Burkitt lymphoma (BL) is a biologically and molecularly defined tumor hallmarked by IG-mediated t(8q24) resulting in up-regulation of MYC.1,2 Recent studies of 59 molecularly defined BL (mBL) identified a novel aberration manifested by a specific 11q-gain/loss pattern in two cases (3%) lacking the MYC translocation.3 The aberration was subsequently detected in 15 MYC-negative high-grade B-cell lymphomas resembling BL and two cell lines derived from high-grade B-cell lymphomas. Further studies defined the minimal gained and lost regions at 11q23.3 and 11q24.1qter, respectively, and identified candidate genes potentially affected by these imbalances: the constantly overexpressed PAFAH1B2/11q23.3, and FLI1 (down-regulated) and ETS1 (recurrently mutated) targeted by a homozygous 11q24 microdeletion.
We provide evidence here that this peculiar 11q-gain/loss aberration is particularly frequent in BL in immunodeficient hosts, as it was identified in three out of seven patients with mBL after solid organ transplantation and under immunosuppressive maintenance therapy. The cases were selected from a cohort of 174 post-transplant patients diagnosed with post-transplant lymphoproliferative disorders (PTLD) between 1989 and 2012 at the University Hospitals of KU Leuven (Leuven, Belgium). The study was approved by the Ethical Committee of the University Hospitals Leuven. The cohort mainly comprised cases of diffuse large B-cell lymphoma (DLBCL, 75%).4,5 Other entities were less frequent and included plasmacytoma and plasmablastic non-Hodgkin lymphoma,6 T-cell non-Hodgkin lymphoma,7 BL, small cell B-non-Hodgkin lymphoma and unspecified cases. The seven post-transplant BL (PT-BL) cases reported here were analyzed using conventional cytogenetics, array comparative genome hybridization (CGH), fluorescence in situ hybridization (FISH), immunohistochemistry, gene expression profiling and bioinformatics (Online Supplementary Information). As controls, we included four cases of typical MYC-translocation-positive BL from immunocompetent hosts (IC-BL). All cases fulfilled the morphological and immunological criteria of BL defined by the 2008 World Health Organization classification8 (Table 1).
Table 1.
Morphology and immunophenotype of the reported post-transplant and immunocompromised BL cases.
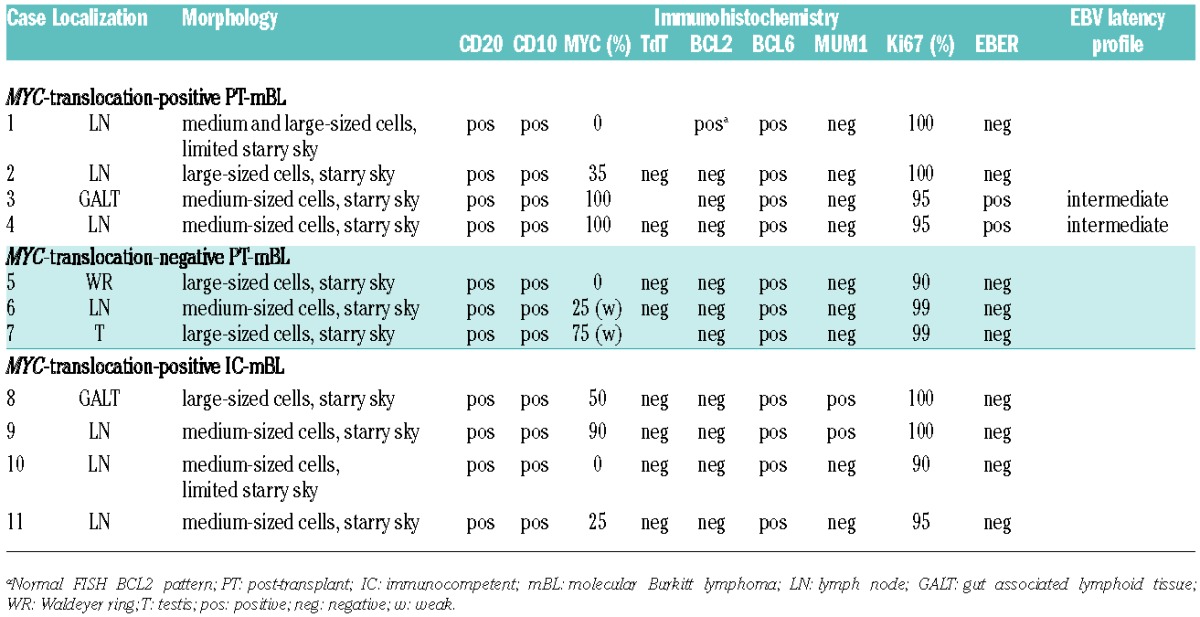
Relevant clinical, pathological and cytogenetic features of the studied cases are summarized in Table 2. The PT-BL patients, all men, had undergone liver (n=3), heart (n=2) or kidney (n=2) transplantation. The median age at time of PTLD was 47.5 years (range, 15–70 years). Five cases were Epstein Barr virus (EBV)-negative and two adolescent cases (n. 3 and 4) were EBV-positive. Patients developed BL a median of 40.7 months (range, 11–66 months) following transplantation. The interval was significantly shorter for EBV-positive cases than EBV-negative cases (12 versus 52.2 months, respectively). All seven patients were treated with either rituximab and/or CHOP. Most of them achieved complete remission but four patients died within 4–11 months after diagnosis (average 6 months), including two patients (n. 6 and 7) who died due to disease-related complications. Three patients are alive, including both EBV-positive patients, and their survival ranges from 34 to 99 months (median 70 months) (Table 2).
Table 2.
Relevant clinical, pathological and genetic data of the reported post-transplant and immunocompetent BL cases.
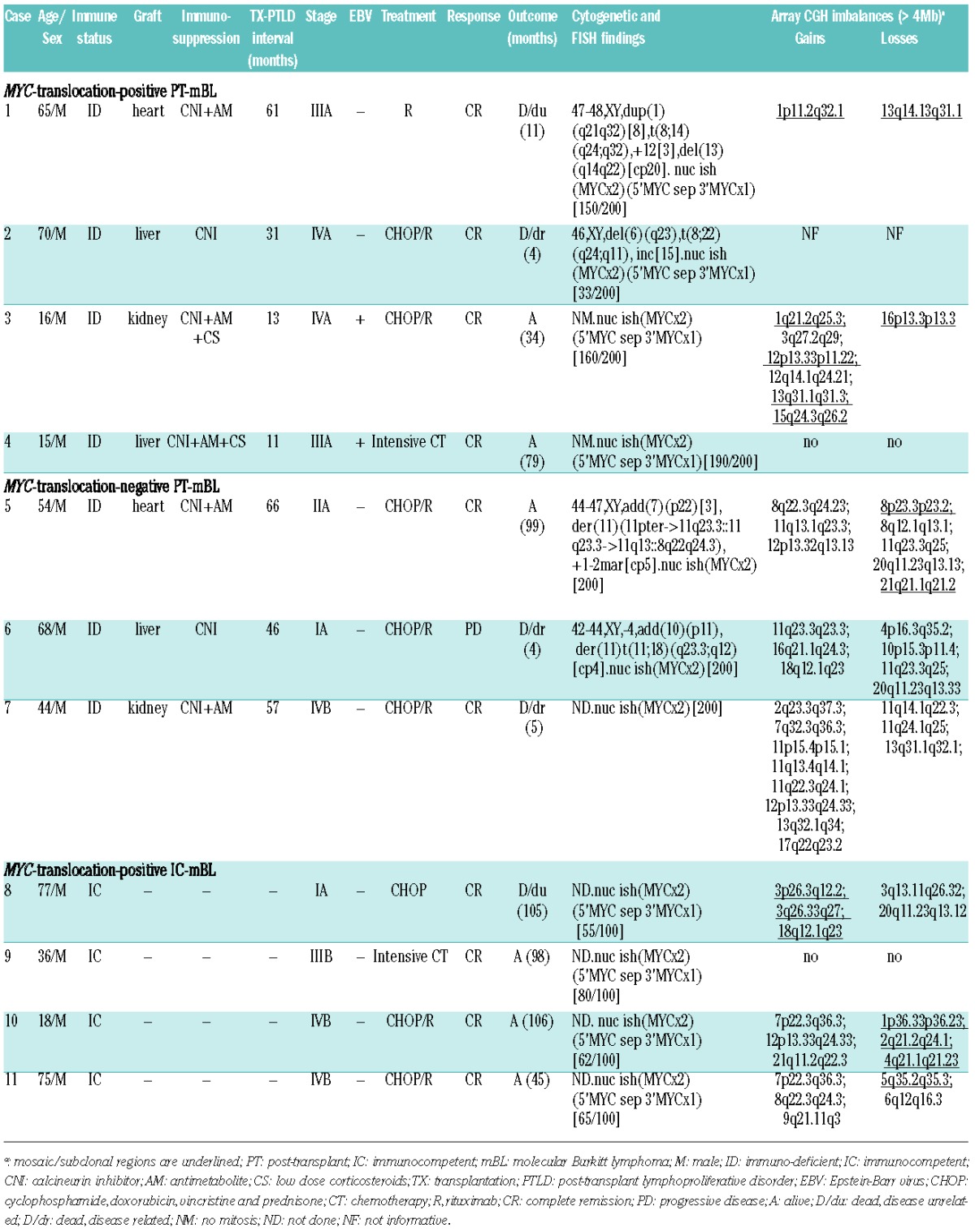
Based on a global gene expression pattern, all seven PT-BL cases (as well as control IC-BL cases) showed the molecular profile of BL and were readily distinguished from PT-/IC-DLBCL5 using two classifiers of mBL1,2 (Online Supplementary Figure S1). Cytogenetics and/or FISH demonstrated t(8q24/MYC) in four PT-mBL, while three cases (n. 5–7, all EBV-negative) appeared to be negative for the MYC translocation. Interestingly, karyotypes of the two latter cases revealed various 11q aberrations, which after additional FISH analysis (data not shown) were described as der(11)(11pter->11q23.3::11q23.3->11q13::8q22q24.3) and der(11)t(11;18)(q23.3;q12) (Figure 1A). Array CGH analysis performed in all 11 cases detected the characteristic 11q-gain/loss pattern in the three MYC-translocation-negative cases (Figure 1B). This pattern was associated with a dup(11)(q13q23.3) in case 5, focal gain-and-amplification of an approximately 4 Mb region at 11q23.3 in case 6 (confirmed by FISH, Figure 1C, Online Supplementary Figure S2) and complex 11q imbalances in case 7 (Table 2). Interestingly, the dup(11)(q13q23.3) identified in case 5 was associated with an inversion, as in several previously reported cases.3,9 Losses, resulting from different non-reciprocal translocations in cases 5 and 6, constantly targeted the 11q23.3q24.1qter region. Homozygous deletions were not detected. The array CGH data enabled definition of the minimal gained region (MGR) (~4 Mb) and minimal lost region (MLR) (~13.5 Mb) which were mapped at 11q23.3 [chr11:116072765-120087526bp (hg19)] and 11q24.1q25 [chr11:121499571-135006516bp (hg19)], respectively. Notably, MYC-translocation-positive PT-mBL cases had either a balanced karyotype, or two to nine additional imbalances, including subclonal gains/losses, similar to the IC-mBL cases (Table 2). The latter ones, however, showed a frequent gain of chromosome 7 (60%), not seen in PT-mBL cases.
Figure 1.
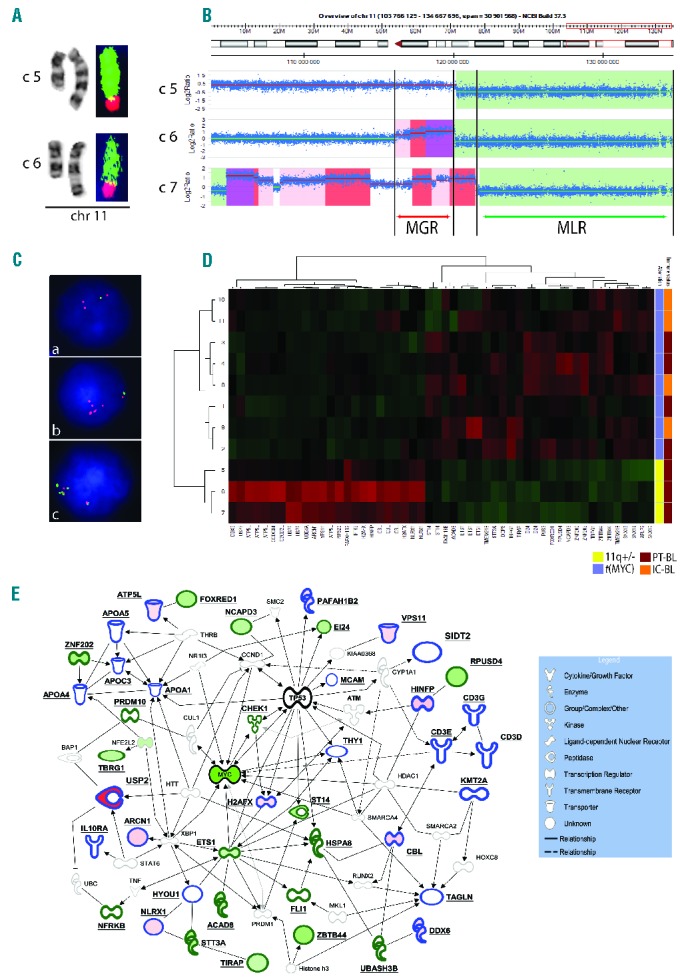
Genomic and transcriptomic data on PT-mBL. (A) Partial karyotype of case 5 (c5) and case 6 (c6) showing chromosome 11 abnormalities and FISH images of both derivative chromosomes painted with WCP11 (green) and WCP8 (red) (case 5), and WCP18 (red) (case 6). The aberrations were eventually described as der(11)(11pter->11q23.3::11q23.3->11q13::8q22q24.3) and der(11)t(11;18)(q23.3;q12), respectively. Note (inverted) duplication of 11q13q23.3 in case 5 and a normal appearance of this region in case 6. (B) Chromosomal view of chromosome 11q23q24 and imbalances identified by array CGH analysis in cases 5–7. Gained regions are highlighted in red-scale (increased intensity reflects an increased amplification level), while lost regions are marked in green. Note a variable level of 11q23.3 gain, a common loss of 11q24qter and the defined MGR (~4 Mb) and MLR (~13.5 Mb). (C) Examples of interphase FISH analysis performed in case 5 (a) and case 6 (b, c). The applied probes include the 11q-MGR/MLR FISH assay (a) (b), and two probes from the duplicated (RP11-284O21-SpectrumOrange) and amplified (RP11-784K23-SpectrumGreen; Online Supplementary Figure S2) area in case 6 (c). Note the duplicated and amplified red/11q23.3 signal in (a) and (c), respectively, and loss of the green/11q24 signal in both cases. In (c), note two red and five green signals in the cluster, illustrating various levels of gain within the MGR. (D) Hierarchical clustering of mBL cases using the dysregulated genes located in the MGR and MLR. (E) Interaction network found by Ingenuity Pathway Analysis involving genes targeted by the 11q-gain/loss aberration (bold edges). Solid and interrupted lines represent direct and indirect interactions, respectively. Notably, most of the interactions in this network are direct protein-protein interactions. The molecules with blue and green edges are encoded by MGR- and MLR-associated genes, respectively. The data obtained by comparison of cases 5–7 (PT-mBL with the 11q-gain/loss pattern) (11q+/−) by cases 1–4 (PT-MYC-translocation-positive mBL) [(t(MYC)] were overlaid in this network. Molecules which are down- and up-regulated in cases with 11q+/− when compared to cases with t(MYC) are filled in green and red, respectively.
In order to identify genes affected by the 11q imbalances, we compared gene expression profiles of cases 5–7 (11q-gain/loss-positive) and cases 1–4 (MYC-translocation-positive), and focused on genes harbored by the MGR (69 genes) and MLR (106 genes). Altogether, 33 genes with a differential expression were identified: 15 in the MGR (all up-regulated) and 18 in the MLR (all down-regulated) (Online Supplementary Table S1). The most significantly up-regulated was USP2 (ubiquitin specific peptidase 2), which showed up to 30.6-fold higher expression in MYC-translocation-negative cases than in MYC-translocation-positive cases (Online Supplementary Figure S3). Differential expression of the remaining MGR genes was much lower (1.35–2.75-fold). Levels of differentially down-regulated genes ranged from -1.32 to -2.35-fold. Notably, both regions harbor several genes which might be implicated in the pathogenesis of MYC-translocation-negative mBL. Particularly interesting are two oncogenes, USP2 and CBL, and the previously discussed PAFAH1B2 located in the MGR.3 USP2, which was the most significantly up-regulated enzyme in 11q-gain/loss-positive mBL, acts as a modulator of tumor necrosis factor-α-induced nuclear factor-κB signaling and prolongs the half-life of targets such as fatty acid synthase, MDM2 and MDM4/MDMX (negative regulators of p53) and cyclin D1 (G1/S transition). The enzyme, like other deubiquitinases, is implicated in cancer, particularly in prostate carcinomas (reviewed by Fraile et al.10). Dysregulated genes in the MLR comprise two candidate tumor suppressor genes, TBRG1 and EI24, and five genes either related to cancer or involved in cancer-related processes, including ETS1, TIRAP, ST14, NCAPD3 and ZNF202. Noteworthy, FLI1, the candidate target gene,3 was not differentially down-regulated in our cases (Online Supplementary Figure S3). Hierarchical clustering of the studied cases using the set of 11q23/q24 dysregulated genes showed that MYC-translocation-negative PT-mBL cases cluster together and separately from MYC-translocation-positive PT-/IC-mBL (Figure 1D).
PT-mBL cases with the 11q-gain/loss pattern revealed a lower expression of MYC mRNA than MYC-translocation-positive cases (Online Supplementary Figure S3). Using immunohistochemistry with MYC antibody (clone Y69; Epitomics, Burlingame, CA, USA), all studied mBL cases showed highly variable expression of MYC protein, which has not necessarily correlated with rearrangement of MYC (Table 1) (Online Supplementary Figure S4). These results, however, remain in line with the recently published data of Chisholm et al.11
To examine whether the 11q-gain/loss aberration also characterizes BL and/or DLBCL cases from immunocompetent patients, among those in our institution, we analyzed by FISH two known cases of MYC-translocation-negative BL and five cases of MYC-translocation-negative DLBCL harboring 11q aberrations [mostly dup(11q)]. Using the designed 11q23/q24 assay covering the MGR and MLR (Online Supplementary Figure S2), the 11q-gain/loss pattern was detected in one of two MYC-translocation-negative BL cases (data not shown). The second BL case showed a normal FISH pattern, while all five DLBCL revealed gain of 11q23.3, without, however, an associated loss of 11q24. These findings confirm the rare occurrence of the 11q-gain/loss pattern in IC-BL/-DLBCL.
To unravel biological consequences of the 11q-gain/loss aberration in mBL, we explored the MGR/MLR-dysregulated genes using Ingenuity Pathway Analysis software (see Online Supplementary Methods). Ingenuity Pathway Analysis showed that the genes are implicated in important biological processes, including cancer, and the majority of them (22/33) are involved in the TP53 and MYC networks, frequently by direct protein-protein interactions (Figure 1E). These findings and the observation that cases with 11q imbalances cluster with MYC-translocation-positive mBL (using two mBL classifiers1,2) suggest that the 11q-gain/loss aberration is a ‘molecular variant’ of t(8q24/MYC) affecting the same or overlapping pathological pathways. A similar phenomenon has recently been described in BCR-ABL-negative Ph-like ALL showing a spectrum of kinase-activating alterations.12 The astonishing consistency of the 11q-gain/loss pattern suggests that the driving potential of this aberration results from a concerted overexpression of the defined 11q23.3 genes, including USP2, CBL and PAFAH1B2, and simultaneous down-regulation of 11q24q25 genes, among others TBRG1, EI24 and ETS1. Interestingly, a similar gain-and-loss pattern has been identified in hepatosplenic T-cell lymphoma characterized by a constant 7p14.1p22.1-loss/7q22.11q31.1-gain pattern,13 which, it is worth noting, is frequently observed in immunosuppressed patients.
PTLD is typically an EBV-driven process and our recent study of 33 PT-DLBCL (72% EBV-positive and 28% EBV-negative) led to the conclusion that EBV-negative PT-DLBCL were coincidental lymphomas in immunosuppressed patients.5 Among the reported PT-mBL, only 30% of cases were EBV-positive and they clustered together with EBV-negative PT-mBL and IC-mBL (Online Supplementary Figure S1). These findings are in line with observations of Piccaluga et al.,14 who found that all three BL subtypes, sporadic, endemic and human immunodeficiency virus-positive, share a common gene expression signature, distinct from other B-cell malignancies, and suggested that not EBV, but the MYC-dependent signature had a major effect on the clustering.
In summary, we confirmed recurrence of the 11q-gain/loss pattern in high-grade B-cell lymphoma and showed that this aberration is significantly more frequent (P<0.007, Fisher exact test) in BL occurring in the setting of transplantation and immunosuppression (43% of all PT-mBL and 60% of EBV-negative PT-mBL) than in immunocompetent patients (3%)1, suggesting that immunosuppression may favor its formation. Further studies of PT-BL are needed to confirm this association. As identification of patients with the 11q-gain/loss aberration is clinically important but cytogenetically challenging, we recommend the designed 11q-MGR/MLR FISH assay as a useful diagnostic tool to evaluate both post-transplant and immunocompetent BL patients.
Acknowledgments
The authors would like to thank Ursula Pluys, Kathleen Doms and Emilie Bittoun for their excellent technical assistance and Rita Logist for editorial help.
Footnotes
Funding: this study was supported by the concerted action grant from the K.U.Leuven n. 3M040406 (TT, PV, and IW) (http://www.kuleuven.be/), research grants from “Stichting tegen Kanker” (PV) (http://www.kanker.be/) and the FWO-Vlaanderen (G081411N to GV and TT). PV is a senior clinical investigator of the FWO-Vlaanderen (http://www.fwo.be/en/).
TT holds a Mandate for Fundamental and Translational Research from the “Stichting tegen Kanker” (2014-083).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Dave SS, Fu K, Wright GW, et al. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med. 2006;354(23):2431–2442. [DOI] [PubMed] [Google Scholar]
- 2.Hummel M, Bentink S, Berger H, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354(23):2419–2430. [DOI] [PubMed] [Google Scholar]
- 3.Salaverria I, Martin-Guerrero I, Wagener R, et al. A recurrent 11q aberration pattern characterizes a subset of MYC-negative high-grade B-cell lymphomas resembling Burkitt lymphoma. Blood. 2014;123(8):1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dierickx D, Tousseyn T, Sagaert X, et al. Single-center analysis of biopsy-confirmed posttransplant lymphoproliferative disorder: incidence, clinicopathological characteristics and prognostic factors. Leuk Lymphoma. 2013;54(11):2433–2440. [DOI] [PubMed] [Google Scholar]
- 5.Morscio J, Dierickx D, Ferreiro JF, et al. Gene expression profiling reveals clear differences between EBV-positive and EBV-negative posttransplant lymphoproliferative disorders. Am J Transplant. 2013; 13(5):1305–1316. [DOI] [PubMed] [Google Scholar]
- 6.Morscio J, Dierickx D, Nijs J, et al. Clinicopathologic comparison of plasmablastic lymphoma in HIV-positive, immunocompetent, and posttransplant patients: single-center series of 25 cases and meta-analysis of 277 reported cases. Am J Surg Pathol. 2014;38(7):875–886. [DOI] [PubMed] [Google Scholar]
- 7.Herreman A, Dierickx D, Morscio J, et al. Clinicopathological characteristics of posttransplant lymphoproliferative disorders of T-cell origin: single-center series of nine cases and meta-analysis of 147 reported cases. Leuk Lymphoma. 2013;54(10):2190–2199. [DOI] [PubMed] [Google Scholar]
- 8.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of tumours of Haematopoietic and Lymphoid Tissues. Lyon: International Agency for Research on Cancer, 2008. [Google Scholar]
- 9.Pienkowska-Grela B, Rymkiewicz G, Grygalewicz B, et al. Partial trisomy 11, dup(11)(q23q13), as a defect characterizing lymphomas with Burkitt pathomorphology without MYC gene rearrangement. Med Oncol. 2011;28(4):1589–1595. [DOI] [PubMed] [Google Scholar]
- 10.Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31(19):2373–2388. [DOI] [PubMed] [Google Scholar]
- 11.Chisholm KM, Bangs CD, Bacchi CE, Kirsch HM, Cherry A, Natkunam Y. Expression profiles of MYC protein and MYC gene rearrangement in lymphomas. Am J Surg Pathol. 2015;39(3):294–303. [DOI] [PubMed] [Google Scholar]
- 12.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014; 371(11):1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finalet FJ, Rouhigharabaei L, Urbankova H, et al. Integrative genomic and transcriptomic analysis identified candidate genes implicated in the pathogenesis of hepatosplenic T-cell lymphoma. PLoS One. 2014;9(7):e102977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piccaluga PP, De FG, Kustagi M, et al. Gene expression analysis uncovers similarity and differences among Burkitt lymphoma subtypes. Blood. 2011;117(13):3596–3608. [DOI] [PubMed] [Google Scholar]


