Abstract
In this report we characterized the ArabidopsisABI1 gene orthologue and Brassica napus gene paralogues encoding protein phosphatase 2C (PP2C, group A), which is known to be a negative regulator of the ABA signaling pathway. Six homologous B. napus sequences were identified and characterized as putative PP2C group A members. To gain insight into the conservation of ABI1 function in Brassicaceae, and understand better its regulatory effects in the drought stress response, we generated transgenic B. napus plants overexpressing A. thalianaABI1. Transgenic plants subjected to drought showed a decrease in relative water content, photosynthetic pigments content and expression level of RAB18- and RD19A-drought-responsive marker genes relative to WT plants. We present the characterization of the drought response of B. napus with the participation of ABI1-like paralogues. The expression pattern of two evolutionarily distant paralogues, BnaA01.ABI1.a and BnaC07.ABI1.b in B. napus and their promoter activity in A. thaliana showed differences in the induction of the paralogues under dehydration stress. Comparative sequence analysis of both BnaABI1 promoters showed variation in positions of cis-acting elements that are especially important for ABA- and stress-inducible expression. Together, these data reveal that subfunctionalization following gene duplication may be important in the maintenance and functional divergence of the BnaABI1 paralogues. Our results provide a framework for a better understanding of (1) the role of ABI1 as a hub protein regulator of the drought response, and (2) the differential involvement of the duplicated BnaABI1 genes in the response of B. napus to dehydration-related stresses.
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-015-0334-x) contains supplementary material, which is available to authorized users.
Keywords: Brassica napus, Drought response, ABI1 protein phosphatase, Gene overexpression, Promoter activity
Introduction
Drought stress induces diverse responses at the cellular and molecular levels including the production of plant phytohormones, especially abscisic acid (ABA) (Swamy and Smith 1999; Zhu 2003). The core ABA signaling pathway consists of three important elements: ABA receptors (PYR/PYL/RCAR proteins), protein phosphatases type 2C (PP2Cs) and SNF1-related protein kinases (SnRK2s) (Yoshida et al. 2006; Ma et al. 2009; Fujita et al. 2009, Umezawa et al. 2010). In Arabidopsis, there are nine group A PP2Cs: ABI1, ABI2, HAB1, HAB2, HAI1, HAI2, HAI3, AHG1 and AHG3 (Gosti et al. 1999; Merlot et al. 2001; Yoshida et al. 2006; Rubio et al. 2009). It is widely accepted that group A PP2Cs have a key role in the ABA signaling pathway (Sheen 1998; Schweighofer et al. 2004; Ludwików 2015 for review). ABA stimulates plant responses to abiotic stresses such as drought, high salt and cold, and several research teams have shown that group A PP2Cs, including ABI1 (ABA Insensitive 1; Leung et al. 1994), are negative regulators of ABA signaling (Gosti et al. 1999; Merlot et al. 2001; Saez et al. 2004). Although it is known that PP2Cs interact with various targets to form a regulatory core (Ludwików 2015 for review) that modulates cellular processes and stress response pathways (Nemhauser et al. 2006; Ludwików et al. 2009; Umezawa et al. 2009; Geiger et al. 2010; Ludwików et al. 2014), their role in the acclimation of plants to drought stress remains unclear. Hence it is important to investigate further the function of ABI1 and other group A PP2Cs in the stress responses of plants, including economically important crops with complex genomes typically carrying multiple gene homologues.
Although group A PP2Cs have been extensively analyzed in Arabidopsis, their orthologues (ancestrally related genes maintained in different species after speciation events) have also been identified and partially characterized in other plant species (Gonzalez-Garcia et al. 2003; Xue et al. 2008; Tougane et al. 2010; Ludwików et al. 2013; Yuan et al. 2013, Jia et al. 2013). In Brassica species, protein-encoding genes are generally triplicated relative to their counterparts in A. thaliana due to whole-genome duplication events (Babula et al. 2003; Parkin et al. 2005; Lysak et al. 2005; Babula et al. 2006; Ziolkowski et al. 2006; Wang et al. 2011) between 7.9 and 14.6 Mya. Despite the high degree of sequence similarity of their transcripts and the corresponding proteins, such paralogous genes often exhibit significant differences in expression patterns. This can result in increased overall gene expression levels and can provide a source of novel gene variants (Udall and Wendel 2006; Whittle and Krochko 2009). The potential for functional divergence among duplicated genes and consequent pleiotropic effects on stress responses, especially to stresses such as drought, is poorly described for crop plants including the Brassica species.
Currently, there is a paucity of information on the role of paralogous ABI1 copies (they are considered in-paralogues, i.e. they result from duplication after the last speciation event; Studer and Robinson-Rechavi 2009) in the regulation of the ABA signaling pathway in the Brassica species. Previous studies have focused on individual ABI1 gene copies in particular species, and have neglected to investigate paralogous examples (Yuan et al. 2013; Zhang et al. 2014).
In this study we have investigated the conservation of the role of the ABI1 gene orthologue in Brassicaceae (Brassicanapus vs. Arabidopsis) and the duplicated ABI1 genes in B. napus with particular reference to the response to drought stress. We generated transgenic B. napus plants overexpressing the A. thalianaABI1 orthologue to study corresponding changes in the drought-stress response, as well as the cumulative effect of all BnaABI1-like genes under drought conditions. Overexpression of AtABI1 resulted in water loss and changes in chlorophyll accumulation indicating that these stress-related physiological responses are controlled by the ABI1-dependent ABA-signaling pathway. Transcript levels of known drought stress marker genes were also downregulated in transgenic plants compared to wild type B. napus. We also observed changes in the transcript levels of the endogenous ABI1 homologues (BnaABI1 s) in wild type (WT) and transgenic B. napus to evaluate their involvement in the drought response. This aspect presents important insight into poorly understood dehydration-induced co-response of duplicated gene homologues occurring typically in plants. To determine whether the duplicated BnaABI1 genes are functionally redundant or whether they have evolved novel subfunctions, we performed a detailed analysis of two evolutionarily distant BnaABI1 paralogues, BnaA01.ABI1.a and BnaC07.ABI1.b, and found evidence for structural and functional diversification. We demonstrated that the promoter activity of these paralogues in A. thaliana was sufficient to control tissue-specific expression, but resulted in different stress-induced expression. The combined results shed new light on diverse roles of the ABI1 gene family in the drought response plasticity in the Brassica polyploid. In-depth knowledge about the functional divergence of BnaABI1 members will facilitate future work on genetic engineering of Brassica species.
Materials and methods
Plant material, growth and stress conditions
Arabidopsis thaliana (L.) Heynh. ecotype Columbia (Col-0) and B. napus microspore-derived embryos of cv. Monolit (HR Strzelce, Poland) were used to generate homozygous transgenic plants. Plant growth conditions, exogenous ABA and drought stress treatments were described previously by Ludwików et al. (2013). The B. napus seeds were germinated in sterilized petri dishes on sand for 3 days at 25 °C. After incubation, good quality seedlings with fully expanded green cotyledon were grown in pots (7 cm × 7 cm × high 8 cm; 2 seedlings per pot; 20 pots per condition) filled with soil in a growth room under long-day conditions (16/8 h of light/dark with 18 °C). They were watered every 2 days for 3 weeks and fertilized weekly with Florovit (0.5 ml/L). The drought stress was induced in well-watered three-week-old plants by withholding water for 5 days. These plants were then re-watered for 5 days after which leaves were collected. In parallel, control plants received water regularly. Relative water content (RWC) was calculated according to the formula: [(fresh weight − dry weight)/(turgid weight − dry weight)] × 100. For wounding treatment all rosette leaves were crushed with forceps, which effectively wounded 40 % of the leaf area (Reymond et al. 2000). All measurements were made on three or more replicates.
Identification and cloning of ABI1 paralogues from the B. napus genome
The A. thaliana ABI1 cDNA sequence (At4g26080) was used as a reference sequence to detect the corresponding B. napus EST and GSS clones. The respective B. napus clone collections were screened by BLASTN search against the ATiDB (The Arabidopsis thaliana Integrated Database) and NCBI (www.ncbi.nlm.nih.gov) databases. The selected EST sequences were used to clone ABI1-related genes with gene-specific primer sets and B. napus genomic DNA as the template. Genomic DNA was extracted using a DNeasy Plant Mini kit (Qiagen). For full-length cDNA amplification, the Rapid Amplification of cDNA Ends (RACE) method was used using a SMARTer™ RACE cDNA Amplification Kit (Clontech) according to the manufacturer’s instructions. All primer sequences are listed in Supplemental Table S1. The resulting PCR amplicons for genomic and full length cDNA sequences corresponding to individual paralogues were subcloned into StrataClone™ PCR Cloning Vector pSC-A (Agilent Technologies) and confirmed by sequencing. The NCBI Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used to identify the PP2C catalytic domain and characteristic motifs within deduced BnaABI1 protein sequences.
Phylogenetic analysis of ABI1-related genes and other group A PP2Cs in A. thaliana and B. napus
The deduced group A PP2C amino acid sequences of B. napus (Brassica Database; http://brassicadb.org/brad/) were used to estimate their phylogenetic relationships. The clone numbers corresponding to individual PP2C are listed in Supplemental Table S2. First, full-length protein sequences were aligned using the ClustalX program with default parameters. Selected amino acid sequences of the conserved PP2C catalytic domain were further imported into MEGA5.1 software (http://www.megasoftware.net/; Tamura et al. 2011). The phylogenetic tree was constructed using the neighbor-joining method with a Poisson model of amino acid substitution and 1000 bootstrap replications.
Plasmid construction for B. napus transgenic plants overexpressing AtABI1
For B. napus transformation, the coding region of the AtABI1 gene (At4g26080) was PCR-amplified using gene-specific primer pairs (Table S1). The amplified AtABI1 coding sequence was cloned into the pKGIB vector between the BamHI and XhoI sites under the control of a 35S-CaMV promoter (Cegielska-Taras et al. 2008). The construct was introduced into B. napus via Agrobacterium-mediated transformation using the method described by Cegielska-Taras et al. (2008). Putative plant transformants were screened on 1/2 MS media including 10 mg/L phosphinotricin. T2 homozygous plants were used for further analysis.
Measurements of physiological parameters
The chlorophyll and carotenoid contents were determined as described in Ludwików et al. (2009). Ten plants per line were evaluated and all tests were repeated three times.
RNA isolation and qPCR analysis
RNA isolation and quantitative real-time PCR (qPCR) was performed as described by Ludwików et al. (2013). Total RNA was extracted from three-week old leaves of the WT and transgenic plants using TRIZOL reagent (Invitrogen) following the manufacturer’s instructions. For quantitative RT-PCR analysis, 1.5 μg DNase-treated RNA was used for the first-strand cDNA synthesis using a First Strand cDNA Synthesis Kit (Fermentas, Gdańsk, Poland) according to manufacturer’s protocol. 2 μL of 10-fold diluted cDNA was used as the template in a 15 μL qRT-PCR reaction using Maxima SYBR Green/ROX/qPCR Master Mix (Fermentas). An 18S rDNA gene was used as the internal control to normalize all data. Each sample was set up with three biological replicates and run twice on separate plates. PCR reactions were conducted in the ABI PRISM 7900 HT sequence detection system (Applied Biosystems). The qRT-PCR amplifications were carried out using the following thermal profile: 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Data analysis was performed using SDS 2.2.1 software (Applied Biosystems). Serial dilutions of genomic DNA (from 100 to 10,000 copies) were used to set up the calibration curve. The results presented here are the mean of the relative gene expression ratios [log2 fold change scale; FC = log2 (ratio/ratio)] of two independent experiments (n = 6). After the PCR, a melting curve was generated to test the amplicon specificity. Primer sequences are listed in Supplemental Table S1.
Promoter activity assay
The regions almost 2.2 kb upstream of the translation start codon for both the BnaA01.ABI1.a and BnaC07.ABI1.b genes were isolated using the GenomeWalker kit (Clontech) following the manufacturer’s instructions. Gene-specific primers for the nested PCR are listed in Supplemental Table S1. For the promoter activity assay, the promoter sequences of the selected BnaABI1 paralogues were subcloned into the pENTR/SD/TOPO cloning vector and confirmed by sequencing. To produce plant transformants, pENTR vector-promoter constructs were recombined with the pMDC162 (::GUS) vector using Gateway® LR Clonase® II Enzyme Mix (Invitrogen). The promoter-reporter fusion construct was transformed into Agrobacterium tumefaciens strain LBA4404 and used for A. thaliana Columbia line plant transformations using the floral dip method (Clough and Bent 1998). Putative plant transformants were screened on 1/2 MS media including 10 mg/L hygromycin. For histochemical analysis of GUS expression, tissue samples were immersed in GUS staining solution (100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.5 mM K4Fe[CN]6, 0.5 mM K3Fe[CN]6, 0.1 % Triton X-100 and 1 mM X-gluc) overnight at 37 °C. After staining, plant samples were rinsed and photographed.
Identification of BnaA01.ABI1.a and BnaC07.ABI1.bcis-regulatory elements
Potential regulatory elements were identified within the 2,180 bp upstream of the BnaA01.ABI1.a and BnaC07.ABI1.b start codons using the MathInspector platform (Genomatix; http://www.genomatix.de/) with the Plant IUPAC Library Version 7.0 (based on PLACE Release 30.0) restricted to A. thaliana and IUPAC search parameters and max. 0 % mismatches. Additionally, stress-responsive regulatory motifs were identified by searching the publicly available cis-acting element databases such as Plant Matrix Family Library (Genomatix) and PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). “Conserved Non-Coding Sequences”, named “CNSs”, including conserved transcription factor binding sites within the BnaA01.ABI1.a and BnaC07.ABI1.b promoters, were defined by sequence similarity (>70 % identity over at least 90 bp). Furthermore, these highly conserved regions were analyzed for the presence of conserved stress-responsive cis-regulatory elements using Plant IUPAC Library Version 7.0 on the MathInspector platform. Regions conserved between both promoters were visualized using VISualization Tool for Alignments (Vista) tools mVista and rVista (http://rvista.dcode.org/).
Results
Response of AtABI1-overexpressing transgenic B. napus lines to drought stress: effect on RWC and photosynthetic pigment levels
To investigate the potential conservation of ABI1 protein phosphatase 2C function in B. napus,AtABI1-overexpressing B. napus lines under the control of a 35S-CaMV promoter were generated. Six independent transgenic plants were produced in which the presence of the transgene was confirmed by PCR assay with NOSt/BAR-specific primers (not shown). Three representative lines containing the 35S:AtABI1 construct were used in subsequent experiments (Fig. S1). The transgenic plants were not morphologically different to wild type plants under normal growth conditions (not shown). Because of the involvement of the ABI1 gene in the abiotic stress response (Leung et al. 1997; Chinnusamy et al. 2004), the effect of AtABI1 overexpression in B. napus plants during drought and rehydration was analyzed. Water status (determined as RWC. i.e. relative water content in leaves) and changes in photosynthetic pigment content were examined in both wild-type plants and transgenic lines grown under well-watered and water-deficient conditions. Measurements of water loss showed no significant differences in RWC between the B. napus control and transgenic plants under normal growth conditions (Fig. 1). However, water content in the leaves of transgenic plants decreased more rapidly during soil drying compared to wild-type plants. In particular, drastic water loss in the leaves of transgenic lines Oex41 (Overexpression line 41) and Oex42 were observed, where RWC decreased by at least 40 %. This demonstrates that overexpression of the AtABI1 orthologue in B. napus plants reduces their water retention capacity, confirming ABI1 role as ABA negative regulator.
Fig. 1.
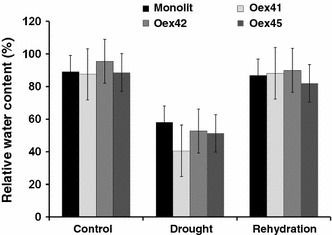
Relative water content (RWC) in B. napus wild-type and transgenic plants during drought and rehydration. The data represent means from five replicates with three biological repeats. Error bars represent SE
These data were complemented by assessment of the content of photosynthetic pigments such as chlorophyll and carotenoids in these plants under well-watered and drought-stress conditions. Initially, chlorophyll a, chlorophyll b, total chlorophyll content and chlorophyll a/b ratio were analyzed. Changes in photosynthetic pigment content were observed in both wild-type plants and transgenic lines under drought-stress conditions (Fig. 2). As expected, chlorophyll a, chlorophyll b and total chlorophyll contents significantly increased in all B. napus transgenic lines overexpressing AtABI1. Chlorophyll a levels were higher in all transgenic plants than in wild-type plants, while remaining unchanged in wild-type plants under both drought-stress and control conditions. Chlorophyll b content remained almost invariable in transgenic line Oex41 and in wild-type plants, while its content in lines Oex42 and Oex45 increased and was higher than in wild-type plants. In general, chlorophyll a levels were higher than chlorophyll b levels in both types of plant. However, the chlorophyll a/b ratio decreased in both wild-type plants and the transgenic lines under drought-stress conditions. The largest decrease in chlorophyll a/b ratio was observed in transgenic line Oex45.
Fig. 2.
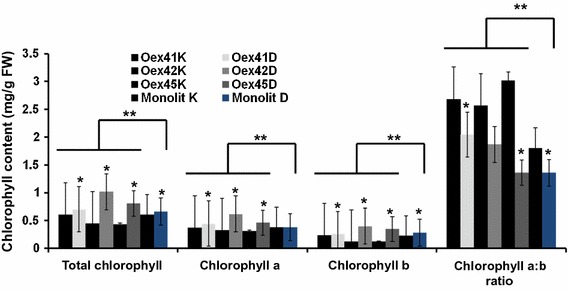
Total chlorophyll content, chlorophyll a, chlorophyll b and chlorophyll a:b ratio in wild type and ABI1-overexpressing transgenic plants under drought conditions. K: control (well-watered) conditions, D: drought conditions. The data represent means from five replicates with three biological repeats. * indicates P < 0.05 between control and experiment (drought); ** indicates P < 0.05 between the wild type (Monolit) and the ABI1-overexpressing lines by t test. Error bars indicate SE
Less variation in the photosynthetic pigment content was observed during rehydration conditions (Fig. 3), but there were, nevertheless, slight differences between particular B. napus transgenic lines overexpressing AtABI1. Line Oex41 showed no changes in chlorophyll a, chlorophyll b and total chlorophyll contents rehydration compared to controls. Chlorophyll b content was invariable in all transgenic lines, but increased in wild type plants under rehydration compared to control, well-watered conditions. On the other hand, total chlorophyll and chlorophyll a contents increased and slightly decreased in lines Oex42 and Oex45, respectively. The chlorophyll a/b ratio increased slightly in all transgenic lines and decreased in the wild-type plants on rehydration.
Fig. 3.
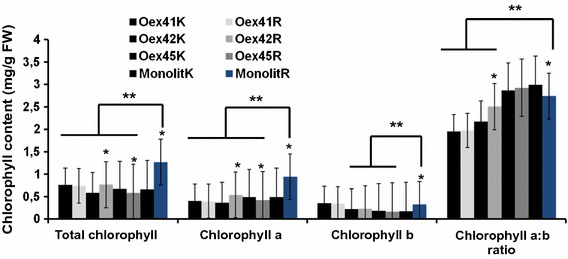
Total chlorophyll content, chlorophyll a, chlorophyll b and chlorophyll a:b ratio in the wild type (Monolit) and ABI1-overexpressing plants after rehydration. K: control (well-watered) conditions, R: drought-treated samples after rehydration. The data represent means from five replicates with three biological repeats. * indicates P < 0.05, between control and experiment (rehydration); ** indicates P < 0.05 between wild type and ABI1-overexpressing lines by t test. Error bars indicate SE
Analysis of carotenoid content during drought stress showed a decrease in both wild-type plants and line Oex41, but an increase in lines Oex42 and Oex45 compared to their respective controls (Fig. 4). On rehydration, carotenoid content returned to the control level in transgenic lines Oex41 and Oex42, but decreased in line Oex45 and wild type plants.
Fig. 4.
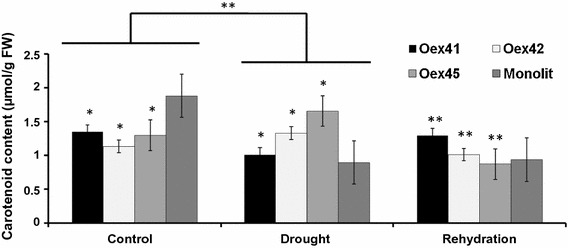
Total carotenoid content of the wild-type (Monolit) and 35S:ABI1 lines under drought and rehydration conditions. K: control (well-watered) conditions, D: drought conditions. The data represent means from five replicates with three biological repeats. ** indicates P < 0.05, between control and experiment (drought); * indicates P < 0.05 between wild type and ABI1-overexpressing lines by t test. Error bars indicate SE
Expression of selected ABA-responsive and drought-responsive genes in WT and transgenic ABI1-overexpressing lines of B. napus
A set of ABA- and/or stress-inducible genes, as well as other components of the ABA signaling pathway selected from publicly available microarray data for A. thaliana (https://www.genevestigator.com/gv/plant.jsp), were included in our analysis. Among them were genes reported to be inducible by exogenous ABA and/or drought such as RAB18, RD19A and MAPKKK18, other members of the PP2C gene family such as ABI2, two copies of HAB2 and genes induced by other factors such as MAPKKK1 (cold and heat responsive) and ERF1 (ethylene responsive). As shown in Fig. 5 for wild type plants and representative transgenic line Oex42, the transcript levels of all genes with the exception of BnaX.ERF1, BnaX.HAB2.c and BnaX.MAPKKK1 increased significantly in wild-type plants and line Oex42 overexpressing AtABI1 in response to drought stress. However, these genes with the exception of BnaX.MAPKKK18 were expressed at a low level in line Oex42. The expression level of most of these genes returned to control values in both wild-type and transgenic plants under rehydration conditions. Interestingly, the two copies of BnaX.HAB2 studied showed different expression profiles, with only one copy being significantly induced under drought stress. This indicates functional divergence of the duplicated BnaHAB2 genes as an example of gene subfunctionalisation in the ABA signaling network.
Fig. 5.
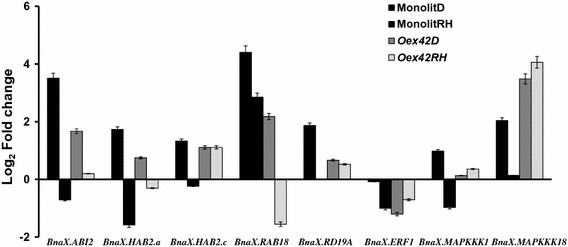
The relative expression of selected ABA-responsive and drought-responsive genes in wild-type plants and AtABI1-overexpressing B. napus transgenic lines during drought and after rehydration. The qPCR results show relative expression of the indicated genes. Gene transcript levels were determined using three replicates and were normalized against 18S rDNA. Each quantification was repeated at least twice with similar results. The results are displayed as mean log2 fold change ± SE (n = 9) of three independent experiments
Taken together, these results confirmed that BnaABI1 genes are negative regulators of the drought response in B. napus. Overexpression of AtABI1 in the B. napus transgenic lines results in decreased tolerance to drought stress.
Structural characteristics and phylogenetic analysis of the ABI1 paralogues in B. napus
Using the ArabidopsisAtABI1 cDNA to query the ATiDB and NCBI databases, six ABI1-related genes were identified in the amphidiploid genome of B. napus. They are paralogous ABI1 copies, which result from the duplication after last speciation event. These six BnaABI1 family members were cloned using paralogue-specific primers and then verified by sequencing (Table S1). They were named according to the nomenclature system proposed for the Brassica genus (Østergaard and King 2008). All BnaABI1-related genes were structurally characterized and their chromosomal localization in the B. napus genome was determined (Babula-Skowronska et al., unpublished). To confirm homology between the ABI1 family members in B. napus and their diploid B. rapa and B.oleracea progenitors, a phylogenetic tree was built with high bootstrap support (Fig. S2). This analysis was performed using amino acid sequences of the conserved PP2C catalytic domain for six, three and three ABI1 sequences from B. napus, B. rapa and B. oleracea, respectively. Additionally, the previously detected ABI1s from B. oleracea (KF577723, corresponding to BolC08.ABI1.c; Yuan et al. 2013) and B. napus (JX122895, corresponding to BnaC01.ABI1.a; Zhang et al. 2014) were included. As expected, there was a high degree of similarity between the ABI1 orthologues derived from the B. napus, B. rapa and B. oleracea genomes, while the paralogues within the same genomes of the individual species showed greater sequence divergence. A phylogenetic analysis of group A PP2Cs of A. thaliana and B. napus also showed that they divided into two subgroups with high bootstrap support. Moreover, the close evolutionary relationship between the BnaA01.ABI1.a and AtABI1 genes was confirmed based on protein sequence alignment of the PP2C catalytic domain (Fig. S3). These results suggest close evolutionary relationships between the respective A. thaliana and B. napus genes.
To investigate the possibility of a functional plasticity of the duplicated BnaABI1 gene copies in response to environmental stress conditions, two evolutionarily distant BnaABI1 genes, BnaA01.ABI1.a and BnaC07.ABI1.b, were chosen for subsequent experiments. The first of these represents the closest homologue to A. thalianaAtABI1, whereas the second is a more distant homologue of AtABI1, dating from the whole-genome duplication that took place after the Arabidopsis and Brassica lineages diverged (Fig. S2). By screening B. rapa (donor of A genome) and B. oleracea (donor of C genome) genomic DNA using BnaABI1 paralogue-specific primers, BnaA01.ABI1.a and BnaC07.ABI1.b were assigned to chromosome A01 in the A genome and chromosome C07 in the C genome, respectively (Fig. S4). Detailed analysis of both BnaABI1 genomic and cDNA sequences revealed a highly conserved gene structure with four exons and three introns (Fig. S5). The lengths of the BnaA01.ABI1.a and BnaC07.ABI1.b genomic sequences are 1588 and 1519 bp, coding sequences are 1281 and 1233 bp and deduced protein sequences are 426 and 410 aa, respectively. The sequence similarity of both BnaABI1 paralogues ranges from 78.6 up to 84.4 % at the genomic and coding sequence levels, respectively. Major sequence differences between both BnaABI1 genes were discovered in the 5′-UTR, first exon and all introns (Fig. S6). As shown in Fig. S6, the most frequent sequence changes between the two BnaABI1 genes are deletion/insertion (indels) of longer fragments. For example, one of the longer indels, which includes a microsatellite motif (CAT), is located in the first exon; additional differences occur in sequences flanking this repeat motif. However, the lengths of exon II, exon III and exon IV, 291, 106 and 359 bp, respectively, were maintained in both BnaABI1 paralogues. To confirm that the selected BnaABI1 genes belong to the PP2C-type phosphatase gene family, the presence of the PP2C catalytic domain in the deduced protein sequences was determined. Both BnaABI1 proteins contained the PP2C catalytic domain at the C-terminus with the same arrangement of 11 motifs; the N-terminal noncatalytic region was variable in both proteins (Figs. S3 and S7).
Expression of ABI1-related endogenes in transgenic ABI1-overexpressing and WT lines of B. napus exposed to drought stress
The plasticity of ABA signaling in response to stress in polyploids or paleopolyploids can be addressed by investigation of the paralogous ABI1 endogenes in B. napus. We analysed their specificity and functional divergence by examining their involvement in the response to drought. We also asked whether AtABI1 overexpression in the B. napus genetic background resulted in co-regulation of transcriptional activity of the endogenous BnABI1 genes. The expression profiles of selected BnaABI1 endogenes were determined using a qPCR assay in wild-type and transgenic plants under control, drought and rehydration conditions (Fig. 6).
Fig. 6.
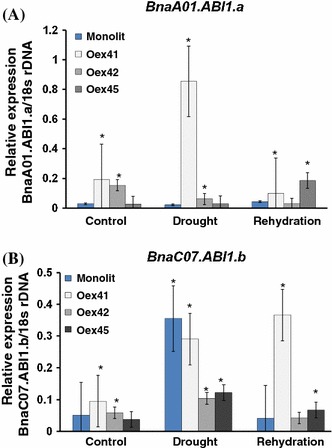
Relative expression of BnaA01.ABI1.a and BnaC07.ABI1.b genes in wild type and ABI1-overexpressing lines. Results were normalized against 18S rDNA expression. Data are representative of three independent experiments. Values represent mean ± SE
First, to identify any functional redundancy or divergence within the BnaABI1 paralogues, two evolutionarily distant examples described above (BnaA01.ABI1.a and BnaC07.ABI1.b), were used. Both genes showed different expression patterns during drought stress in WT and in the transgenic plants generated (Fig. 6). Under well-watered conditions, transcript levels of both BnaA01.ABI1.a and BnaC07.ABI1.b in WT plants and transgenic lines were low. However, significant changes in the induction of these genes were observed when the plants were subjected to drought stress. In general, BnaA01.ABI1.a expression did not change in either wild-type or transgenic plants, and remained suppressed; an exception was Oex41, which showed a significant increase in AtABI1 (and possibly BnaA01ABI1.a) expression compared to the wild-type plants. In contrast, BnaC07.AB11.b was upregulated in both wild-type plants and all transgenic lines exposed to drought. Following rehydration, the expression of BnaC07.ABI1.b returned to control levels in wild-type plants and transgenic lines (Fig. 6).
Analysis of BnaA01.ABI1.a and BnaC07.ABI1.b promoter activity
To further investigate the expression patterns of the BnaA01.ABI1.a and BnaC07.ABI1.b genes, their 2.2 kb promoter fragments were fused to the GUS reporter gene and the resulting ProBnaA01.ABI1.a:GUS and ProBnaC07.ABI1.b:GUS constructs were used to generate transgenic A. thaliana plants. Five independent transgenic lines for each construct were tested for GUS activity. All construct-specific lines showed the same expression pattern under any conditions tested. Histochemical staining of plants harboring the ProBnaA01.ABI1.a:GUS and ProBnaC07.ABI1.b:GUS constructs showed that there were no major differences in GUS activity in different organs (Fig. 7). Both promoters induced GUS activity in leaves, flowers, siliques and pollen (Fig. 7c–f) and in response to exogenous ABA treatment and wounding (Fig. 8). Interestingly, only BnaC07.ABI1.b was induced in the leaves and stems during drought (Fig. 8). This observation confirmed the earlier findings indicating different expression patterns of two BnaABI1 paralogues under drought stress conditions (Fig. 6). To better understand transcriptional regulation of the BnaA01.ABI1.a and BnaC07.ABI1.b genes, a de novo search for ABA- and water deficit-responsive cis-regulatory elements within the 2.2 kb regions upstream of their translation start codons was performed. Based on data from different databases (PLACE, Plant Matrix Family Library at Genomatix platform and PlantCare) it was shown that the BnaA01.ABI1.a and BnaC07.ABI1.b promoters contained 20 and 14 ABA- and osmotic stress-responsive cis-elements within the 2.2 kb upstream of the translation start site, respectively (Table S3). They include known cis-acting plant elements such as ABREs, MYB recognition sites, NYC recognition sites and SALTs, which are arranged in several copies throughout the upstream regions. The sequence similarity between the BnaA01.ABI1.a and BnaC07.ABI1.b promoter regions is 56.1 % which as expected is much lower than between their protein-coding sequences (Fig. S8). Comparison of both promoter sequences allowed us to identify six conserved regions with almost 70 % sequence similarity over at least 90 bp (Figs. 9 and S8). These regions covered over 52 % of each promoter and included some common regulatory elements located at similar relative positions. Detailed comparative studies of regulatory motifs involved in abscisic acid responsiveness such as ABRE, NAC, MYB and SALT revealed variations in their numbers and genome localization in the two promoter regions (Table S3; Fig. S8). Only four common protein-binding elements were identified in the conserved regions (Fig. 9). These data imply that sequence variation in the BnaA01.ABI1.a and BnaC07.ABI1.b promoter regions, outside the conserved regions, may be responsible for changes in their induction under drought stress (Figs. 6 and 8). Especially that some of drought-responsive elements such as MYB were identified in 4 copies within 600–800 bp promoter region of drought-inducible BnaC07.ABI1.b paralogue; meaningfully, drought-insensitive BnaC01.ABI1.a paralogue misses that regulatory boxes in the corresponding promoter region.
Fig. 7.

Localization of ProBnaA01.ABI1.a:GUS and ProBnaC07.ABI1.b:GUS activity (blue staining) in transgenic A. thaliana plants. A-mock, B-whole plant, C-leaves, D-flowers, E-siliques, F-pollen. Experiments were repeated (n = 3) with similar results and representative data are shown
Fig. 8.
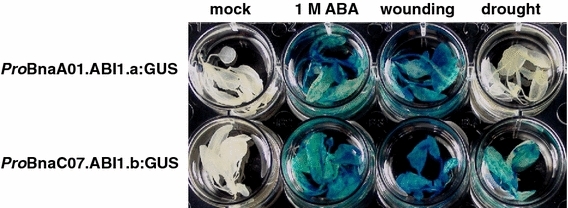
Histochemical visualization of GUS activity (blue staining) directed by BnaA01.ABI1.a and BnaC07.ABI1.b upstream regions in A. thaliana transgenic plants. Experiments were repeated (n = 3) with similar results and representative data are shown
Fig. 9.
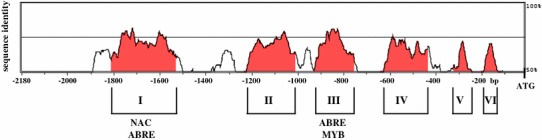
Conservation of the BnaA01.ABI1.a and BnaC07.ABI1.b promoter regions in B. napus. Six conserved regions named “Conserved Non-Coding Sequences” were defined by sequence similarity at the level of >70 % over a 90 bp interval. The sequences conserved between both promoters were visualized using VISualization Tool for Alignments (Vista) tools mVista and rVista (http://rvista.dcode.org/)
Discussion
In this report, we show that the role of the ABI1 orthologous gene (unique gene copy from the Arabidopsis genome) is conserved in Brassicaceae. We investigated the effect of AtABI1 overexpression in B. napus in drought stress conditions. Our results on the role of ABA signaling in drought stress, in particular the involvement of ABI1 protein phosphatase, highlight the complexity of the B. napus stress response mechanism. The transgenic B. napus lines overexpressing AtABI1 were characterized in detail with respect to their drought stress response. When overexpressed in B. napus, AtABI1 negatively influences several important cellular processes such as plant water content (for example, line Oex41, which has a high ABI1 transcript level, suffered water loss of 40 %), chlorophyll accumulation and the expression profile of several ABA- and/or dehydration stress-inducible genes (Figs. 1, 2 and 5). Intriguingly, most genes with the exception of BnaA01.ABI1.a and BnaX.MAPKKK18 were transcriptionally induced in control plants in a similar way to counterparts in A. thaliana. The expression profiles of selected PP2Cs, such as BnaA01.ABI1.a, BnaC07.ABI1.b, BnaX.ABI2,BnaX.HAB2.a and BnaX.HAB2.c, were also similar to those found in B. oleracea exposed to drought stress (Ludwików et al. 2013). Thus, our results highlight the regulatory functions of the ABI1 PP2C, which is highly conserved within the Brassicaceae family, and suggest that it plays a role as a regulatory hub protein (for review Ma et al. 2009) for dehydration stress responses in plants. Our results showing that the expression levels of several ABA-regulated genes were reduced in each transgenic line exposed to drought stress are consistent with such a role for ABI1 as a regulator of the drought stress response. Our previous results, which showed that ABI1 negatively regulates expression of RAB18, MAPKKK18 and other drought-induced genes (Ludwików et al. 2009), supports this hypothesis.
Understanding the biological role of duplicated genes, and particularly whether they have redundant or divergent functions in stress responses, is currently a high priority, although it is a challenging task in crop plant studies. We demonstrate differential involvement of the B. napusABI1 paralogues in the water-stress response. There are six ABI1-related genes in the B. napus genome, consistent with previous genetic studies indicating the presence of several ABI1 gene copies in the Brassica genomes (Sadowski and Quiros 1998; Quiros et al. 2001; Ludwików et al. 2013). By analysis of the genome sequences of B. rapa and B. oleracea and of different sets of B. napus EST clones, the six copies disclosed represent the complete set of BnaABI1 paralogues in B. napus. Prior to the current work, single ABI1-like gene had been described in the B. napus genome, corresponding to that identified and characterized herewith as BnaC01.ABI1.a (Zhang et al. 2014; Fig. S2). As expected, a phylogenetic analysis of group A PP2Cs in A. thaliana and B. napus confirmed their close evolutionary relationship (Fig. S2). Generally, the number of multiple copies of BnaABI1 correspond proportionally to the increase in total gene number in B. napus polyploid genome. This suggests that a repertoire of protein targets recognized by numerous BnaABI1 s is enlarged. Despite the difference in the number of genes in both species, they divided into two subgroups as in other species (Xue et al. 2008; Komatsu et al. 2009).
The maintenance of several duplicated copies of key regulatory genes over long periods of evolution is consistent with the gene balance hypothesis, which assumes that elements of complex signaling networks are preferentially protected against gene loss (Blanc and Wolfe 2004; Wang et al. 2011). Certainly, if such duplicated gene copies acquire different functions, they are more likely to be stably retained in a given genome. Studies on different organisms reveal that members of gene families may respond differently to environmental conditions as a result of either sub- and/or neofunctionalization (for example, Whittle and Krochko 2009; Liu and Adams 2010). Using a custom-made Arabidopsis cDNA macroarray, we previously revealed differences in the gene expression profiles of PP2C (group A) members under drought conditions and after ABA treatment in B. oleracea and A. thaliana (Ludwików et al. 2013). Interestingly, the BolABI1 gene (detected by AtABI1-gene-specific primers) was downregulated during drought stress in B. oleracea; this suggests that it is not essential for the drought stress response. On the other hand, subsequent studies indicate that one of the BolABI1 genes in B. oleracea corresponding to BnaC08.ABI1.c in B. napus is induced following exogenous ABA treatment and in the early stages of drought stress, confirming its involvement in the ABA signaling pathway (Yuan et al. 2013).
To determine the degree of conservation of the transcriptional regulation of the BnaABI1 paralogues, BnaA01.ABI1.a and BnaC07.ABI1.b, which represent the closest and a more distant homologues of the A. thalianaAtABI1 gene, these genes were characterized in detail (Fig. S2). Both BnaABI1 paralogues have a similar gene structure, with four exons and three introns, a C-terminal PP2C catalytic domain architecture with the same arrangement of 11 motifs, and identical localization of the NLS-like motif (Figs. S3, S5 and S7). Multiple alignments of genomic, cDNA and predicted protein sequences showed a high degree of sequence similarity between BnaA01.ABI1.a and BnaC07.ABI1.b of up to almost 85 %. However, our analysis reveals sequence variability in the 5′ upstream regions including the promoter, exon I and intron sequences, enabling gene-specific analyses at both the transcriptional and proteomic levels (Fig. S6). These data confirmed previous results indicating reduced sequence conservation of these upstream regions among members of the same gene family, which may reflect changes in the expression pattern of the respective genes (Chen et al. 2011). Our finding that the transcriptional expression patterns of selected BnaABI1 endogenous paralogues, i.e. BnaA01.ABI1.a and BnaC07.ABI1.b, significantly differ in response to drought stress, while showing similar transcriptional patterns after treatment with other stimuli (Figs. 6, 8), is entirely novel. Both genes appeared to be responsive to exogenous ABA, but only BnaC07.ABI1.b is induced by drought. Thus, BnaA01.ABI1.a and BnaC07.ABI1.b endogenes may have developed new subfunctions that improve the fine-scale response to drought stress. It remains possible that further subfunctions have evolved in other B. napusABI1 and PP2C group A homologues. Similarly, the different expression patterns under drought were found for two BnaHAB2 gene copies in WT plants and AtABI1 overexpressed lines of B. napus, revealing gene subfunctionalisation in the ABA signaling network (Fig. 5). This is a promising area of investigation as a comparative analysis of the B. rapa transcriptome (Lee et al. 2008) indicates that B. rapa homologues corresponding to BnaA01.ABI1.a and BnaC07.ABI1.b also differ in the response to salt stress: the B. rapa homologue of BnaA01.ABI1.a was found to be responsive to salt stress while, in contrast, a homologue of BnaC07.ABI1.b was not. In light of our observations, we assume that BnaABI1 homologues are potentially drought- as well as ABA-responsive, as these two stimuli positively induces all ABI1 phosphatases studied so far in different species.
Presently, little is known about the transcriptional regulation of the PP2C gene family in plants. A previous study revealed that the A. thalianaABI1 promoter is active in guard cells and root meristem (Leung et al. 2006). To approximate a mechanism controlling induction of both BnaABI1 promoters under ABA treatment and drought stress, in silico identification of ABA-inducible and stress-responsive elements within these regulatory regions was performed. Previous studies on the mechanisms of transcriptional regulation of ABA-inducible genes identified specific cis-acting elements associated with the responses to ABA, osmotic and water stresses (Narusaka et al. 2003; Yamaguchi-Shinozaki and Shinozaki 2005, 2006). Among the best described are the ABRE (ABA-responsive element), MYB, MYC, NAC, DRE, SALT and W-box elements recognized by the AREB/ABF, MYB, MYC, NAC, DREB, Alphin and WRKY family transcription factors, respectively (Abe et al. 2003; Yamaguchi-Shinozaki and Shinozaki 2005; Wang et al. 2009). Some reports document the interaction between ABI1 and transcription factors in Arabidopsis (Himmelbach et al. 2002; Cui et al. 2013). Thus, AtMYB20, which binds to the MYB recognition sequence (TAACTG) and the ACGT core element in the ABI1 and AtPP2CA promoter regions, acts as a negative regulator of PP2C genes, thereby enhancing salt tolerance (Cui et al. 2013). Interestingly, sequence comparison of AtABI1 and both BnaABI1 promoter regions showed that these cis-acting elements binding AtMYB20 are only present in the BnaA01.ABI1.a gene (data not shown). This suggests that AtABI1 and one, at least, of the B. napus BnaABI1-like genes may be involved in the same NaCl-stress-responsive pathways. An earlier study also demonstrated physical interaction between the homeodomain transcription factor AtHB6 and ABI1, which was positively correlated with PP2C activity (Himmelbach et al. 2002). To identify potentially all cis-acting elements associated with the response to ABA and/or drought stress within the BnaA01.ABI1.a and BnaC07.ABI1.b promoters, plant databases such as Plant Matrix Family Library, PLACE and PlantCare were used, revealing several ABA-inducible and stress-responsive elements (Table S3). Comparative sequence analysis of the BnaA01.ABI1.a and BnaC07.ABI1.b promoters showed significant differences in number and localization of ABA- and stress-inducible regulatory elements, despite the close relationship of these genes (Table S3 and Fig. S8). Interestingly, most elements are located from −1180 to −1680 bp and from −680 to −1180 bp in the BnaA01.ABI1.a and BnaC07.ABI1.b promoters, respectively, i.e. outside the most conserved regions. Only four of the regulatory motifs detected are located in the CNSs of both promoters (Fig. 9). In summary, comparison of the two promoters suggests that the lack of induction of BnaA01.ABI1.a by drought could result from the loss of several cis-elements in the BnaA01.ABI1.a promoter during the course of evolution. This indicates that specific expression patterns of these homologous genes, resulting in their subfunctionalization, may be modulated through mutation and possibly also through epigenetic changes within their respective promoter regions.
Electronic supplementary material
List of primers used in this study (DOC 49 kb)
List of clones used to a phylogenetic analysis (DOC 35 kb)
List of cis-acting elements involved in ABA and drought stress identified in BnaA01.ABI1.a and BnaC07.ABI1.b promoter regions based on PLACE (http://www.dna.affrc.go.jp/PLACE/), Plant Matrix Family Library (MathInspector, Plant IUPAC Library Version 7.0 restricted to A. thaliana with IUPAC search parameters: max. 0% mismatches; http://www.genomatix.de) and PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) databases (DOC 76 kb)
AtABI1 transcript levels in the wild-type Monolit and AtABI1-overexpressing B. napus transgenic lines. Expression of AtABI1 was measured by qRT-PCR (n = 3, ± SE) in leaves of control (well-watered) plants. AtABI1 mRNA abundance was normalized against 18S rDNA expression. Error bars represent SE (PPT 79 kb)
Phylogenetic analysis of ABI1 proteins of B. napus and its diploid B. rapa and B. oleracea progenitors. ABI1 proteins of the Brassica species were named according to the nomenclature system proposed for the Brassica genus (Østergaard and King 2008). Additionally, the protein sequences derived from JX122895 (Zhang et al. 2014) and KF577723 (Yuan et al. 2013) clones were included. The root tree was constructed using ClustalX to generate alignments of the studied amino acid sequences and selected fragments corresponding to the PP2C catalytic domain. Evolutionary analyses were conducted in MEGA5.1. The percentage of replicate trees is shown on the branches and is calculated according to the bootstrap test (1000 replicates) (PDF 13 kb)
Multiple sequence alignment of full-length ABI1 proteins from A. thaliana and B. napus. Sequences were aligned using the ClustalX program with default parameters. Gaps for optimal alignment are indicated by dashes. Asterisks beneath sequences indicate identical amino acid residues. The gray background and red/blue letters mark the catalytic domain and 11 characteristic motifs, respectively, as assigned by Bork et al. (1996), which are highly conserved across the ABI1 gene family in A. thaliana and B. napus. The amino acid position is given on the right of each sequence. The NLS-like (monopartite nuclear localization signal) motif is underlined (DOC 24 kb)
Alignments of (A) BnaA01.ABI1.a (B. napus), BraA01.ABI1.a (B. rapa) and BolC01.ABI1.a (B. oleracea) genomic DNA sequences; (B) BnaC07.ABI1.b (B. napus), BraA03.ABI1.b (B. rapa) and BolC07.ABI1.b (B. oleracea) genomic DNA sequences. These sequences were obtained using the same BnaABI1 gene-specific primer pair. Introns are shown in gray. The names of B. napus and B. rapa, and B. napus and B. oleracea, orthologous sequences are marked in red. Asterisks indicate identical residues (DOC 33 kb)
Schematic diagram representing the structure of the BnaA01.ABI1.a and BnaC07.ABI1.b genes in B. napus. The exons are shown as thick yellow boxes, the introns as lines and the 3’ and 5’ UTRs as thick blue boxes. The exon/intron structure of each BnaABI1 gene was defined by comparison of their genomic and cDNA sequences. The 5′ and 3’ end of BnaABI1 genes was predicted by five independent 5′ and 3’ RACE reactions. The scale is shown below in base pairs. The gene structure of both BnaABI1 genes was illustrated using the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/) (PNG 8 kb)
Sequence alignment of BnaA01.ABI1.a and BnaC07.ABI1.b fragment genomic DNA including exon I and intron I. Introns are shown in gray. Asterisks indicate identical residues. Translation initiation codons (ATG) are shown (DOC 24 kb)
Localization of the conserved PP2C catalytic domain in deduced BnaABI1 protein sequences. The PP2C domain family was defined and mapped in BnaABI1 protein sequences using The Conserved Domain Database of the NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The scale is shown at the top in number of amino acids (TIFF 37 kb)
Alignment of BnaA01.ABI1.a and BnaC07.ABI1.b promoter regions. The six CNS (Conserved Non-Coding Sequence) regions were identified using VISualization Tool for Alignments (Vista) tools mVista and rVista (http://rvista.dcode.org/) with sequence similarity at the level of >70% over a 90 bp region; they are highlighted in red. The putative regulatory elements identified by public database searches are boxed in green and named above the core sequence. Gaps for optimal alignment are indicated by dashes. Asterisks beneath sequences indicate identical nucleotide residues (DOC 48 kb)
Acknowledgments
We thank Assist. Prof. Tomasz Pniewski for the pKGIB vector. This work was supported by the Polish Committee for Scientific Research Grants PBZ-MNiSW-2/3/2006/19/IGR/1, PBZ-MNiSW-2/3/2006/19/IGR/3, MNiSW-2/3/2006/19/IGR/4 and N N303 568339.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Danuta Babula-Skowrońska and Agnieszka Ludwików have contributed equally to this article.
Contributor Information
Danuta Babula-Skowrońska, Phone: (+ 48 61) 65 50 237, Email: dbab@igr.poznan.pl.
Jan Sadowski, Phone: (+ 48 61) 829 59 63, Email: jsad@amu.edu.pl.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babula D, Kaczmarek M, Barakat A, Delseny M, Quiros CF, Sadowski J. Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: complexity of the comparative map. Mol Genet Genomics. 2003;268:656–665. doi: 10.1007/s00438-002-0782-2. [DOI] [PubMed] [Google Scholar]
- Babula D, Misztal LH, Jakubowicz M, Kaczmarek M, Nowak W, Sadowski J. Genes involved in biosynthesis and signalisation of ethylene in Brassica oleracea and Arabidopsis thaliana: identification and genome comparative mapping of specific gene homologues. Theor Appl Genet. 2006;112:410–420. doi: 10.1007/s00122-005-0136-7. [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Brown NP, Hegyi H, Schultz J. The protein phosphatase 2C (PP2C) superfamily: detection of bacterial homologues. Protein Sci. 1996;5:1421–1425. doi: 10.1002/pro.5560050720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegielska-Taras T, Pniewski T, Szała L. Transformation of microspore-derived embryos of winter oilseed rape (Brassica napus L.) by using Agrobacterium tumefaciens. J Appl Genet. 2008;49:343–347. doi: 10.1007/BF03195632. [DOI] [PubMed] [Google Scholar]
- Chen X, Truksa M, Snyder CL, El-Mezawy A, Shah S, Weselake RJ. Three homologous genes encoding sn-glycerol-3-phosphate acyltransferase 4 exhibit different expression patterns and functional divergence in Brassica napus. Plant Physiol. 2011;155:851–865. doi: 10.1104/pp.110.169482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Schumaker K, Zhu J-K. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot. 2004;55:225–236. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cui MH, Yoo KS, Hyoung S, Nguyen HTK, Kim YY, Kim HJ, Ok SH, Yoo SD, Shin JS. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 2013;587:1773–1778. doi: 10.1016/j.febslet.2013.04.028. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KA, Grill E, Romeis T, Hedrich R. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2 + affinities. Proc Natl Acad Sci USA. 2010;107:8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Garcia MP, Rodriguez D, Nicolas C, Rodriguez PL, Nicolas G, Lorenzo O. Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiol. 2003;133:135–144. doi: 10.1104/pp.103.025569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell. 1999;11:1897–1910. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Hohener B, Grill E. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 2002;21:3029–3038. doi: 10.1093/emboj/cdf316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H-F, Lu D, Sun J-H, Li C-L, Xing Y, Qin L, Shen Y-Y. Type 2C protein phosphatase ABI1 is a negative regulator of strawberry fruit ripening. J Exp Bot. 2013;64:1677–1687. doi: 10.1093/jxb/ert028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K, Nishikawa Y, Ohtsuka T, Taji T, Quatrano RS, Tanaka S, Sakata Y. Functional analyses of the ABI1-related protein phosphatase type 2C reveal evolutionarily conserved regulation of abscisic acid signaling between Arabidopsis and the moss Physcomitrella patens. Plant Mol Biol. 2009;70:327–340. doi: 10.1007/s11103-009-9476-z. [DOI] [PubMed] [Google Scholar]
- Lee SC, Lim MH, Kim JA, Lee SI, Kim JS, Jin M, Kwon SJ, Mun JH, Kim YK, Kim HU, Hur Y, Park BS. Transcriptome analysis in Brassica rapa under the abiotic stresses using Brassica 24 K oligo microarray. Mol Cells. 2008;26:595–605. [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. The Arabidopsis ABSCISIC ACID-INSENSlTIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Orfanidi S, Chefdor F, Mészaros T, Bolte S, Mizoguchi T, Shinozaki K, Giraudat J, Bögre L. Antagonistic interaction between MAP kinase and protein phosphatase 2C in stress recovery. Plant Sci. 2006;171:596–606. doi: 10.1016/j.plantsci.2006.06.009. [DOI] [Google Scholar]
- Liu SL, Adams KL. Dramatic change in function and expression pattern of a gene duplicated by polyploidy created a paternal effect gene in the Brassicaceae. Mol Biol Evol. 2010;27:2817–2828. doi: 10.1093/molbev/msq169. [DOI] [PubMed] [Google Scholar]
- Ludwików A. Targeting proteins for proteasomal degradation—a new function of Arabidopsis ABI1 protein phosphatase 2C. Front Plant Sci. 2015;6:310. doi: 10.3389/fpls.2015.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwików A, Kierzek D, Gallois P, Zeef L, Sadowski J. Gene expression profiling of ozone-treated Arabidopsis abi1td insertional mutant: protein phosphatase 2C ABI1 modulates biosynthesis ratio of ABA and ethylene. Planta. 2009;230:1003–1017. doi: 10.1007/s00425-009-1001-8. [DOI] [PubMed] [Google Scholar]
- Ludwików A, Babula-Skowrońska D, Szczepaniak M, Belter N, Dominiak E, Sadowski J. Expression profiles and genomic organisation of group A protein phosphatase 2C genes in Brassicaoleracea. Annals Appl Biol. 2013;163:124–134. doi: 10.1111/aab.12039. [DOI] [Google Scholar]
- Ludwików A, Cieśla A, Kasprowicz-Maluśki A, Mituła F, Tajdel M, Gałgański Ł, Ziółkowski PA, Kubiak P, Małecka A, Piechalak A, Szabat M, Górska A, Dąbrowski M, Ibragimow I, Sadowski J. Arabidopsis protein phosphatase 2C ABI1 interacts with type I ACC synthases and is involved in the regulation of ozone-induced ethylene biosynthesis. Mol Plant. 2014;7:960–976. doi: 10.1093/mp/ssu025. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Koch MA, Pecinka A, Schubert I. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005;15:516–525. doi: 10.1101/gr.3531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001;25:295–303. doi: 10.1046/j.1365-313x.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003;34:137–148. doi: 10.1046/j.1365-313X.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Østergaard L, King GJ. Standardized gene nomenclature for the Brassica genus. Plant Methods. 2008;4:10. doi: 10.1186/1746-4811-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin IAP, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics. 2005;171:765–781. doi: 10.1534/genetics.105.042093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros CF, Grellet F, Sadowski J, Suzuki T, Li G, Wroblewski T. Arabidopsis and Brassica comparative genomics: sequence, structure and gene content in the ABI1-Rps2-Ck1 chromosomal segment and related regions. Genetics. 2001;157:1321–1330. doi: 10.1093/genetics/157.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell. 2000;12:707–719. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim T, et al. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 2009;50:1345–1355. doi: 10.1104/pp.109.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski J, Quiros CF. Organization of an Arabidopsis thaliana gene cluster on chromosome 4 including the RPS2 gene, in the Brassica nigra genome. Theor Appl Genet. 1998;96:468–474. doi: 10.1007/s001220050763. [DOI] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signaling. Plant J. 2004;37:354–369. doi: 10.1046/j.1365-313X.2003.01966.x. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Heribert Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 2004;9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Sheen J. Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci. 1998;95:975–980. doi: 10.1073/pnas.95.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer RA, Robinson-Rechavi M. How confident can we be that orthologs are similar, but paralogs differ? Trends Gen. 2009;25:210–216. doi: 10.1016/j.tig.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Swamy PM, Smith B. Role of abscisic acid in plant stress tolerance. Current Sci. 1999;76:1220–1227. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tougane K, Komatsu K, Bhyan SB, Sakata Y, Ishizaki K, Yamato KT, et al. Evolutionarily conserved regulatory mechanisms of abscisic acid signaling in land plants: characterization of ABSCISIC ACID INSENSITIVE1-like type 2C protein phosphatase in the liverwort Marchantiapolymorpha. Plant Physiol. 2010;152:1529–1543. doi: 10.1104/pp.110.153387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall JA, Wendel JF. Polyploidy and crop improvement. Crop Sci. 2006;46:S-3–S-14. doi: 10.2135/cropsci2006.07.0489tpg. [DOI] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayash S, Myouga F, Yamaguchi-Shinozaki K, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 2010;51:1821–1839. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhu Y, Wang L, Liu X, Liu Y, Phillips J, Deng X. A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta. 2009;230:1155–1166. doi: 10.1007/s00425-009-1014-3. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Whittle CA, Krochko JE. Transcript profiling provides evidence of functional divergence and expression networks among ribosomal protein gene paralogs in Brassica napus. Plant Cell. 2009;21:2203–2219. doi: 10.1105/tpc.109.068411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T, Wang D, Zhang S, Ehlting J, Ni F, Jakab S, Zheng C, Zhong Y. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics. 2008;9:550. doi: 10.1186/1471-2164-9-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T. ABA-hypersensitive germination 3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006;140:115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F, Wang M, Hao H, Zhang Y, Zhao H, Guo A, Xu H, Zhou X, Xie CG. Negative regulation of abscisic acid signaling by the Brassica oleracea ABI1 ortholog. Biochem Biophys Res Commun. 2013;442:202–208. doi: 10.1016/j.bbrc.2013.11.035. [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu W-Z, Zhang Y, Deng M, Niu F, Yang B, Wang X, Wang B, Liang W, Deyholos MK, Yiang Y-Q. Identification, expression and interaction analyses of calcium-dependent protein kinase (CPK) genes in canola (Brassica napus L.) BMC Genomics. 2014;15:211. doi: 10.1186/1471-2164-15-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2003;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowski PA, Kaczmarek M, Babula D, Sadowski J. Genome evolution in Arabidopsis/Brassica: conservation and divergence of ancient rearranged segments and their breakpoints. Plant J. 2006;47:67–74. doi: 10.1111/j.1365-313X.2006.02762.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers used in this study (DOC 49 kb)
List of clones used to a phylogenetic analysis (DOC 35 kb)
List of cis-acting elements involved in ABA and drought stress identified in BnaA01.ABI1.a and BnaC07.ABI1.b promoter regions based on PLACE (http://www.dna.affrc.go.jp/PLACE/), Plant Matrix Family Library (MathInspector, Plant IUPAC Library Version 7.0 restricted to A. thaliana with IUPAC search parameters: max. 0% mismatches; http://www.genomatix.de) and PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) databases (DOC 76 kb)
AtABI1 transcript levels in the wild-type Monolit and AtABI1-overexpressing B. napus transgenic lines. Expression of AtABI1 was measured by qRT-PCR (n = 3, ± SE) in leaves of control (well-watered) plants. AtABI1 mRNA abundance was normalized against 18S rDNA expression. Error bars represent SE (PPT 79 kb)
Phylogenetic analysis of ABI1 proteins of B. napus and its diploid B. rapa and B. oleracea progenitors. ABI1 proteins of the Brassica species were named according to the nomenclature system proposed for the Brassica genus (Østergaard and King 2008). Additionally, the protein sequences derived from JX122895 (Zhang et al. 2014) and KF577723 (Yuan et al. 2013) clones were included. The root tree was constructed using ClustalX to generate alignments of the studied amino acid sequences and selected fragments corresponding to the PP2C catalytic domain. Evolutionary analyses were conducted in MEGA5.1. The percentage of replicate trees is shown on the branches and is calculated according to the bootstrap test (1000 replicates) (PDF 13 kb)
Multiple sequence alignment of full-length ABI1 proteins from A. thaliana and B. napus. Sequences were aligned using the ClustalX program with default parameters. Gaps for optimal alignment are indicated by dashes. Asterisks beneath sequences indicate identical amino acid residues. The gray background and red/blue letters mark the catalytic domain and 11 characteristic motifs, respectively, as assigned by Bork et al. (1996), which are highly conserved across the ABI1 gene family in A. thaliana and B. napus. The amino acid position is given on the right of each sequence. The NLS-like (monopartite nuclear localization signal) motif is underlined (DOC 24 kb)
Alignments of (A) BnaA01.ABI1.a (B. napus), BraA01.ABI1.a (B. rapa) and BolC01.ABI1.a (B. oleracea) genomic DNA sequences; (B) BnaC07.ABI1.b (B. napus), BraA03.ABI1.b (B. rapa) and BolC07.ABI1.b (B. oleracea) genomic DNA sequences. These sequences were obtained using the same BnaABI1 gene-specific primer pair. Introns are shown in gray. The names of B. napus and B. rapa, and B. napus and B. oleracea, orthologous sequences are marked in red. Asterisks indicate identical residues (DOC 33 kb)
Schematic diagram representing the structure of the BnaA01.ABI1.a and BnaC07.ABI1.b genes in B. napus. The exons are shown as thick yellow boxes, the introns as lines and the 3’ and 5’ UTRs as thick blue boxes. The exon/intron structure of each BnaABI1 gene was defined by comparison of their genomic and cDNA sequences. The 5′ and 3’ end of BnaABI1 genes was predicted by five independent 5′ and 3’ RACE reactions. The scale is shown below in base pairs. The gene structure of both BnaABI1 genes was illustrated using the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/) (PNG 8 kb)
Sequence alignment of BnaA01.ABI1.a and BnaC07.ABI1.b fragment genomic DNA including exon I and intron I. Introns are shown in gray. Asterisks indicate identical residues. Translation initiation codons (ATG) are shown (DOC 24 kb)
Localization of the conserved PP2C catalytic domain in deduced BnaABI1 protein sequences. The PP2C domain family was defined and mapped in BnaABI1 protein sequences using The Conserved Domain Database of the NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The scale is shown at the top in number of amino acids (TIFF 37 kb)
Alignment of BnaA01.ABI1.a and BnaC07.ABI1.b promoter regions. The six CNS (Conserved Non-Coding Sequence) regions were identified using VISualization Tool for Alignments (Vista) tools mVista and rVista (http://rvista.dcode.org/) with sequence similarity at the level of >70% over a 90 bp region; they are highlighted in red. The putative regulatory elements identified by public database searches are boxed in green and named above the core sequence. Gaps for optimal alignment are indicated by dashes. Asterisks beneath sequences indicate identical nucleotide residues (DOC 48 kb)


