relative proportion of particles which appeared to have at least one flattened edge increased significantly after acid-treatment (Fisher’s exact test, P = 0.0001; Fig.8 A–C). In some flat-edged particles, the interior RNP appeared to have a crisp edge, which was more distant from the membrane than RNP in native particles and M had the blurred appearance characteristic of MCOMPACT (Fig.8B). Radial density plots and M tail to body ratios demonstrate that the altered M which was more frequently found at flat edges of acid-treated particles strongly resembled MCOMPACT from native MHV particles (Fig.8D and E). From this we concluded that transient acidification can partly convert MLONG to MCOMPACT, but other factors that occur during purification can have a more dramatic effect on particle shape.
Fig. 8.
Conversion between MLONG and MCOMPACT. Purified MHV was imaged by cryo-EM after incubation at pH 7 (A), or a 5 min pulse at pH 5 followed by re-neutralization to pH 7 (B). Arrows in panel B point to flattened edges of the particle with the appearance of MCOMPACT. The proportion of flat-sided virions before and after acidification is shown in table form (C). Radial density maps (D) and M tail to body ratios (E) of the altered regions from acidified MHV, native EM VLPs and native EMN VLPs are shown alongside MHV MLONG and MCOMPACT data from Fig. 3h-i.
We next investigated how the expression of N affected M conformation. We compared M tail to body ratios for whole MHV VLPs to regions of MHV MLONG and MCOMPACT and found that M in EMN VLPs was nearly identical to MLONG from native MHV (T-test, P = 0.94), while M from EM VLPs was intermediate between MLONG and MCOMPACT (Fig.8E). This suggests that intracellular N plays an important role in the formation or packaging of MLONG.
3.7. Relationship between spikes and morphology
Previous studies have shown that spikes are dispensable for assembly but essential for infectivity. It was therefore surprising to find that spikeless EM and EMN VLPs were significantly larger than virions (Supplementary Fig.S1) and rare viral particles on which no spikes were visible were significantly larger than spike-decorated virions (Fig.9 A), suggesting that spike incorporation was linked to envelope size. Since the size of the virus envelope is fixed at the scission stage of the budding process, we inferred that spike incorporation is linked to factors which produce small virions.
Fig. 9.
Relationship of S protein to morphology. (A) The ratio of the longest to the shortest diameter of coronavirus particles from micrographs is plotted against the percentage of particles which had fewer than half the expected number of spikes visible at the particle edge. Each datapoint depicts the average and standard deviation for a group of 20 similar-sized particles. Curves were fitted by logistic regression to datasets in which the association between spike decoration and size was statistically significant (ANOVA, P = 6.71 × 10−8 for FCoV and 0.046 for MHV). (B) SARS-CoV particles were classified according to shape and spike distribution. The three shape categories refer to particles with dMAX/dMIN less than 1.1 (round), 1.1–1.4 (slightly oval) or greater than 1.4 (elongated). The number of particles of each type that were classified is listed.
To determine which factors were linked to particle size, we measured MHV VLPs in cryo-EM images and MHV virions in tomograms, where spike decoration could be assessed in three dimensions. MHV EM VLPs (93 nm average diameter) were larger than EMN VLPs (91 nm) and spikeless MHV in tomograms (91 nm) and spike-decorated MHV (88 nm). Viral particle size decreased as particles approached the full complement of proteins and RNP (Supplementary Fig.S1), suggesting that small virions are produced as a result of the interplay between all viral components during assembly.
A few SARS-CoV particles appeared to have clusters of spikes at one or two spots on the viral envelope. Spike clusters were significantly associated with the curved ends of ellipsoidal particles (Fig.9B), which we had previously found to be marked by MLONG. We therefore concluded that MLONG is a marker of spike decoration.
4. Discussion
Here we demonstrate that a network of M has intrinsic membrane-bending properties, as recently demonstrated for the HIV-1 Gag polyprotein (Carlson et al., 2008). We report that M is functionally dimeric in viral particles, and the membrane-altering properties of M depend on interactions with other viral components. Our analysis suggests that two types of M–M interactions should be considered in future structural studies: interactions which maintain the M dimer and may occur throughout the protein, and interactions between dimers which are probably mediated by the endodomains, which form a matrix-like layer underneath the membrane. In several ways, the function of coronavirus M appears be analogous to that of influenza A virus M1 protein, which shows pH-specific differences in membrane-bending (Ruigrok et al., 1992). Crystallography studies have also revealed that M1 proteins have similar structures at acidic and neutral pH, but differ in the way protein monomers interact (Harris et al., 2001).
M of all coronaviruses appears to adopt an N-ecto/C-endo topology (Armstrong et al., 1984), but transmissible gastroenteritis coronavirus M also adopts an alternate N-endo/C-ecto topology (Risco et al., 1995). We initially considered that the difference between MLONG and MCOMPACT might be due to altered topology. The N-ecto/C-ecto topology proposed for TGEV M involves part of the endodomain forming a fourth transmembrane span, with part of the C-terminus exposed on the virion surface. Since the pre-transmembrane region of M is much smaller than the post-transmembrane region, an upside-down topology should be visually distinctive. However, the endodomains of MLONG and MCOMPACT were of similar size in class averages. Instead, we prefer an interpretation in which MCOMPACT and MLONG are conformationally distinct homodimers of N-ecto/C-endo topology, and the difference in appearance is due to a conformational change that either stretches (MLONG) or collapses (MCOMPACT) the structure of the endodomain.
Prior to this study, the existence of reduction-sensitive complexes with a molecular weight consistent with approximately two, four and eight copies of M had been reported for HCoV-229E (Arpin and Talbot, 1990), but formation of a discrete M oligomer had not been demonstrated for any other coronavirus. The coronavirus M dimer would appear to be functionally equivalent to the heterodimeric complex of the triple-spanning membrane proteins GP5 and M which is essential for assembly of Equine arteritis virus (Snijder et al., 2003), a distantly related nidovirus. Further study of nidovirus ultrastructure is needed, but these observations coupled with the presence of one or more predicted three-transmembrane protein genes in every known coronavirus (M, but also SARS 3A and FCoV 3C), arterivirus (M and GP5), bafinivirus (GP4/M) and torovirus (M) genome suggests that dimeric complexes of triple-spanning membrane proteins may be a hallmark of nidovirus assembly.
While examining virions, we noticed that particles were most often either entirely spike-decorated or entirely spikeless. Further examination revealed that spikeless particles were significantly larger than spike-decorated particles, demonstrating that spikelessness resulted from a defect in assembly, not by accidental shearing or fusion activation. This led to a closer examination of SARS-CoV particles with patchy spike decoration. Spike patches were most often found at the ends of elongated particles, where MLONG is common, suggesting that MLONG mediates spike incorporation. Fig.10 A shows a conceptual model of an elongated SARS-CoV particle with spikes decorating both ends, where MLONG is plentiful. The association of small clusters of S with MLONG was unexpected, but we were unable to determine whether the missing spikes were not incorporated, or were present in an elongated disordered conformation, as would be expected after fusion activation. Another possible explanation for this phenomenon is that incorporated spikes stabilize MLONG.
Fig. 10.
Interpretation of coronavirus structure and assembly. (A) A schematic cutaway of an elongated SARS-CoV particle (size ≈ 85 nm, dMAX/dMIN ≈ 1.4) is shown to demonstrate how interactions between MLONG (black), MCOMPACT (white) and the ribonucleoprotein influence particle morphology. Spikes are shown in red and E protein oligomers in yellow. (B, C) Model for how MLONG could be related to the spacing of spikes and RNP densities in edge view (B) and axial view (C). Hollow shapes mark the nearest potential attachment points for an additional spike or RNP density. (D) A flowchart presents a model for the role of M conformation in assembly. An additional path suggests and explanation for how particle morphology can be altered during purification.
There is some evidence to link the endodomain, which appears to undergo the most noticeable change between MLONG and MCOMPACT, to incorporation of S protein. While the transmembrane region of M is important for M–S interactions, one residue of the M endodomain has been implicated in incorporation of S (de Haan et al., 1999). Mutation of a conserved tyrosine residue at position 211, near the N interaction site, does not affect VLP production but prevents S incorporation (de Haan et al., 1999). The C-terminus of the M endodomain shows the most profound structural difference between MLONG and MCOMPACT. This suggests that tyrosine-211 may be important for MLONG stability or for conversion between conformers. Further study is needed to determine the structural basis for coronavirus M, N and S protein interactions.
By comparing M proteins on virus particles and VLPs, we identified N, S and genomic RNA as factors that increase the ratio of MLONG to MCOMPACT. Properties attributed specifically to MLONG, which include membrane rigidity, uniform curvature and spike incorporation, appear to be geared to assembly of infectious virus, and appear to be opposed by the properties of MCOMPACT. We would therefore hypothesize that factors or treatments which increase the relative abundance of MLONG predict structural success, marked by increased virion fitness. Further work is needed to address the relationship between internal virion structure and infectivity.
4.1. Assessment of model quality
Protein spacing data presented here and previously (Neuman et al., 2006) suggests that eight dimeric M densities can accommodate a maximum of four N proteins and one trimeric spike protein, thus forming an 8M2:4N:1S3 unit at the virion surface (see the boxed region in Figs.10B and C). We previously reported that the minimum spacing between spikes on highly decorated SARS-CoV particles was ∼14–15 nm, which is about 4–5 nm farther apart than would be expected based on the ∼10 nm width of each spike (Neuman et al., 2006). Fourier transformation revealed a ∼13–18 nm−1 frequency signal in tomographic projections of spike-decorated MHV, but not spikeless MHV, confirming that spike spacing for MHV-sp1 and SARS-CoV is similar (data not shown).
The diagrams in Figs.10B and C show a packing model in which adjacent spikes are incorporated at anchor points across the MLONG network. Each M dimer could potentially interact with a spike, but the closest packing that could be achieved in this model would be ∼14–15 nm apart, based on the bulk of each spike and the nearest available anchor point. To test the quality of this model, we counted the number of spikes on three-dimensional reconstructions of MHV particles. These particles showed an average of 74 spikes per particle, which gives an approximate inter-spike spacing of 17 nm. Our model predicts ∼90 spikes per particle. Low spike incorporation in areas of MCOMPACT may explain some of the difference between predicted and actual spike count.
The protein spacing model in Fig.10B and C shows an average spacing of 4–5 nm between M protein dimers. Using the M spacing data for each virus (Fig.6C), this would give ∼1100 M2 molecules per average SARS-CoV, MHV and FCoV particle. We are unable to directly count the number of M features per particle because of the resolution of the reconstructed tomograms. However, the ratio of M to N in purified coronavirus particles has been well studied, and provides an indirect way to test the validity of our proposed M2 spacing. Estimated ratios of M to N protein in purified coronaviruses range from about 3M:1N (Cavanagh, 1983, Escors et al., 2001b) to 1M:1N (Hogue and Brian, 1986, Liu and Inglis, 1991), giving 730–2200 N molecules per virion. That works out to one N protein per 14–40 nucleotides of the genome. By comparison, the nucleoproteins of rabies virus and vesicular stomatitis virus are of similar molecular weight to coronavirus N proteins, and have been demonstrated to bind nine nucleotides per nucleoprotein (Albertini et al., 2006, Green et al., 2006). If, as suggested by Barcena et al. (2009), M dimers are spaced 6.5 nm apart, results of 60–200 nt/N molecule are obtained.
4.2. Model for coronavirus assembly
In the model of coronavirus assembly shown in Fig.10D, M is shown as a mixed population of MLONG and MCOMPACT in the endoplasmic reticulum membrane. M is then transported and interacts with other viral membrane proteins at the site of budding. The arrival of the ribonucleoprotein acts as a catalyst for the conversion of MCOMPACT to MLONG. The MLONG densities bend the membrane to form a sphere around the ribonucleoprotein, with the size of the sphere inversely related to the relative abundance of MLONG. Supplementary Fig.S5 relates virus particle size to membrane curvature per M dimer. We hypothesize that factors that interact with M such as N, S and genomic RNA could decrease particle size by limiting variation membrane curvature. After budding and release, environmental stress could convert some regions of MLONG back to MCOMPACT, allowing the RNP to form longer helices and causing particle elongation.
The function of E in virogenesis remains poorly understood. It is interesting that the function and packaging of E and S are dependent on palmitin acylation (Boscarino et al., 2008, Lopez et al., 2008, Thorp et al., 2006), and further research will be needed to test whether M densities may contain one or more palmitin binding regions. Point mutations in E have been shown to result in the assembly of large, elongated, thermolabile MHV particles (Fischer et al., 1998). In light of the model proposed here, we would speculate that these characteristics can be attributed to an overabundance of packaged MCOMPACT, suggesting that E plays a role in promoting MLONG formation or incorporation. This interpretation could be tested by cryo-EM and tomography of viruses carrying mutations in M and E.
5. Conclusions
In this study we described two functionally distinct forms of coronavirus M protein. Both types of M visible on viral particles were larger than expected for a single M protein, but consistent with the expected size of two M proteins. We demonstrated that M protein endodomains can self-assemble into oligo-dimeric complexes at 37 °C. We showed that formation of a convex, rigidified viral envelope is dependent on the presence of one form of M, which we have called MLONG. Statistical evidence suggests that spike incorporation is linked to particle size, and that spikes cluster in regions where MLONG is common. To explain these observations, we proposed a model in which locally-ordered networks of MLONG, stabilized by S, N and possibly E proteins, control particle size and the efficiency of assembly.
Acknowledgments
The authors wish to thank Mark Yeager, Kelly Dryden, Joel D. Quispe, Craig Yoshioka and Jessica del Rio for technical assistance with imaging and analysis.
This research was funded by the NIH/NIAID contract HHSN-266200400058C, AI-059799, AI-25913, AI-29984 and AI-72493, and by the Danish Research Council. Some of the electron microscopy shown here was conducted at the National Resource for Automated Molecular Microscopy which is supported by the National Institutes of Health though the National Center for Research Resources’ P41 program (RR17573).
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jsb.2010.11.021.
Appendix A. Supplementary data
References
- Albertini A.A., Wernimont A.K., Muziol T., Ravelli R.B., Clapier C.R., Schoehn G., Weissenhorn W., Ruigrok R.W. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- Armstrong J., Niemann H., Smeekens S., Rottier P., Warren G. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature. 1984;308:751–752. doi: 10.1038/308751a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpin N., Talbot P.J. Molecular characterization of the 229E strain of human coronavirus. Adv. Exp. Med. Biol. 1990;276:73–80. doi: 10.1007/978-1-4684-5823-7_11. [DOI] [PubMed] [Google Scholar]
- Barcena M., Oostergetel G.T., Bartelink W., Faas F.G., Verkleij A., Rottier P.J., Koster A.J., Bosch B.J. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc. Nat. Acad. Sci. USA. 2009;106:582–587. doi: 10.1073/pnas.0805270106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniac D.R., Andonov A., Grudeski E., Booth T.F. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 2006;13:751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscarino J.A., Logan H.L., Lacny J.J., Gallagher T.M. Envelope protein palmitoylations are crucial for murine coronavirus assembly. J. Virol. 2008;82:2989–2999. doi: 10.1128/JVI.01906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson L.A., Briggs J.A., Glass B., Riches J.D., Simon M.N., Johnson M.C., Muller B., Grunewald K., Krausslich H.G. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4:592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus IBV: further evidence that the surface projections are associated with two glycopolypeptides. J. Gen. Virol. 1983;64:1787–1791. doi: 10.1099/0022-1317-64-8-1787. [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Chang C.K., Chang Y.W., Sue S.C., Bai H.I., Riang L., Hsiao C.D., Huang T.H. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H.A., Macnaughton M.R. Comparison of the morphology of three coronaviruses. Arch. Virol. 1979;59:25–33. doi: 10.1007/BF01317891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Vennema H., Rottier P.J. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 2000;74:4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A., Smeets M., Vernooij F., Vennema H., Rottier P.J. Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. J. Virol. 1999;73:7441–7452. doi: 10.1128/jvi.73.9.7441-7452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escors D., Ortego J., Laude H., Enjuanes L. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 2001;75:1312–1324. doi: 10.1128/JVI.75.3.1312-1324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escors D., Camafeita E., Ortego J., Laude H., Enjuanes L. Organization of two transmissible gastroenteritis coronavirus membrane protein topologies within the virion and core. J. Virol. 2001;75:12228–12240. doi: 10.1128/JVI.75.24.12228-12240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Ooi A., Tan Y.W., Wang S., Fang S., Liu D.X., Lescar J. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure. 2005;13:1859–1868. doi: 10.1016/j.str.2005.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F., Stegen C.F., Masters P.S., Samsonoff W.A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 1998;72:7885–7894. doi: 10.1128/jvi.72.10.7885-7894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Gallagher T.M., Escarmis C., Buchmeier M.J. Alteration of the pH dependence of coronavirus-induced cell fusion: effect of mutations in the spike glycoprotein. J. Virol. 1991;65:1916–1928. doi: 10.1128/jvi.65.4.1916-1928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green T.J., Zhang X., Wertz G.W., Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- Harpaz Y., Gerstein M., Chothia C. Volume changes on protein folding. Structure. 1994;2:641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- Harris A., Forouhar F., Qiu S., Sha B., Luo M. The crystal structure of the influenza matrix protein M1 at neutral pH: M1–M1 protein interfaces can rotate in the oligomeric structures of M1. Virology. 2001;289:34–44. doi: 10.1006/viro.2001.1119. [DOI] [PubMed] [Google Scholar]
- Harris A., Cardone G., Winkler D.C., Heymann J.B., Brecher M., White J.M., Steven A.C. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. USA. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogue B.G., Brian D.A. Structural proteins of human respiratory coronavirus OC43. Virus Res. 1986;5:131–144. doi: 10.1016/0168-1702(86)90013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogue B.G., Machamer C.E. Coronavirus structural proteins and virus assembly. In: Perlman S., editor. Nidoviruses. ASM Press; p: 2007. pp. 179–200. [Google Scholar]
- Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., Hajduk P., Mack J., Fesik S.W., Olejniczak E.T. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- Jayaram H., Fan H., Bowman B.R., Ooi A., Jayaram J., Collisson E.W., Lescar J., Prasad B.V. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J., Locker J.K., Meijer A., Horzinek M.C., Geuze H.J., Rottier P.J. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kunding A.H., Mortensen M.W., Christensen S.M., Stamou D. A fluorescence-based technique to construct size distributions from single-object measurements: application to the extrusion of lipid vesicles. Biophys. J. 2008;95:1176–1188. doi: 10.1529/biophysj.108.128819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 2002;76:4987–4999. doi: 10.1128/JVI.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.N., Chen L.K., Ma H.C., Yang H.H., Li H.P., Lo S.Y. Thermal aggregation of SARS-CoV membrane protein. J. Virol. Methods. 2005;129:152–161. doi: 10.1016/j.jviromet.2005.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Liu D.X., Inglis S.C. Association of the infectious bronchitis virus 3c protein with the virion envelope. Virology. 1991;185:911–917. doi: 10.1016/0042-6822(91)90572-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Yoshikawa-Iwata N., Ito N., Watts D.M., Wyde P.R., Wang N., Newman P., Kent Tseng C.T., Peters C.J., Makino S. Chimeric coronavirus-like particles carrying severe acute respiratory syndrome coronavirus (SCoV) S protein protect mice against challenge with SCoV. Vaccine. 2008;26:797–808. doi: 10.1016/j.vaccine.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez L.A., Riffle A.J., Pike S.L., Gardner D., Hogue B.G. Importance of conserved cysteine residues in the coronavirus envelope protein. J. Virol. 2008;82:3000–3010. doi: 10.1128/JVI.01914-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke S.J., Baldwin P.R., Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Machamer C.E., Rose J.K. A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. J. Cell Biol. 1987;105:1205–1214. doi: 10.1083/jcb.105.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle J.F., Tristram-Nagle S. Structure of lipid bilayers. Biochim. Biophys. Acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nal B., Chan C., Kien F., Siu L., Tse J., Chu K., Kam J., Staropoli I., Crescenzo-Chaigne B., Escriou N., van der Werf S., Yuen K.Y., Altmeyer R. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Chen C.J., Maeda J., Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 2003;77:2922–2927. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Adair B.D., Yeager M., Buchmeier M.J. Purification and electron cryomicroscopy of coronaviruses. In: Cavanagh D., editor. SARS and Other Coronaviruses: Laboratory Protocols. Humana Press; New York: 2008. pp. 129–138. [Google Scholar]
- Neuman B.W., Adair B.D., Burns J.W., Milligan R.A., Buchmeier M.J., Yeager M. Complementarity in the supramolecular design of arenaviruses and retroviruses revealed by electron cryomicroscopy and image analysis. J. Virol. 2005;79:3822–3830. doi: 10.1128/JVI.79.6.3822-3830.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Adair B.D., Yoshioka C., Quispe J.D., Orca G., Kuhn P., Milligan R.A., Yeager M., Buchmeier M.J. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 2006;80:7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Joseph J.S., Saikatendu K.S., Serrano P., Chatterjee A., Johnson M.A., Liao L., Klaus J.P., Yates J.R., 3rd, Wüthrich K., Stevens R.C., Buchmeier M.J., Kuhn P. Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J. Virol. 2008;82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V.P., Hogue B.G. Protein interactions during coronavirus assembly. J. Virol. 1997;71:9278–9284. doi: 10.1128/jvi.71.12.9278-9284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opstelten D.J., Raamsman M.J., Wolfs K., Horzinek M.C., Rottier P.J. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K., Tan E., Parthasarathy K., Lin X., Jiang F.L., Yu D., Vararattanavech A., Soong T.W., Liu D.X., Torres J. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009;5:e1000511. doi: 10.1371/journal.ppat.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramjeesingh M., Huan L.J., Garami E., Bear C.E. Novel method for evaluation of the oligomeric structure of membrane proteins. Biochem. J. 1999;342:119–123. [PMC free article] [PubMed] [Google Scholar]
- Risco C., Anton I.M., Enjuanes L., Carrascosa J.L. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J. Virol. 1996;70:4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C., Anton I.M., Sune C., Pedregosa A.M., Martin-Alonso J.M., Parra F., Carrascosa J.L., Enjuanes L. Membrane protein molecules of transmissible gastroenteritis coronavirus also expose the carboxy-terminal region on the external surface of the virion. J. Virol. 1995;69:5269–5277. doi: 10.1128/jvi.69.9.5269-5277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottier P.J., Rose J.K. Coronavirus E1 glycoprotein expressed from cloned cDNA localizes in the Golgi region. J. Virol. 1987;61:2042–2045. doi: 10.1128/jvi.61.6.2042-2045.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruigrok R.W., Hewat E.A., Wade R.H. Low pH deforms the influenza virus envelope. J. Gen. Virol. 1992;73:995–998. doi: 10.1099/0022-1317-73-4-995. [DOI] [PubMed] [Google Scholar]
- Saikatendu K.S., Joseph J.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J. Virol. 2007;81:3913–3921. doi: 10.1128/JVI.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G. Characterization of a small plaque mutant of the A59 strain of mouse hepatitis virus defective in cell fusion. Adv. Exp. Med. Biol. 1987;218:169–174. doi: 10.1007/978-1-4684-1280-2_21. [DOI] [PubMed] [Google Scholar]
- Schutze H., Ulferts R., Schelle B., Bayer S., Granzow H., Hoffmann B., Mettenleiter T.C., Ziebuhr J. Characterization of white bream virus reveals a novel genetic cluster of nidoviruses. J. Virol. 2006;80:11598–11609. doi: 10.1128/JVI.01758-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Dobbe J.C., Spaan W.J. Heterodimerization of the two major envelope proteins is essential for arterivirus infectivity. J. Virol. 2003;77:97–104. doi: 10.1128/JVI.77.1.97-104.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A., Grigorieff N. Noise bias in the refinement of structures derived from single particles. Ultramicroscopy. 2004;102:67–84. doi: 10.1016/j.ultramic.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Sticht J., Humbert M., Findlow S., Bodem J., Muller B., Dietrich U., Werner J., Krausslich H.G. A peptide inhibitor of HIV-1 assembly in vitro. Nat. Struct. Mol. Biol. 2005;12:671–677. doi: 10.1038/nsmb964. [DOI] [PubMed] [Google Scholar]
- Sturman L.S., Holmes K.V., Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 1980;33:449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C.S., Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tang C., Loeliger E., Kinde I., Kyere S., Mayo K., Barklis E., Sun Y., Huang M., Summers M.F. Antiviral inhibition of the HIV-1 capsid protein. J. Mol. Biol. 2003;327:1013–1020. doi: 10.1016/s0022-2836(03)00289-4. [DOI] [PubMed] [Google Scholar]
- Thorp E.B., Boscarino J.A., Logan H.L., Goletz J.T., Gallagher T.M. Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. J. Virol. 2006;80:1280–1289. doi: 10.1128/JVI.80.3.1280-1289.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., Godeke G.J., Rossen J.W., Voorhout W.F., Horzinek M.C., Opstelten D.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q., Langereis M.A., van Vliet A.L., Huizinga E.G., de Groot R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. USA. 2008;105:9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong-can O.Y., Helfrich W. Instability and deformation of a spherical vesicle by pressure. Phys. Rev. Lett. 1987;59:2486–2488. doi: 10.1103/PhysRevLett.59.2486. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.