Abstract
Aim
We sought to discover endogenous urinary biomarkers of human CYP2D6 activity.
Patients & methods
Healthy pediatric subjects (n = 189) were phenotyped using dextromethorphan and randomized for candidate biomarker selection and validation. Global urinary metabolomics was performed using liquid chromatography quadrupole time-of-flight mass spectrometry. Candidate biomarkers were tested in adults receiving fluoxetine, a CYP2D6 inhibitor.
Results
A biomarker, M1 (m/z 444.3102) was correlated with CYP2D6 activity in both the pediatric training and validation sets. Poor metabolizers had undetectable levels of M1, whereas it was present in subjects with other phenotypes. In adult subjects, a 9.56-fold decrease in M1 abundance was observed during CYP2D6 inhibition.
Conclusion
Identification and validation of M1 may provide a noninvasive means of CYP2D6 phenotyping.
Keywords: biomarker, clinical, CYP2D6, dextromethorphan, endogenous, fluoxetine, metabolomics, pediatric, phenotype
In various pediatric populations, a substantial percentage (10–65%) [1] of drugs are prescribed off-label, primarily because of lack of study information in this age group [2]. Although the number of drugs with some pediatric dosing information doubled from 22% in 1975 to 41% in 2009 [3], available information remains inadequate to provide effective safety and dosing guidances for all pediatric age groups. Studies conducted in the past two decades show major deficiencies with pediatric dosing methods built on allometry [4–8]. Optimal drug safety and efficacy in this vulnerable population requires an increased understanding of the impacts of physiological maturation and genetic variation on drug-metabolizing enzymes [9–11]. One such enzyme, CYP2D6, participates in the elimination of approximately 15% of clinically used drugs metabolized by cytochrome P450s [12], including antipsychotics and antidepressants, beta-blockers, opioid analgesics [13,14] and the antitussive dextromethorphan (DM) [15].
CYP2D6 activity is determined, in significant part, by genetic variation. More than 100 allelic variants and subvariants of this highly polymorphic enzyme are defined [16], including gene deletions, and nonfunctional, reduced-function and multiple-copy-number alleles [17,18]. The various allele combinations result in a continuum from little or no activity and poor metabolizer (PM) phenotype, reduced function and intermediate metabolizer phenotype, ‘wild-type’ activity and extensive metabolizer phenotype to, in the case of gene duplications, increased CYP2D6 expression and ultrarapid metabolizer phenotype [18–20]. Gaedigk et al. proposed using these genetic factors to predict individual CYP2D6 activity scores [19,21].
Substantial interindividual variation in CYP2D6 activity exists within each category of CYP2D6 phenotype [21–23]. Factors such as ontogeny, drug–drug interactions (DDIs), diet and disease may contribute to patient variation in drug disposition. Thus, phenotyping may be necessary to determine CYP2D6 activity when personalizing drug therapy [24] or evaluating DDIs [25]. Traditional phenotyping studies involve the administration of a probe drug or drug cocktail. The administration of such drugs to children, pregnant women and the elderly may be a concern.
Phenotyping using endogenous biomarkers is an alternative method of assessing CYP2D6 activity that may eliminate risks of exogenous drug administration in CYP2D6 DDI studies or when evaluating altered activity in special patient populations. Additionally, retrospective analyses of banked samples may be conducted without prior knowledge of an individual's CYP2D6 phenotype. If biomarker measurements from a spot urine sample could be used, that could eliminate the need for prolonged clinical visits in which the patient must provide timed sample collections. For example, CYP2D6 activity can be measured using a 4-h urine collection following DM administration. Several endogenous substrates have been proposed as CYP2D6 substrates. They include 5-methoxy-N,N-dimethyltryptamine [26], pinoline [27], progesterone [28], anandamide [29,30] and a number of compounds up- or downregulated in CYP2D6-transgenic mice [31]. However, the identification and validation of endogenous biomarkers for CYP2D6 phenotyping, particularly in humans, is immature.
The goal of this study was to discover endogenous biomarkers of CYP2D6 activity. Children were phenotyped for CYP2D6 activity using a urinary dextromethorphan-to-dextrorphan metabolic ratio (DM/DX) [21,32], and a global metabolomics approach was used to detect endogenous, urinary metabolites capable of predicting DM/DX in a pediatric training set group. A targeted analytical approach was used to further analyze the relationship between a candidate biomarker and CYP2D6 activity, and these results were validated in urine from a second group of children. Furthermore, the change in metabolite levels and CYP2D6 activity was assessed in adults during CYP2D6 inhibition.
Patients & methods
Subjects
Healthy pediatric subjects (n = 189) between 6 and 15 years of age on the date of study enrollment were recruited at Children's Mercy Hospitals and Clinics (CMH), Kansas City, MO, USA for a longitudinal study of CYP2D6 activity. Results from the first study visit are presented. Following an overnight fast, subjects received a single oral 0.5 mg/kg dose of DM (Robitussin® Pediatric). Urine was collected predose (spot sample) and throughout the next 4 h following DM administration (timed sample). Samples were stored at −80°C until analysis. Subjects were randomized into training and validation sets based on their CYP2D6 phenotype (PM or non-PM) so there was equal representation between the two sets.
Studies were approved by the Institutional Review Boards at CMH and the University of Washington (UW), Seattle, WA, USA. The study population was slightly skewed towards males due to an additional study aim to recruit children diagnosed with attention-deficit hyperactivity disorder, an aim not explored here. Exclusion criteria included: current therapy with medications metabolized by or known to inhibit CYP2D6 (although atomoxetine was permitted for the attention-deficit hyperactivity disorder component of the overall study); inability or unwillingness to fast 4 h prior to the study session; existence of diagnoses that may influence absorption and gastric emptying, such as reflux, inflammatory bowel disease or Crohn's disease; a demonstrated adverse reaction to previous DM exposure; impaired hepatic or renal activity or physical examination as determined by pediatrician sub-investigator's discretion; pregnancy; body mass index (BMI) <5th or >95th percentile. Subjects were given a complete medical examination including assessment of Tanner stage and blood samples were taken for liver function testing and DNA testing at the screening visit.
Ten healthy adults (five females and five males) were enrolled at UW in a study to evaluate the complex DDI of fluoxetine on CYP2D6, CYP3A4 and CYP2C19 activities. A detailed description of the study design, demographics and results has been reported elsewhere [25]. All subjects included in the study were genotypic CYP2D6 extensive metabolizers. Each subject was given an oral probe drug cocktail, which included 30 mg DM, at baseline (day 1) and during a 2-week multiple-dose fluoxetine treatment (day 16). Urine was collected 0–12 and 12–24 h on days 1–4 and 16–19. Secondary use of the samples in this study was approved by the UW Human Subjects Review Board.
Genotype analysis & assignment of activity score of pediatric subjects
CYP2D6 genotype analysis of greater than 20 allelic variants was performed at CMH. Genomic DNA was isolated from whole blood with a QIAamp DNA Blood Mini kit (Qiagen, CA, USA). Genotype analysis was performed using long-range (XL) PCR coupled with commercially available TaqMan (Life Technologies, CA, USA) and restriction fragment length polymorphism assays. The following allelic variants were assessed: CYP2D6*2, *3, *4, *5 (gene deletion), *6,*7, *9, *10, *11, *12, *13 (2D7/6 hybrid genes), *15,*17, *29, *31, *35, *36, *41, *42, *45 or *46 and *59. Analysis included XL-PCR-based detection of gene deletions, duplications/multiplications and CYP2D7/6 hybrid gene arrangements. Gene duplications were characterized for their allelic variation (*1xN, *2xN, *4xN, etc.) and gene copy number assessed at four gene loci by quantitative multiplex PCR. Additional XL-PCR testing in conjunction with DNA sequencing was applied to resolve complex cases or cases with ambiguous results.
Genotyping procedures and assignment of CYP2D6 activity scores have been described previously [21,33–36]. Nonfunctional, reduced function, fully functional and gain-of-function CYP2D6 alleles were given values of 0, 0.5, 1 and 1.5, respectively; alleles with duplications received double the value assigned to the single counterparts. The CYP2D6 activity score was obtained by summing the two alleles for each individual.
Analysis of DM & metabolites
Pediatric urine samples were analyzed for DM and its metabolites at CMH [37]. Briefly, urine samples were adjusted to a pH of 4.5–5.0 and subsequently incubated with β-glucuronidase at 37°C overnight prior to analysis. Concentrations of DM and total deconjugated dextrorphan (DX) were determined using reverse-phase HPLC with fluorescence detection. The urinary DM metabolic ratio was calculated as the molar ratio of DM/DX. Based on the DM metabolic ratio (DM/DX), subjects were assigned to PM (DM/DX ≥ 0.3) or non-PM phenotypes (DM/DX < 0.3) as described previously [21,32]. Analytical details for the urinary analysis of DM and its metabolites in the adult study are described by Sager et al. [25].
Determination of creatinine concentrations
Creatinine was quantified on an Agilent 1100 series HPLC in line with an Agilent 1050 series UV detector set to monitor absorbance at 234 nm. Urine was diluted 100-fold with 10 mM potassium phosphate buffer, pH 6.5 and 10 μl were injected onto a Waters Symmetry C18 5 μm, 4.6 × 1.5 mm, 300 Å analytical column using 10 mM potassium phosphate buffer, pH 6.5, with 0.1% acetonitrile for mobile phase A and 100% acetonitrile for mobile phase B. The flow rate was 0.5 ml/min with a linear gradient consisting of 0% B until 5 min, a linear increase to 100% B from 5.0 min until 5.5 min, 100% B until 6.5 min, a linear decrease to 0% B from 6.5 to 7 min and re-equilibration with 0% B for 8 min.
Global metabolomics analysis of pediatric training set by LC-QTOF
Samples were prepared for metabolomics analysis by adding 800 μl of ice-cold acetonitrile to 200 μl of urine to precipitate proteins. Following centrifugation (20,000 × G for 10 min, 4°C), the supernatant was evaporated under nitrogen gas. The resulting residue was reconstituted in 40 μl of methanol followed by 40 μl of 0.4% (v/v) acetic acid.
Global metabolomic analyses of samples were performed using an Agilent (Santa Clara, CA, USA) 1200 HPLC coupled to an Agilent 6520 QTOF mass spectrometer. Samples (2-μl injection) were separated chromatographically using a 3.5 μm, 2.1 × 30 mm Agilent Zorbax SB-C8 guard column and a 1.8 μm, 2.1 × 50 mm Agilent Zorbax SB-Aq analytical column heated to 60°C. The flow rate was 0.6 ml/min, and the mobile phase consisted of A: 0.2% acetic acid in water and B: 0.2% acetic acid in methanol utilizing the following gradient profile: 2% B at 0 min, 2–98% B in 13 min, 98% B until 19 min followed by re-equilibration for 6.5 min. The source temperature was maintained at 350°C with a nitrogen gas flow rate of 12 l/min and a capillary voltage of 3500 V. Scans were obtained between m/z 100 and 1000 at an acquisition rate of 3 spectra/s. Data were collected in centroid mode using positive and negative electrospray ionization (ESI+ and ESI−, respectively).
Global metabolomics data processing & statistical analyses
For the pediatric training set, raw LC quadrupole TOF (LC-QTOF) mass spectral data from each ionization mode were aligned using the Bioconductor R-package XCMS [38,39]. Raw data files were exported to mzData format using MassHunter Qualitative Analysis (Agilent, B.05.00) with a minimum peak height of 1000 counts. Feature detection was performed using the Bioconductor R-package XCMS [38,39]. Peak picking was performed using the centWave algorithm, requiring within-peak m/z deviations of less than 15 ppm, five consecutive scans above 500 counts and peak widths between 4 and 12 s. The default peak integration method was used, and the peaks were fit to a Gaussian shape with a Mexican-hat wavelet for integration. The peaks were grouped using the ‘density’ method with a mass accuracy requirement of 0.007 m/z and a peak width at half height of 4 s (before retention time correction) or 2 s (after retention time correction). A retention time correction was performed using the method ‘obiwarp,’ and peaks were recursively filled. Subsequently, ion signals were normalized to the sum of all mass feature abundances in that sample as a surrogate marker for urine concentration, and finally log transformed.
Statistical analyses were performed on each spot and timed dataset and ionization mode independently. Log-transformed DM/DX was regressed on each ion with no additional covariates. To account for possible dependencies among siblings and to avoid assuming homoscedasticity, we used a generalized estimating equation approach implemented in the R package ‘gee’ [40]. We adjusted the p-values for multiple-hypothesis testing using the Benjamini–Hochberg method as implemented in the R stats package's p.adjust function. This methodology addresses the multiple hypothesis testing problem without using excessively conservative approaches to control the family-wise error, such as a Bonferroni correction. An adjusted p-value <0.01 was considered to be significant.
LC-QTOF spectral fragmentation
Ions of interest were analyzed as described for global metabolomics except that the quadrupole selected the precursor ion and the TOF mass analyzer scanned product ions. Selected precursor ions (1.3 amu isolation width, 0.5 min allowable retention time shift) were fragmented at fixed collision energies of 10, 20, 40 and 80 V in ESI+ mode. The MS/MS scan rate was 4 spectra/s.
Database queries
For significant ions, we attempted to determine identities by querying major metabolomics databases using the accurate mass (within 20 ppm), and MS/MS fragmentation spectra or retention time, when available. Databases queried included the Scripps METLIN Metabolite Database [41,42], the Human Metabolome Database [43] (version 3.6 [44]) and in-house databases.
Relative quantification of M1 by LC-QqQ
MRM MS was performed on an Agilent 1290 Infinity HPLC coupled to an Agilent 6460 Triple Quadrupole (QqQ) to quantify the relative abundance of M1 in urine samples. A similar method as described for LC-QTOF was employed but with a shortened total run time of 6 min and a fixed collision energy of 40 V in ESI+ mode. We injected 5 μl and up to 15 μl for pediatric non-PM and PM samples, respectively. Two non-PM samples from the pediatric timed urine set were not analyzed due to insufficient sample volume. Peak areas for the mass transition of m/z 444.3→98.1 were divided by creatinine concentration (mM) to account for urine concentration differences. A value of 5, approximately the value of noise, was assigned to missing values. For the adult samples, creatinine normalized signals (m/z 444.3→98.1) for 0–12 and 12–24 h urine samples from days 3–4 (without fluoxetine) and days 18–19 (with fluoxetine) were summed to obtain a composite 0–24 h value.
General statistical analyses
GraphPad Prism (version 5.04, GraphPad Software, CA, USA) was used for general statistical analyses. Univariate regression was used to compare log(creatinine/M1) and log(DM/DX). Univariate regression was also used to model log M1 given creatinine, age, or urinary pH; and creatinine given age or urinary pH. A two-tailed t-test was used to determine whether log(DM/DX), log M1 or creatinine differed between genders. One-way analysis of variance (ANOVA) was used to determine the relationship between log(DM/DX) and Tanner stage or activity score, and to determine the relationship between log(creatinine/M1) and activity score. Spearman rank correlation was used to determine the relationship between log(DM/DX) and log(creatinine/M1) in adult subjects. A p-value <0.05 was considered to be significant.
Results
Demographic characteristics of pediatric subjects
Healthy pediatric volunteers (n = 189), some of whom were related, were recruited for a longitudinal study of CYP2D6 activity. A spot urine sample was collected predose and a timed urine sample was collected from 0–4 h after DM administration for determination of DM/DX. Based on DM/DX, ten subjects were phenotypic CYP2D6 PMs and the remaining 179 were classified as non-PMs (intermediate, extensive or ultrarapid metabolizers). PMs and non-PMs were randomly assigned to training or validation sets. Demographics for the training (n = 94, 5 PMs) and validation sets (n = 95, 5 PMs) are presented in Table 1. The subjects ranged from 7 to 16 years of age. Pubertal development was determined by the assignment of Tanner stage scores on a scale of 1 (prepubertal) to 5 or 6 (full maturation) for breast size and pubic hair, respectively. Log(DM/DX) did not differ by gender, age or measures of pubertal development (data not shown [Gaedigk A, Pearce RE, Tay-Sontheimer J et al. Effect of age and genotype on CYP2D6 Activity in children and adolescents (2014), Manuscript in preparation]). However, there was a negative correlation between log(DM/DX) and urinary pH (p < 0.0001, n = 189).
Table 1.
Demographics of the pediatric study population.
| Characteristic | All subjects (n = 189) | Training set (n = 94) | Validation set (n = 95) | ||
|---|---|---|---|---|---|
| PM (n = 5) | Non-PM (n = 89) | PM (n = 5) | Non-PM (n = 90) | ||
| Age (years) | 11.2 ± 2.5 (7–16) | 11.3 ± 2.4 (9–14) | 11.2 ± 2.4 (7–16) | 11.0 ± 1.6 (9–14) | 11.1 ± 2.5 (7–16) |
| Tanner stage | |||||
| – Breast size | 2.4 ± 1.5 (1–5) | 2.4 ± 1.7 (1–5) | 2.4 ± 1.5 (1–5) | 1.6 ± 0.5 (1–2) | 2.5 ± 1.5 (1–5) |
| – Pubic hair | 2.4 ± 1.5 (1–6) | 2.4 ± 1.7 (1–5) | 2.3 ± 1.5 (1–6) | 1.4 ± 0.5 (1–2) | 2.5 ± 1.5 (1–5) |
| Gender | |||||
| – Male (%) | 61 | 60 | 66 | 80 | 56 |
| Race (%) | |||||
| – African–American | 43 | 0 | 43 | 20 | 47 |
| – Caucasian (Hispanic) | 49 (5) | 100 (0) | 46 (4) | 80 (0) | 47 (6) |
| – Mixed | 8 | 0 | 11 | 0 | 7 |
| Urinary DM/DX ratio | 0.078 ± 0.3 (9.0 × 10−5–3.3) | 1.4 ± 1 (0.63–3.3) | 0.014 ± 0.03 (9.0 × 10−5–0.20) | 1.1 ± 0.7 (0.28–1.8) | 0.013 ± 0.02 (3.0 × 10−4–0.14) |
Age at visit 1, Tanner stage at enrollment and DM/DX are shown as the mean ± standard deviation (range).
DM/DX: Dextromethorphan-to-dextrorphan metabolic ratio; Non-PM: CYP2D6 intermediate, extensive or ultrarapid metabolizer phenotype subjects; PM: CYP2D6 poor metabolizer phenotype subjects.
Selection of CYP2D6 endogenous biomarkers in pediatric subjects
Identification of CYP2D6 biomarker candidates by global metabolomics in the pediatric training set
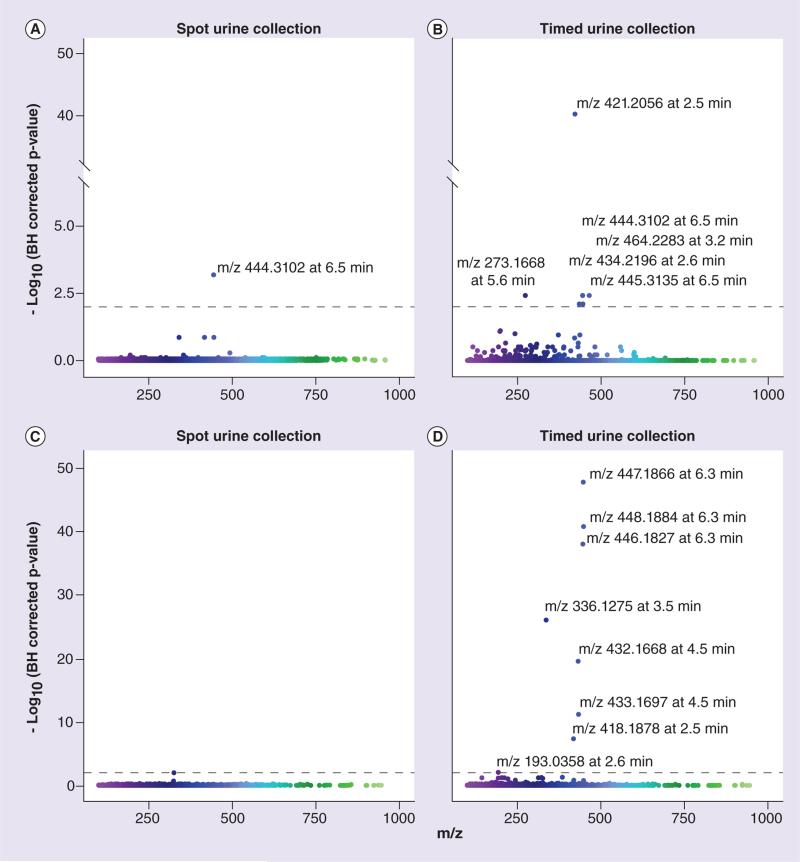
Spot and timed urine collection samples in the pediatric training set were analyzed by global metabolomic profiling using LC-QTOF MS in ESI+ and ESI− modes. Ions were aligned, normalized and log-transformed as described in the ‘Patients & methods’ section. The LC-QTOF data contained 3839 and 2883 ions in ESI+ and ESI− modes, respectively. For selecting ions associated with CYP2D6 phenotype, a generalized- estimating equation approach was used to linearly regress log ion intensities against log(DM/DX). An independent correlation structure was used in this approach to account for subject relatedness. As summarized in Figure 1 & Table 2, in ESI+ data, one and six ions (Benjamini–Hochberg [BH] corrected p-value <0.01) were found in the spot and timed urine samples, respectively; in ESI− data, eight ions (BH corrected p < 0.01) were observed in the timed urine samples. As listed in Table 2, some significant ions found only in timed urine samples are likely DM metabolites, such as DX glucuronide and oxo-dextrorphan-glucuronide [45]. Significant ions found in both urine-collection time points would likely result in fewer false positives and would exclude DM metabolites. For these reasons, only significant ions found both before and after DM administration (spot and timed urine samples, respectively) were further investigated. One ESI+ ion with a m/z of 444.3102 and eluting at 6.5 min, which we will refer to as M1, was found to be significant in both spot and timed urine samples. We focused on this candidate biomarker in subsequent experiments. A negative correlation existed between M1 abundance and log(DM/DX) (Table 2) suggesting that it may be the product of a reaction catalyzed by CYP2D6.
Figure 1. Manhattan plots of pediatric training dataset ions.
The ion abundances were regressed against log(dextromethorphan-to-dextrorphan metabolic ratio). Corresponding p-values were obtained and corrected using the BH method. The corresponding -log(BH corrected p-values) are shown for ESI+ mode (A & B) and ESI− mode (C & D) from LC quadrupole TOF analysis. Results from spot and 0–4 h timed urine samples are presented in (A & C) and (B & D), respectively. Ion m/z and retention time are explicitly shown for significant ions. The BH corrected significance threshold is indicated by the dashed line (p = 0.01). BH: Benjamini–Hochberg.
Table 2.
Summary of significant (Benjamini–Hochberg corrected p-value <0.01) m/z ions by ionization mode associated with log(dextromethorphan-to-dextrorphan metabolic ratio) following global metabolomics analysis of pediatric training set samples by LC-QTOF mass spectrometry.
| m/z | RT (min) | Possible identity [Ref.] | Slope | r2 | p-value | BH corrected p-value |
|---|---|---|---|---|---|---|
| ESI+ using spot urine samples | ||||||
| 444.3102 | 6.5 | – † | −0.79 | 0.34 | 1.69 × 10−7 | 6.50 × 10−4 |
| ESI+ using timed urine samples | ||||||
| 421.2056 | 2.5 | – † | −1.70 | 0.67 | 1.31 × 10−44 | 5.03 × 10−41 |
| 444.3102 | 6.5 | – † | −0.79 | 0.27 | 2.33 × 10−6 | 3.80 × 10−3 |
| 464.2283 | 3.2 | – † | −0.78 | 0.40 | 2.97 × 10−6 | 3.80 × 10−3 |
| 273.1668 | 5.6 | – † | 0.73 | 0.24 | 3.97 × 10−6 | 3.81 × 10−3 |
| 434.2196 | 2.6 | Dextrorphan glucuronide [49] | −1.51 | 0.56 | 1.09 × 10−5 | 7.85 × 10−3 |
| 445.3135 | 6.5 | – † | −0.65 | 0.25 | 1.23 × 10−5 | 7.85 × 10−3 |
| ESI− using timed urine samples‡ | ||||||
| 447.1866 | 6.3 | – † | −2.05 | 0.70 | 9.55 × 10−52 | 2.75 × 10−48 |
| 448.1884 | 6.3 | Hydroxy-dextrorphan-glucuronide [41] | −2.23 | 0.68 | 1.87 × 10−44 | 2.70 × 10−41 |
| 446.1827 | 6.3 | Oxo-dextrorphan-glucuronide [41] | −1.91 | 0.71 | 1.49 × 10−41 | 1.43 × 10−38 |
| 336.1275 | 3.5 | Dextrorphan sulfate [49] | −1.49 | 0.57 | 1.63 × 10−29 | 1.18 × 10−26 |
| 432.1668 | 4.5 | Oxo-hydroxymorphinan-glucuronide [41] | −1.60 | 0.56 | 5.77 × 10−23 | 3.33 × 10−20 |
| 433.1697 | 4.5 | – † | −1.73 | 0.51 | 1.47 × 10−14 | 7.08 × 10−12 |
| 418.1878 | 2.5 | 3-hydroxymorphinan-glucuronide [49] | −1.14 | 0.41 | 1.21 × 10−10 | 4.97 × 10−8 |
| 193.0358 | 2.6 | – † | 1.03 | 0.093 | 2.52 × 10−5 | 9.08 × 10−3 |
The possible identity of m/z ions was assigned based on mass.
Identity unknown.
No ions were found to be significant in the spot urine collection samples in ESI− mode.
BH: Benjamini–Hochberg; RT: Retention time.
Fragmentation of M1
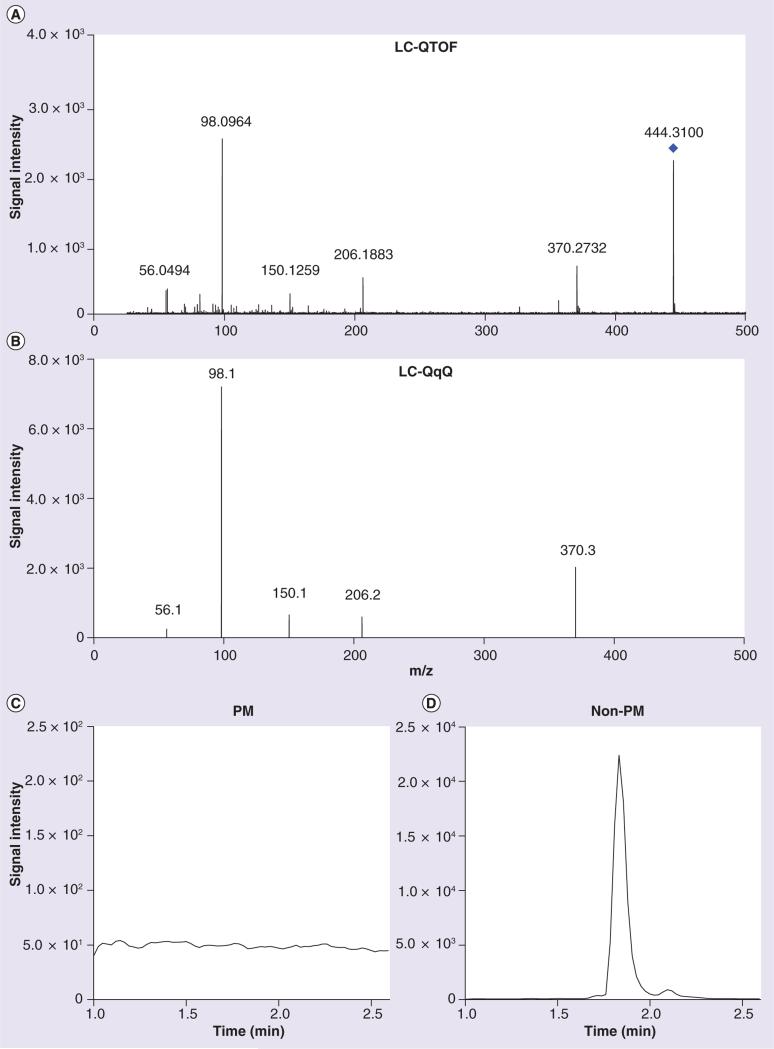
Fragmentation of M1 in a representative urine sample at 20 V by LC-QTOF yielded the MS/MS spectrum shown in Figure 2A. The most abundant product ions were, in order of decreasing abundance, m/z 98.0964 > 370.2732 > 206.1883 > 56.0494 > 55.0550 > 150.1259 > 81.0692. Major metabolomics databases were queried for the potential identity of M1 based on the parent mass and product ions, but no matches were found.
Figure 2. Product ion spectra of M1 (m/z 444.3102) from a representative pediatric spot urine sample.
Spectra were obtained in ESI+ mode via (A) LC-QTOF, precursor mass indicated by a diamond and (B) LC-QqQ, precursor mass not shown. A peak for the mass transition of m/z 444.3→98.1, the most abundant product ion, was (C) undetectable in a representative PM subject after a 15 μl injection and (D) clearly observable in a urine sample in a representative non-PM subject after a 5 μl injection. LC-QqQ: LC triple-quadrupole; LC-QTOF: LC quadrupole TOF; PM: Poor metabolizer.
Association & validation of endogenous biomarkers with CYP2D6 activity in pediatric subjects
Semiquantitative analysis of M1 in pediatric training set samples
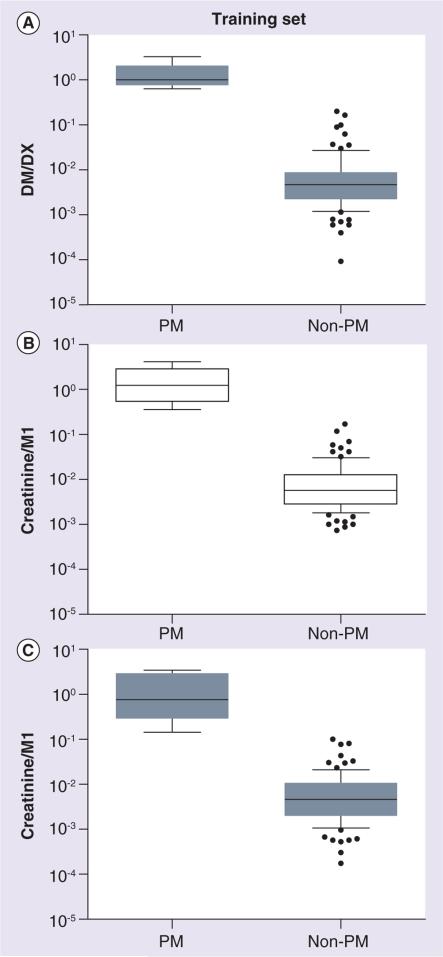
To improve sensitivity compared with the LC-QTOF method, we developed a targeted semiquantitative LC-QqQ based assay. Using multiple-reaction monitoring (MRM), we monitored the mass transitions (precursor to product ions) of m/z 444.3→370.3, 444.3→206.2, 444.3→150.1, 444.3→98.1 and 444.3→56.1 (Figure 2B). The mass transition of m/z 444.3→98.1, the most abundant product ion, was selected to determine the relative abundance of M1 in the urine samples. All training set non-PM subjects analyzed had quantifiable levels of M1 in both spot and timed urine samples (Figure 2D). In contrast, M1 levels were undetectable in all but one timed PM sample, even with an injection volume threefold higher than that used for non-PM samples (Figure 2C). For reference, when urine with approximately the mean M1 abundance was diluted 264-fold, a peak was still visually observable (data not shown). In samples with undetectable M1 signals, a low value was assigned as the M1 abundance. M1 peak intensities were normalized by urinary creatinine concentration to account for differences in urine concentration for each sample. As M1 may be the product of a reaction catalyzed by CYP2D6, we expressed the ratio as creatinine/M1 to match the DM/DX, where the parent (DM) is normalized to the metabolite (DX). Compared to non-PM subjects, PM subjects had 105-fold higher DM/DX and 123- and 147-fold higher creatinine/M1 ratios in spot and timed urine samples, respectively (Figure 3).
Figure 3. Phenotypic measures of CYP2D6 in the pediatric training set for poor metabolizer (n = 5) and non-poor metabolizer (n = 89) groups.
(A) DM/DX was determined in timed urine samples. Creatinine/M1 was measured by LC-MS/MS in (B) spot and (C) timed urine samples. A low value was assigned for samples with undetectable M1 abundances. Lines depict the median and boxes represent the 25th and 75th percentiles. The 10th and 90th percentiles are indicated by the whiskers. As PM and non-PM status was determined by DM/DX, the p-value is not reported in (A). Similarly, as M1 was identified as a marker of CYP2D6 using DM/DX in this same set of urine samples, the p-values are not reported in (B & C). Open boxes show spot urine samples and gray shaded boxes show timed urine samples. DM/DX: Dextromethorphan-to-dextrorphan metabolic ratio; PM: Poor metabolizer.
Confirmation of CYP2D6 biomarker candidate in the pediatric validation set
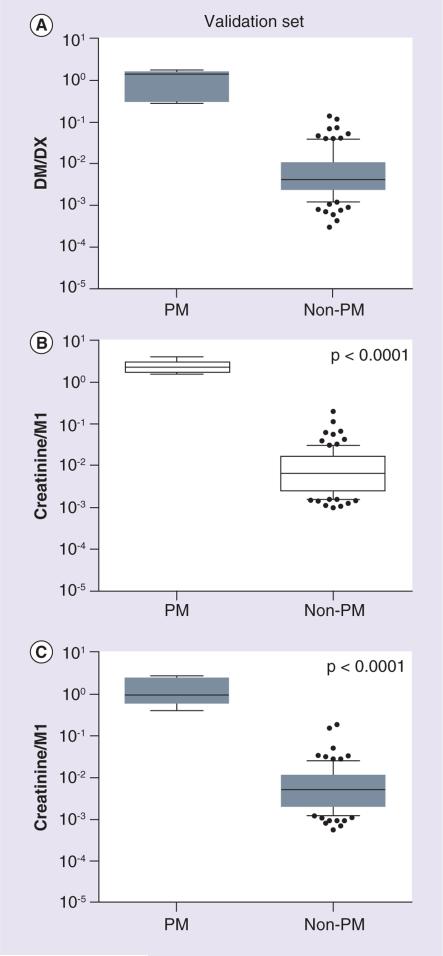
Similar results were found in the pediatric validation set. M1 abundance was undetectable in all PM subjects and a low value was assigned to M1 in these cases. A significant and positive relationship was observed between log(creatinine/M1) and log(DM/DX) in spot and timed urine samples (r2 = 0.31 and 0.28, respectively; p < 0.0001 for each). Compared to non-PM subjects, PM subjects had 82-fold higher DM/DX and 167- and 120-fold higher creatinine/M1 (p < 0.0001) in spot and timed urine samples, respectively (Figure 4).
Figure 4. Phenotypic measures of CYP2D6 in the pediatric validation set for poor metabolizer (n = 5) and non-poor metabolizer (n = 90) subjects.
(A) DM/DX was determined in timed urine samples. Creatinine/M1 was measured by LC-MS/MS in (B) spot and (C) timed urine samples. A low value was assigned for samples with undetectable M1 abundances. Lines depict the median and boxes represent the 25th and 75th percentiles. The 10th and 90th percentiles are indicated by the whiskers. Statistical analyses were performed using unpaired t-tests. As PM and non-PM status was determined by DM/DX, the p-value is not reported in (A). Open boxes show spot urine samples and gray shaded boxes show timed urine samples. DM/DX: Dextromethorphan-to-dextrorphan metabolic ratio; PM: Poor metabolizer.
Relationship between CYP2D6 activity score & phenotypic activity as determined by DM/DX and M1 in pediatric subjects
Assigning an activity score to each child allowed us to group individuals by genotypes with comparable levels of function. In both the training and validation sets, 5% of the subjects had an activity score of 0 (PMs), 91–92% of the subjects had activity scores between 0.5 and 2 owing to the presence of partially or fully functional alleles and 3% of subjects had activity scores greater than 2. Despite the large interindividual variability observed in each activity score group, both DM/DX and creatinine/M1 differed among the activity score groups (p < 0.0001 for all sets, Figure 5).
Figure 5. The relationship between phenotypic measures of CYP2D6 activity and CYP2D6 activity score.
DM/DX and creatinine/M1 as measured by LC triple-quadrupole in subjects from the (A & C) training (n = 94) and (B & D) validation (n = 95) datasets are shown. Lines depict the median and boxes represent the 25th and 75th percentiles. The 10th and 90th percentiles are indicated by the whiskers. Spot and timed urine samples are indicated with open and shaded boxes, respectively. DM/DX and creatinine/M1 ratios differed by activity score (p < 0.0001) as determined by one-way ANOVA. DM/DX: Dextromethorphan-to-dextrorphan metabolic ratio; PM: Poor metabolizer.
Biomarker response to CYP2D6 inhibition in adult subjects
Effect of CYP2D6 inhibition on M1 in adult subjects
The ability of M1 to reflect alterations in CYP2D6 activity was tested in adults. As part of a DDI study, 10 subjects received multiple doses of oral fluoxetine, a potent CYP2D6 inhibitor. Urine was collected before and during fluoxetine treatment. The urinary DM/DX increased 218-fold during fluoxetine treatment (p < 0.0001, Figure 6A) compared with baseline. Creatinine/M1 ratios were increased by 9.56-fold during the treatment phase (p = 0.029, Figure 6B). In addition, log(DM/DX) and log(creatinine/M1) were correlated (Spearman r = 0.55, p = 0.012; Figure 6C).
Figure 6. The effect of CYP2D6 inhibition by fluoxetine treatment on phenotypic markers of CYP2D6 activity in adult subjects.
CYP2D6 activity was measured by (A) DM/DX or (B) creatinine/M1 in subject urines. (C) The relationship between log(DM/DX) and log(creatinine/M1). Open diamonds indicate subjects without fluoxetine treatment, gray shaded diamonds show subjects with fluoxetine treatment. Statistical analyses were performed using paired t-tests or the Spearman rank correlation. DM/DX: Dextromethorphan-to-dextrorphan metabolic ratio.
Discussion
To our knowledge, this is the first study using global metabolomics to identify endogenous biomarkers of CYP2D6 activity in humans. Using this approach, we obtained a list of unique ions (m/z and retention time) in urine samples from a cohort of pediatric subjects and selected the ion m/z 444.3102 (M1) from our ESI+ training set data because its abundance correlated significantly with DM/DX parent-to-metabolite ratios in both spot and timed urine samples in these subjects. We were unable to identify M1 based on the parent mass and product ion fragmentation spectra in metabolomics databases. By restricting our investigation to the ions present in both spot (predose) and timed (postdose) urine samples, DM and its metabolites were excluded as endogenous biomarker candidates. As expected, DM metabolites were significantly correlated with DM/DX in timed but not spot samples (Table 2). Additional significant ions that were present only in timed samples may be uncharacterized metabolites of DM.
The mass transition of M1 to the most abundant product ion (m/z 444.3 → 98.1) in ESI+ mode was monitored using a targeted approach on a QqQ instrument. In the pediatric training set, M1 was present in all non-PM individuals and absent in all but one PM individuals, even with increased injection volumes for PM subjects. Admittedly, the lower limit of detection of our targeted assay is unknown without a standard, but an M1 peak diluted over 250-fold below the mean training set M1 abundance was visually observable. For our statistical comparisons, we assigned a low value of M1 for samples with experimentally undetectable levels, allowing us to conservatively and semiquantitatively compare the two phenotypic groups. The creatinine/M1 ratio clearly distinguished PMs from non-PMs.
We verified that creatinine/M1 was associated with DM/DX in both spot and timed urine samples in the validation set of urine collected from an additional 95 children. PMs have lower M1 abundances than non-PMs (p < 0.0001), suggesting that M1 is likely a product of a reaction catalyzed by CYP2D6. However, much of the variance in the relationship between log-transformed DM/DX and creatinine/M1 is still unexplained (r2 ~ 0.3). A stronger correlation with DM/DX would likely be observed if M1 were normalized by its unidentified precursor in a parent-to-metabolite molar ratio.
The creatinine/M1 ratio differed among activity score groups; however, there was appreciable overlap among non-PMs (activity scores between 0.5 and 3; Figure 5). The differentiation between activity scores may improve if the parent-to-M1 molar ratio can be used. In addition, fine-tuning CYP2D6 genotype by taking recently described enhancer SNPs into account [46] may alter activity score assignments. These enhancer SNPs appear to impact the regulation of CYP2D6 expression and thereby modulate an individual's activity.
Although age-dependent increases in CYP2D6 mRNA, and microsomal protein and activity levels have been reported in human fetal and pediatric liver [47,48], we observed no correlation between log M1 abundance and age in our study cohort (data not shown). However, in vivo pharmacokinetic data and longitudinal phenotyping studies indicate that genetic variation in CYP2D6 is a more important determinant of interindividual variability in activity than ontogeny [10,11,37]. Moreover, neither log M1 abundance nor creatinine concentration differed by gender or urinary pH. Creatinine was used to normalize for urine concentrations in our study, and urinary creatinine concentrations were not significantly correlated with log M1 in any dataset except for the spot urine samples of the training set (p = 0.01). Urinary creatinine concentrations have been reported to change with age, [49] and a significant association between creatinine concentrations and age was observed in the training set (p = 0.036 and < 0.0001 in the spot and timed urine collections, respectively) but not in the validation set. Admittedly, creatinine concentrations may influence the interpretation of the data in the training set, but it does not appear to be an important confounder in the validation set.
In addition to distinguishing CYP2D6 phenotype in pediatric subjects, the creatinine/M1 ratio was sensitive to inhibition of CYP2D6 in adults, increasing by approximately ninefold in adults during fluoxetine treatment as compared with control. Creatinine/M1 was less sensitive to CYP2D6 inhibition than DM/DX, once again reinforcing the need to account for the parent of M1.
M1 may join the growing list of endogenous CYP2D6 substrates and products. CYP2D6 catalyzed the O-demethylation of 5-methoxy-N,N-dimethyltryptamine, pinoline and 5-methoxytryptamine to bufotenine, 6-hydroxy-1,2,3,4-tetrahydro-β-carboline and serotonin, respectively, in human recombinant CYP2D6 and CYP2D6-transgenic mouse liver microsomes [26,50]. Additionally, CYP2D6 has been shown to hydroxylate and epoxygenate anandamide in human recombinant CYP2D6 and brain microsomal and mitochondrial preparations [29]. Recently, Cheng et al. reported that serotonin, 5-hydroxyindoleacetic acid, L-carnitine, acetyl-L-carnitine, pantothenic acid, 2′-deoxycytidine diphosphate, anandamide, N-acetylglucosaminylamine and stearoyl-L-carnitine concentrations differed significantly in brain homogenate and cerebrospinal fluid between wild-type and CYP2D6-transgenic mice [31]. Using chemical standards, many of these compounds were screened by LC-QTOF MS, but they were undetectable in our urine samples (data not shown). These urinary compound concentrations may be below our detection limit or elute with the column void volume (e.g., bufotenine). Many previously identified compounds were detected in specific tissues or biofluids, such as the brain or cerebrospinal fluid, and may not be excreted to an appreciable extent in the urine. Further investigation of these endogenous compounds as possible human CYP2D6 biomarkers may require the development of specific and sensitive targeted urinary assays or sampling of other biofluids.
In the field of metabolomics, the identification of complete unknowns from complex biological samples presents a substantial challenge. A limitation of our study is our current inability to identify M1. Unlike traditional metabolite identification, we have few clues regarding the structure of M1 since the precursor to M1 is also unknown. Preliminary efforts to obtain a high yield and pure isolate of M1 from urine for NMR analysis were unsuccessful. Despite the lack of structural information for M1, we would propose that the use of global metabolomics to identify endogenous biomarkers is akin to genome-wide association studies to identify SNPs associated with a particular disease. For example, the identified SNPs associated with a particular disease can be located in the protein-coding region of genes or are in linkage disequilibrium with SNPs in those regions. However, the majority of the identified SNPs (approximately 80%) are located in intergenic regions or noncoding introns [51]. Despite the apparent nonfunctional nature of these significantly associated SNPs, validation, replication and efforts to identify the causal SNPs have resulted in continued use of genome-wide association studies. Similarly, the relationship between M1 and CYP2D6 activity is still valid, even if the structure is currently unknown. Although structural identification of M1 is beyond the scope of the study at present, the identification of M1 and its parent will increase our understanding of the endogenous role of CYP2D6. The possibility that M1 or its precursor is a dietary component or a product of the microbiome cannot be excluded, but it is worth noting that the pediatric and adult studies were conducted independently and at two geographic sites, suggesting that regional or site-specific exposures can be ruled out as sources of M1.
Conclusion
In conclusion, we demonstrated that discovery of human biomarkers of CYP2D6 can be facilitated using a global metabolomics approach. We found that a positive ion with m/z of 444.3102 was associated with the urinary DM/DX ratio, an established method of CYP2D6 phenotyping, in two cohorts of children and was altered following CYP2D6 inhibition in adults. The ability of this ion to distinguish between CYP2D6 metabolizer phenotypes is currently limited to PM and non-PM phenotypes. Clearly, future structural identification of M1 and its parent are needed and may lead to a combined marker of CYP2D6 pheno type that may be more sensitive than the metabolite alone.
Future perspective
The findings of this study extend our current knowledge of CYP2D6 and demonstrate that metabolomics methods may be useful in revealing new biomarkers of drug metabolizing enzymes. In this study, the exploration of urinary metabolites led to the detection of a novel ion, M1, which may, lead to its use as an endogenous marker alone or in combination with other biomarkers of CYP2D6 activity. Future studies of M1 could include validation in different ethnic groups, additional pediatric and adult studies and may be useful in studying CYP2D6 activity in pregnant women and patients with kidney or liver disease. Upon further in vitro and in vivo validation, the clinical implementation of a noninvasive, endogenous biomarker test predictive of CYP2D6 phenotype could advance personalized medicine by potentially replacing current phenotyping or genotyping methods. Furthermore, validated endogenous biomarkers would make retrospective analyses of clinical samples possible and may aid in first-in-man studies where there is a concern for DDIs.
Executive summary.
Current knowledge on CYP2D6 phenotyping
CYP2D6 is an important drug-metabolizing enzyme encoded by a highly polymorphic and complex gene locus. Commonly used methods of CYP2D6 phenotyping require administration of a probe drug, such as dextromethorphan (DM).
Aim
In this study, global metabolomics was used to identify an endogenous biomarker of CYP2D6 activity in pediatric subjects.
Association & validation of endogenous biomarkers with CYP2D6 activity in pediatric subjects
Spot and timed urine samples were collected in pediatric subjects before and after a single oral dose of DM, respectively.
Following global metabolomics analysis of the urine samples in the training set (n = 94, five poor metabolizers), over 6500 mass features were regressed against the urinary DM metabolic ratio.
An unknown endogenous ion (with m/z 444.3102 at 6.5 min, referred to as M1) detected in ESI+ mode was significantly correlated with the urinary DM metabolic ratio (Benjamini–Hochberg corrected p < 0.01).
Biomarker relationship with CYP2D6 activity in pediatric subjects
Targeted LC-MS/MS analysis of M1 showed that the ion was able to discriminate poor metabolizers from intermediate, extensive and ultrarapid metabolizers in separate training and validation sets of pediatric subjects.
M1 differed among CYP2D6 activity scores (p < 0.0001 for all sets in a one-way ANOVA), albeit with large interindividual variability in each group.
Major metabolomics databases were queried for parent and product ions (following fragmentation), but no matches were found.
Biomarker response to CYP2D6 inhibition in adult subjects
In adult subjects, CYP2D6 inhibition by fluoxetine resulted in over ninefold reduction of urinary abundance of M1 along with decreased dextrorphan metabolite formation from DM.
Conclusion
M1 may be a product of a reaction catalyzed by CYP2D6.
The current study is limited to the ability of M1 to discriminate poor metabolizers from other phenotypes.
Future structural elucidation needs to be performed to identify M1 and its parent compound. The urinary ratio of the parent to the metabolite may provide a more sensitive measure of CYP2D6 activity than M1 alone.
Upon validation as a biomarker, this endogenous metabolite may be used to improve personalized medicine by conveniently and noninvasively ascertaining CYP2D6 phenotype without the use of an exogenous probe in various populations.
Acknowledgements
The authors would like to thank K Kerr (University of Washington, Seattle, WA, USA) for her statistical consultation.
Footnotes
Financial & competing interests disclosure
This research was supported in part by the NIH R01 HD058556 (to JS Leeder and YS Lin), Pharmacological Sciences Training grant T32 GM007750 (to J Tay-Sontheimer), NIEHS Center grants P30 ES07033 (to RP Beyer and YS Lin), Clinical and Translational Science Award UL1TR000423 (pilot funds to YS Lin), CTSA KL2 TR000421 (to YS Lin) and University of Washington School of Pharmacy DMTPR funding. Clinical support for the study was provided by a supplement from CTSA UL1 TR000001 (formerly UL1 RR033179), Heartland Institute for Clinical and Translational Research (HICTR). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as:
• of interest; •• of considerable interest
- 1.Kimland E, Odlind V. Off-label drug use in pediatric patients. Clin. Pharmacol. Ther. 2012;91(5):796–801. doi: 10.1038/clpt.2012.26. [DOI] [PubMed] [Google Scholar]
- 2.Kimland E, Nydert P, Odlind V, Bottiger Y, Lindemalm S. Paediatric drug use with focus on off-label prescriptions at Swedish hospitals – a nationwide study. Acta Paediatr. 2012;101(7):772–778. doi: 10.1111/j.1651-2227.2012.02656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sachs AN, Avant D, Lee CS, Rodriguez W, Murphy MD. Pediatric information in drug product labeling. JAMA. 2012;307(18):1914–1915. doi: 10.1001/jama.2012.3435. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez W, Selen A, Avant D, et al. Improving pediatric dosing through pediatric initiatives: what we have learned. Pediatrics. 2008;121(3):530–539. doi: 10.1542/peds.2007-1529. [DOI] [PubMed] [Google Scholar]
- 5.Johnson TN. The problems in scaling adult drug doses to children. Arch. Dis. Child. 2008;93(3):207–211. doi: 10.1136/adc.2006.114835. [DOI] [PubMed] [Google Scholar]
- 6.Bouzom F, Walther B. Pharmacokinetic predictions in children by using the physiologically based pharmacokinetic modelling. Fundament. Clin. Pharmacol. 2008;22(6):579–587. doi: 10.1111/j.1472-8206.2008.00648.x. [DOI] [PubMed] [Google Scholar]
- 7.Mahmood I. Prediction of drug clearance in children from adults: a comparison of several allometric methods. Br. J. Clin. Pharmacol. 2006;61(5):545–557. doi: 10.1111/j.1365-2125.2006.02622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abernethy DR, Burckart GJ. Pediatric dose selection. Clin. Pharmacol. Ther. 2010;87(3):270–271. doi: 10.1038/clpt.2009.292. [DOI] [PubMed] [Google Scholar]
- 9.Laer S, Barrett JS, Meibohm B. The in silico child: using simulation to guide pediatric drug development and manage pediatric pharmacotherapy. J. Clin. Pharmacol. 2009;49(8):889–904. doi: 10.1177/0091270009337513. [DOI] [PubMed] [Google Scholar]
- 10•.Leeder JS, Kearns GL, Spielberg SP, Van Den Anker J. Understanding the relative roles of pharmacogenetics and ontogeny in pediatric drug development and regulatory science. J. Clin. Pharmacol. 2010;50(12):1377–1387. doi: 10.1177/0091270009360533. [Discusses strategies of designing pediatric studies and understanding the relative roles of ontogeny and genetic variation for a given chemical entity] [DOI] [PubMed] [Google Scholar]
- 11.Allegaert K, Rochette A, Veyckemans F. Developmental pharmacology of tramadol during infancy: ontogeny, pharmacogenetics and elimination clearance. Paediatr. Anaesth. 2011;21(3):266–273. doi: 10.1111/j.1460-9592.2010.03389.x. [DOI] [PubMed] [Google Scholar]
- 12.Zanger UM, Turpeinen M, Klein K, Schwab M. Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal. Bioanal. Chem. 2008;392(6):1093–1108. doi: 10.1007/s00216-008-2291-6. [DOI] [PubMed] [Google Scholar]
- 13.Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin. Pharmacokinet. 2009;48(11):689–723. doi: 10.2165/11318030-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 14.Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part II. Clin. Pharmacokinet. 2009;48(12):761–804. doi: 10.2165/11318070-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 15.Abduljalil K, Frank D, Gaedigk A, et al. Assessment of activity levels for CYP2D6*1, CYP2D6*2, and CYP2D6*41 genes by population pharmacokinetics of dextromethorphan. Clin. Pharmacol. Ther. 2010;88(5):643–651. doi: 10.1038/clpt.2010.137. [DOI] [PubMed] [Google Scholar]
- 16.The Human Cytochrome P450 (CYP) Allele Nomenclature Database. www.cypalleles.ki.se/cyp2d6.htm.
- 17•.Gaedigk A. Complexities of CYP2D6 gene analysis and interpretation. Int. Rev. Psychiatry. 2013;25(5):534–553. doi: 10.3109/09540261.2013.825581. [Highlights the complexities of the CYP2D6 gene and the complications that may arise when interpreting phenotype from genotype] [DOI] [PubMed] [Google Scholar]
- 18.Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin. Pharmacol. Ther. 2012;91(2):321–326. doi: 10.1038/clpt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hicks JK, Swen JJ, Gaedigk A. Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr. Drug Metabol. 2014;15(2):218–232. doi: 10.2174/1389200215666140202215316. [DOI] [PubMed] [Google Scholar]
- 20.Teh LK, Bertilsson L. Pharmacogenomics of CYP2D6: molecular genetics, interethnic differences and clinical importance. Drug Metabol. Pharmacokinet. 2012;27(1):55–67. doi: 10.2133/dmpk.dmpk-11-rv-121. [DOI] [PubMed] [Google Scholar]
- 21••.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin. Pharmacol. Ther. 2008;83(2):234–242. doi: 10.1038/sj.clpt.6100406. [Proposes the CYP2D6 activity score system which simplifies phenotype interpretation from genotype] [DOI] [PubMed] [Google Scholar]
- 22.Jones AE, Brown KC, Werner RE, et al. Variability in drug metabolizing enzyme activity in HIV-infected patients. Eur. J. Clin. Pharmacol. 2010;66(5):475–485. doi: 10.1007/s00228-009-0777-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llerena A, Dorado P, Ramirez R, et al. CYP2D6 genotype and debrisoquine hydroxylation phenotype in Cubans and Nicaraguans. Pharmacogenomics J. 2012;12(2):176–183. doi: 10.1038/tpj.2010.85. [DOI] [PubMed] [Google Scholar]
- 24.Wu AH. Drug metabolizing enzyme activities versus genetic variances for drug of clinical pharmacogenomic relevance. Clin. Proteomics. 2011;8(1):12. doi: 10.1186/1559-0275-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Sager JE, Lutz JD, Foti RS, Davis C, Kunze KL, Isoherranen N. Fluoxetine and norfluoxetine mediated complex drug-drug interactions: in vitro to in vivo correlation of effects on CYP2D6, CYP2C19 and CYP3A4. Clin. Pharmacol. Ther. 2014;95(6):653-62–662. doi: 10.1038/clpt.2014.50. [Primary analysis of the fluoxetine drug–drug interaction study from which adult urine samples were included in this study; includes demographic information and demonstrates potent CYP2D6 inhibition by fluoxetine in the study population] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Yu AM, Idle JR, Herraiz T, Kupfer A, Gonzalez FJ. Screening for endogenous substrates reveals that CYP2D6 is a 5-methoxyindolethylamine O-demethylase. Pharmacogenetics. 2003;13(6):307–319. doi: 10.1097/01.fpc.0000054094.48725.b7. [In vitro study screening for endogenous substrates of CYP2D6 using recombinant CYP2D6, CYP2D6-transgenic mouse liver microsomes and human liver microsomes] [DOI] [PubMed] [Google Scholar]
- 27.Jiang XL, Shen HW, Yu AM. Pinoline may be used as a probe for CYP2D6 activity. Drug Metabol. Dispos. 2009;37(3):443–446. doi: 10.1124/dmd.108.025056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiroi T, Kishimoto W, Chow T, Imaoka S, Igarashi T, Funae Y. Progesterone oxidation by cytochrome P450 2D isoforms in the brain. Endocrinology. 2001;142(9):3901–3908. doi: 10.1210/endo.142.9.8363. [DOI] [PubMed] [Google Scholar]
- 29.Snider NT, Sikora MJ, Sridar C, Feuerstein TJ, Rae JM, Hollenberg PF. The endocannabinoid anandamide is a substrate for the human polymorphic cytochrome P450 2D6. J. Pharmacol. Exp. Ther. 2008;327(2):538–545. doi: 10.1124/jpet.108.141796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Sridar C, Snider NT, Hollenberg PF. Anandamide oxidation by wild-type and polymorphically expressed CYP2B6 and CYP2D6. Drug Metabol. Dispos. 2011;39(5):782–788. doi: 10.1124/dmd.110.036707. [In vitro study of anandamide metabolism by wild-type and polymorphically expressed CYP2B6 and CYP2D6, suggesting that alterations in CYP2D6 activity may impact endocannabinoid system signaling] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Cheng J, Zhen Y, Miksys S, et al. Potential role of CYP2D6 in the central nervous system. Xenobiotica. 2013;43(11):973–984. doi: 10.3109/00498254.2013.791410. [A metabolomic analysis that reports differences in several endogenous compounds between brain and cerebrospinal fluid of CYP2D6 transgenic and wild-type mice] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Etiology B, Bircher J, Preisig R, Kupfer A. Polymorphic dextromethorphan metabolism: co-segregation of oxidative O-demethylation with debrisoquin hydroxylation. Clin. Pharmacol. Ther. 1985;38(6):618–624. doi: 10.1038/clpt.1985.235. [Reports the now widely accepted urinary dextromethorphan metabolic ratio of 0.3 as the cut-off or antimode between CYP2D6 poor metabolizers and other phenotypes] [DOI] [PubMed] [Google Scholar]
- 33.Gaedigk A, Fuhr U, Johnson C, Berard LA, Bradford D, Leeder JS. CYP2D7–2D6 hybrid tandems: identification of novel CYP2D6 duplication arrangements and implications for phenotype prediction. Pharmacogenomics. 2010;11(1):43–53. doi: 10.2217/pgs.09.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaedigk A, Jaime LK, Bertino JS, Jr., et al. Identification of novel CYP2D7–2D6 Hybrids: non-functional and functional variants. Front. Pharmacol. 2010;1:121. doi: 10.3389/fphar.2010.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaedigk A, Twist GP, Leeder JS. CYP2D6, SULT1A1 and UGT2B17 copy number variation: quantitative detection by multiplex PCR. Pharmacogenomics. 2012;13(1):91–111. doi: 10.2217/pgs.11.135. [DOI] [PubMed] [Google Scholar]
- 36.Gaedigk A, Isidoro-Garcia M, Pearce RE, et al. Discovery of the nonfunctional CYP2D6 31 allele in Spanish, Puerto Rican, and US Hispanic populations. Eur. J. Clin. Pharmacol. 2010;66(9):859–864. doi: 10.1007/s00228-010-0831-4. [DOI] [PubMed] [Google Scholar]
- 37.Blake MJ, Gaedigk A, Pearce RE, et al. Ontogeny of dextromethorphan O- and N-demethylation in the first year of life. Clin. Pharmacol. Ther. 2007;81(4):510–516. doi: 10.1038/sj.clpt.6100101. [DOI] [PubMed] [Google Scholar]
- 38.Smith CA, Want EJ, O'maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78(3):779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 39.Tautenhahn R, Bottcher C, Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinform. 2008;9:504. doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.gee Generalized Estimation Equation solver. http://cran.r-project.org/web/packages/gee/index.html.
- 41.Tautenhahn R, Cho K, Uritboonthai W, Zhu Z, Patti GJ, Siuzdak G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 2012;30(9):826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.METLIN Metabolite Database. http://metlin.scripps.edu.
- 43.Wishart DS, Knox C, Guo AC, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The Human Metabolome Database. www.hmdb.ca.
- 45.Lutz U, Bittner N, Lutz RW, Lutz WK. Metabolite profiling in human urine by LC-MS/MS: method optimization and application for glucuronides from dextromethorphan metabolism. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2008;871(2):349–356. doi: 10.1016/j.jchromb.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 46.Wang D, Poi MJ, Sun X, Gaedigk A, Leeder JS, Sadee W. Common CYP2D6 polymorphisms affecting alternative splicing and transcription: long-range haplotypes with two regulatory variants modulate CYP2D6 activity. Hum. Mol. Genet. 2014;23(1):268–278. doi: 10.1093/hmg/ddt417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yokoi T. Essentials for starting a pediatric clinical study (1): Pharmacokinetics in children. J. Toxicol. Sci. 2009;34(Suppl. 2):SP307–SP312. doi: 10.2131/jts.34.sp307. [DOI] [PubMed] [Google Scholar]
- 48.Stevens JC, Marsh SA, Zaya MJ, et al. Developmental changes in human liver CYP2D6 expression. Drug Metabol. Dispos. 2008;36(8):1587–1593. doi: 10.1124/dmd.108.021873. [DOI] [PubMed] [Google Scholar]
- 49.Gu H, Pan Z, Xi B, et al. 1H NMR metabolomics study of age profiling in children. NMR Biomed. 2009;22(8):826–833. doi: 10.1002/nbm.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu AM, Idle JR, Byrd LG, Krausz KW, Kupfer A, Gonzalez FJ. Regeneration of serotonin from 5-methoxytryptamine by polymorphic human CYP2D6. Pharmacogenetics. 2003;13(3):173–181. doi: 10.1097/01.fpc.0000054066.98065.7b. [DOI] [PubMed] [Google Scholar]
- 51.Manolio TA. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 2010;363(2):166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]