Abstract
We have developed a simple NMR-based method to determine the turnover of nucleotides and incorporation into RNA by stable isotope resolved metabolomics (SIRM) in A549 lung cancer cells. This method requires no chemical degradation of the nucleotides or chromatography. During cell growth, the free ribonucleotide pool is rapidly replaced by de novo synthesized nucleotides. Using [U-13C]-glucose and [U-13C,15N]-glutamine as tracers, we showed that virtually all of the carbons in the nucleotide riboses were derived from glucose, whereas glutamine was preferentially utilized over glucose for pyrimidine ring biosynthesis, via the synthesis of Asp through the Krebs cycle. Incorporation of the glutamine amido nitrogen into the N3 and N9 positions of the purine rings was also demonstrated by proton-detected 15N NMR. The incorporation of 13C from glucose into total RNA was measured and shown to be a major sink for the nucleotides during cell proliferation.
This method was applied to determine the metabolic action of an anti-cancer selenium agent (methylseleninic acid or MSA) on A549 cells. We found that MSA inhibited nucleotide turnover and incorporation into RNA, implicating an important role of nucleotide metabolism in the toxic action of MSA on cancer cells.
Keywords: RNA biosynthesis, nucleotide pools, SIRM, Selenium
1. Introduction
Stable isotope tracer approaches are indispensible to mapping metabolic pathways and fluxes in detail, which have been applied in studies of cells, tissues, organisms, and even human subjects (Fan et al. 1986; Zwingmann et al. 2003; Delgado et al. 2004; Mancuso et al. 2005; Vizan et al. 2005; Mendes et al. 2006; Bak et al. 2007; DeBerardinis et al. 2007; Mason et al. 2007; Fan et al. 2008; Lane et al. 2008; Beger et al. 2009; Fan et al. 2009).
We have previously noted that the free nucleotide pools of proliferating cells supplied with 13C-glucose are rapidly replaced by 13C-enriched nucleotides. In particular, the ribose moiety became essentially completely labeled, indicating its synthesis via the sequence of glycolysis and the pentose phosphate pathway (Fan et al. 2005; Lane et al. 2007; Telang et al. 2007; Fan et al. 2008; Moseley et al. 2011). The nucleobase carbons also became labeled from 13C-glucose, which is consistent with active synthesis of pyrimidine bases. As the total concentration of free nucleotide remains essentially unchanged during the labeling period, it was assumed that, the free nucleotides are turned over for other uses, and must be replenished by de novo synthesis. Notable sinks for nucleotides in proliferating cells are RNA and nucleotide modified proteins, which are not normally recovered by methods designed to extract polar metabolites, or is invisible to NMR in vivo for their large size.
Dividing cells also require nucleotide triphosphates (NTPs) for driving numerous anabolic processes such as the synthesis of lipids, proteins, RNA, and DNA. The regulation of free nucleotide biosynthesis is thus critically important in cells (Murray et al. 2009). De novo nucleotide biosynthesis requires the coordinate activity of the pentose phosphate pathways (for ribose) and of nucleobase synthesis. For pyrimidines, the base synthesis involves orotate dehydrogenase which requires functioning electron transport chain in the mitochondria (Nelson et al. 2005). Quantitatively, the synthesis of RNA is the most important process that incorporates free nucleotides into macromolecules, thereby necessitating de novo nucleotide synthesis in proliferating cells. Thus, in order to understand how nucleotide biosynthesis is regulated, knowledge of both the turnover of the free nucleotide pools and the major sink, RNA is required.
Total RNA can be easily extracted from cultured cells (Centelles et al. 2007), and the radiotracer approach has been popular because incorporation of radioisotopes can be readily detected. Radioisotope labeling however damages the nucleic acids (Hu et al. 2002), and radioactivity counting provides no information about the position of the labeled atoms in individual nucleotides, which is essential to reconstructing the various pathways leading to the nucleotide synthesis. More recently, Cascante and coworkers have developed a mass-spectrometry based method for determining stable isotope incorporation into RNA. This involves extraction of RNA, hydrolysis to the nucleotides, and then further chemical processing to distinguish between label in the ribose and heterocyclic base moieties (Boren et al. 2001; Vizan et al. 2005; Centelles et al. 2007; Vizan et al., 2007).
An alternative approach is to utilize NMR to determine directly isotope distributions at individual atoms in both the ribose and base moieties simultaneously without chemical degradation or chromatographic separation. We have therefore developed such NMR-based method for determining positional 13C or 15N labeling patterns in RNA recovered from cancer cells grown in 13C-glucose or 13C,15N-Gln. With modern NMR instruments, the method is sufficiently sensitive to determine isotope incorporation into a few μg RNA extracted from cells, which is comparable to the amounts typically used for gene microarray analysis.
We have applied the method to probe the effect of anti-cancer selenium compounds on nucleotide and RNA metabolism. Various selenium compounds have been shown to be cytotoxic or cytostatic to tumor cells (Ip et al. 2002), which was the basis for their potential use as chemopreventive or therapeutic agents (Combs 2004). From the early clinical trials with selenium supplementation at supranutritional levels (Clark et al. 1996), certain selenium compounds in the selenized yeast were effective against prostate cancer (Combs 2004). It is now clear that the anti-cancer efficacy is highly dependent on both the chemical form of the selenium and on the target site (Ganther 1999; Fan et al. 2005; Fan et al. 2006; Lippman et al. 2009). This is particularly illustrated by the recent conclusion of the human prostate cancer trials (SELECT) that selenomethionine (SeM) was ineffective against prostate cancer prevention (Lippman et al. 2009). In contrast, monomethylated selenium compounds such as methylseleninic acid (MSA) are highly toxic to lung cancer cells (Fan et al. 2005; Fan et al. 2006). We have been investigating the biochemical mechanisms of MSA action on inhibiting the proliferation of lung cancer cells (Fan et al. 2005; Fan et al. 2006) among others.
Here we demonstrate the technique for measuring the time course of incorporation of 13C from [U-13C]-glucose into the RNA of cultured human lung adenocarcinoma A549 cells which was directly compared with the time course of labeling of the free nucleotide pools, and the effect of MSA on nucleotide biosynthesis.
2. Materials and Methods
Materials
Pure yeast RNA, nucleoside monophosphates and methylseleninic acid were obtained from Sigma Aldrich (St. Louis, MO). P1 microccocal nuclease was purchased from US Biological. All other reagents were of analytical grade.
Methods
Cell growth
Human lung adenocarcinoma A549 cells (ATCC) were grown on 10 cm plates at 37 °C in a 5% CO2 atmosphere in glucose and glutamine-free RPMI supplemented with 10 mM [U-13C]-glucose (Sigma Isotec) and 2 mM unlabeled glutamine as previously described (Fan et al. 2005). In parallel, cells were also grown in 2 mM [U-13C,15N]-glutamine (Cambridge Isotope Laboratories, MA) plus 10 mM unlabeled glucose (Lane et al. 2009). Cells were harvested by trypsinization at different times after exposure to the labeled glucose and washed 3 times in cold PBS to remove medium components as described previously (Fan et al. 2005; Fan et al. 2008). Polar extracts were made using 60% cold acetonitrile or trichloracetic acid (w/v ≥ 40:1) as previously described (Fan et al. 2008; Fan et al. 2010).
MSA treatment
Time courses of A549 cells grown in 5 mM [U-13C]-glucose were obtained in the absence or presence of 5 μM MSA, which is the IC50 for these cells grown under these conditions (T.W-M. Fan, unpublished data). Cells were harvested and processed as described above, and the extracts analyzed by NMR as described below.
RNA preparation
RNA was extracted and purified from the cells using the TRIzol reagent and RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Purified RNA was assayed by absorption at 260 and the ratio 260/280 nm using a Nanodrop UV/Vis spectrometer (ThermoFisher, Waltham, MA). Typically 100 μg RNA (length > 200 nt) was obtained from one 10 cm plate, and 30 μg were used for RNA hydrolysis.
The extracted RNA was lyophilized and redissolved in 0.1 M sodium acetate buffer containing 1 mM ZnCl2, pH 5.3. The conditions for digestion were determined using unlabeled Sigma RNA. Two units of P1 nuclease were added to 1 ml RNA solution in the assay buffer (A260 = 0.5 to 1 unit) in a capped quartz cuvette, and incubated at 50 °C. The increase in A260 was monitored as a function of time for the progress of hydrolysis in a Cary 300 spectrophotometer. After the reaction had stopped, another aliquot of the enzyme was added to ensure complete digestion. The solution was then lyophilized, and redissolved in 350 μL D2O containing 30 nmol DSS-d6 for analysis by NMR.
NMR data collection
NMR spectra were recorded at 20°C on a Varian Inova spectrometer operating at 14.1 T equipped with a cold probe. One-dimensional 1H NMR spectra were recorded with a 2 s acquisition time and a 3 second relaxation delay during which the transmitter was used to saturate the residual water signal. One-dimensional 1H-{13C}-HSQC spectra were recorded using an acquisition time of 0.15 s and a recycle time of 1.5 s, with the 13C decoupling set to 80 ppm, and the evolution delay set for 160 Hz.
Two-dimensional TOCSY spectra were recorded using a spin lock duration of 50 ms with a B1 field strength of 9 kHz, and acquisition times of 0.34 s in t2 and 0.043 s in t1. Data were zero-filled to 8192 by 2048 points, apodized with an unshifted Gaussian and a 1 Hz line broadening exponential in both dimensions prior to Fourier transformation.
One-dimensional 1H-{15N}-HSQC spectra were recorded using an acquisition time of 0.15 s and a recycle time of 1.5 s, with the 15N GARP decoupling set to 120 ppm, and the evolution delay set for 10 Hz corresponding approximately to 2JNH (Ippel et al. 1996). 2D 1H-{15N}-HSQC spectra were recorded with acquisition times of 0.15 s in t2 and 0.021 s in t1 with a recycle time of 1.6 s. The data were zerofilled in t2, linear predicted and zerofilled in t1 to a final 8192 by 2048 points, apodized with an unshifted Gaussian and a 1 Hz line broadening exponential in both dimensions as previously described (Fan et al. 2011).
Assignments were made according to the chemical shifts and scalar coupling patterns in TOCSY and by reference to our databases (Fan et al. 2008; Fan et al. 2011). The chemical shifts of the nucleotide in the P1 nuclease digestion buffer were verified against standards in the same buffer system.
The fractional incorporation (F) for specific 13C positional isotopomers was determined by normalizing the areas or volume of 13C satellite peaks in 1-D or the TOCSY spectra against the total integrated area or volume of central plus satellite peaks, with appropriate corrections for differential relaxation as previously described (Lane and Fan 2007; Fan and Lane 2008; Lane et al. 2008) according to Eq. 1.
| (1) |
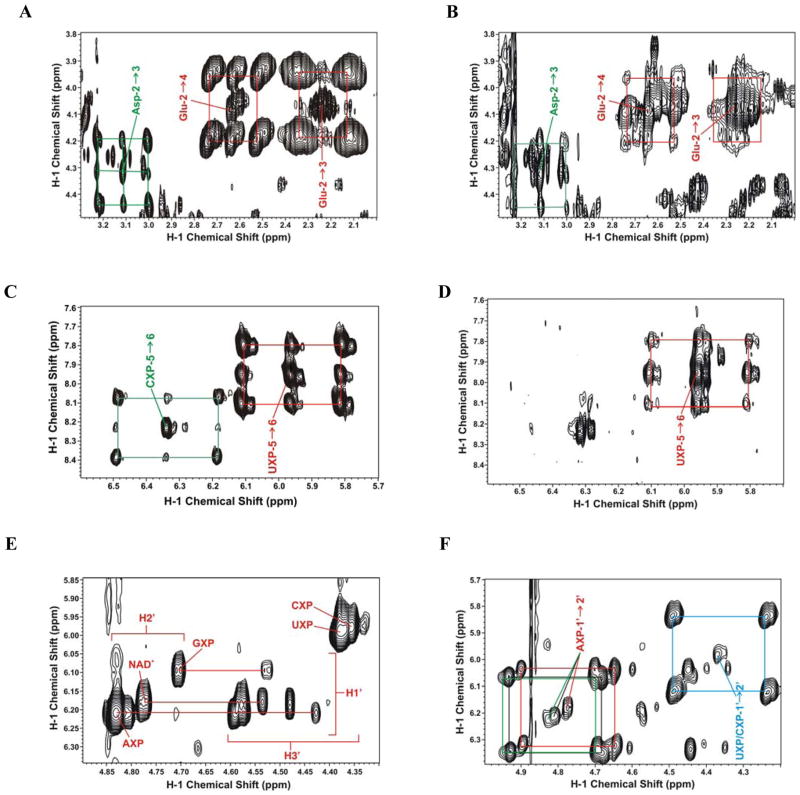
A is the peak area (1D) or peak volume (2D TOCSY). 13C satellite peaks arise from the one-bond 13C-1H J coupling, which splits the resonance into two components separated by the coupling constant J, as shown by the boxes in Figure 1. Peak areas or volume were determined using Varian VNMR or MResNova software.
Figure 1. 2-D 1H TOCSY NMR spectra of A549 cell extracts labeled with [U-13C]-Glucose or [U-13C,15N]-Glutamine.
A549 cells were grown in the presence of [U-13C]-Glucose + unlabeled Gln or unlabeled glucose (Glc) + [U-13C,15N]-glutamine (Gln) for 24 h and extracted as described in the Methods. TOCSY spectra were recorded as described in the Methods.
A. 13C Gln labeling showing 13C satellites for aspartate C2-C3 (green lines) and glutamate and glutathione C2-C3 and C2-C4 cross peaks, respectively (red lines).
B. 13C Glc labeling showing the same region as in A. The 13C labeling from Glc is much less than from 13C Gln (panel A).
C. 13C Gln labeling showing the 13C satellites of the base rings C5-C6 cross-peaks of cytosine nucleotides (CXP) (green box) and uracil nucleotides (UXP) (red box).
D. 13C Glc labeling showing the same region as in C. The 13C labeling from Glc is much less than from 13C Gln (panel C).
E. 13C Gln labeling showing C1′-C2′ cross-peaks of the ribose subunits of the free nucleotides, and the absence of 13C satellites. AXP, GXP: adenine and guanine nucleotides; UXP, CXP: as in panel C.
F. 13C Glc labeling showing the same region as in E. 13C satellites for AXP (red and black) and UXP+CXP (cyan) are much more intense than the 12C cross peak, indicating essentially all of the ribose carbons were derived from glucose via the pentose phosphate pathway.
3. Results and Discussion
De novo ribonucleotide biosynthesis
A549 cells proliferated with a doubling time of 18–20 h under the growth conditions. Cells grown in the presence of [U-13C]-glucose or [U-13C,15N]-glutamine were grown for 6 or 24 h before harvest and extracted as described in the methods. The incorporation of 13C into nucleotides and nucleotide precursors was determined by 2-D 1H TOCSY NMR, as shown in Figure 1. 13C deriving from [U-13C,15N]-glutamine entered the pools of free Glu, glutathione (GSH), aspartate (Fig 1A) and the pyrimidine rings of the free nucleotides, which were extensively 13C-labeled by 24 h. In contrast these molecules showed much less labeling from [U-13C]-glucose as the source (Figure 1B). The % enrichments at specific carbon positions of these molecules are given in Table 1. With [U-13C]-glucose as the source, only 15–20% of the Glu, Asp and pyrimidine rings contained 13C, whereas with [U-13C,15N]-glutamine as source, these metabolites were more than 75% labeled by 24 h. Furthermore, the degree of label scrambling was lower for glutamine than for glucose (Table 1 and Fig. 1). These data show that Gln is preferred over glucose as the carbon source for the synthesis of Glu, glutamate in GSH, aspartate, and pyrimidine rings.
Table 1. SIRM analysis of A549 Cells by NMR.
Cells grown for 24 h in RPMI containing 5 mM [U-13C]-glucose + 4 mM unlabeled Gln or 5 mM unlabeled glucose + 4 mM [U-13C,15N]-Gln.
Each isotopomer species with the carbon isotope at the designated positions was quantified from the corresponding cross-peaks in the TOCSY recorded at 14.1 T, 20 °C on trichloracetic acid extracts. Enrichments have been corrected for differential relaxation of protons attached to 13C or 12C (Lane and Fan 2007). UXP, CXP, and AXP: uracil, cytosine, and adenine nucleotides, respectively.
| Isotopomer species | [U-13C]-Glc + unlabeled Gln | unlabeled glucose + [U-13C,15N]-Gln | [U-13C] Glc + 5 μM MSA |
|---|---|---|---|
| % species | % species | % species | |
| Uracil in UXP | |||
| 12C6-12C5 | 81±4 | 24 ±3 | 93±3 |
| 13C6-12C5 | 6±2 | 9±2 | 3±1 |
| 12C6-13C5 | 10±4 | 9±2 | 4±1 |
| 13C6-13C5 | 3±1 | 58±2 | nd a |
| Cytosine in CXP | |||
| 12C6-12C5 | >80 | 35±3 | >95 |
| 13C6-12C5 | nd | 6±2 | nd |
| 12C6-13C5 | nd | 6±2 | nd |
| 13C6-13C5 | <20 | 53±2 | nd |
| Ribose in AXP | |||
| 12C1-12C2 | 7±2 | ≈99 | 30±2 |
| 13C1-13C2 | 93±3 | ≈1 | 70±3 |
| Ribose in UXP+CXP | |||
| 12C1-12C2 | 5±2 | ≈99 | 38±3 |
| 13C1-13C2 | 95±2 | ≈1 | 62±3 |
| Lactate | |||
| 12C212C3 | 50±2 | >98 | 44±2 |
| 13C213C3 | 50±2 | <2 | 56±2 |
| Alanine | |||
| 12C212C3 | 48±3 | 82±3 | nd a |
| 13C213C3 | 52±3 | 18±2 | nd a |
| Aspartate | |||
| 12C212C3 | 84±4 | 15±2 | >90 |
| 12C213C3 | 3±1 | 2±1 | nd |
| 13C212C3 | 9±2 | 21±2 | nd |
| 13C213C3 | 4±1 | 68±3 | nd |
| Glutamate | |||
| 12C212C4 | 77±3 | 11±2 | >90 |
| 12C213C4 | 18±2 | 1±1 | nd |
| 13C212C4 | 2±1 | 21±2 | nd |
| 13C213C4 | 2±1 | 68±3 | nd |
| 12C212C3 | 96±3 | 9±2 | >90 |
| 12C213C3 | 1.5±1 | 2±1 | nd |
| 13C212C3 | 2.5±1 | 2±1 | nd |
| 13C213C3 | <1 | 88±3 | <10 |
nd not determined because of weak or overlapped signals
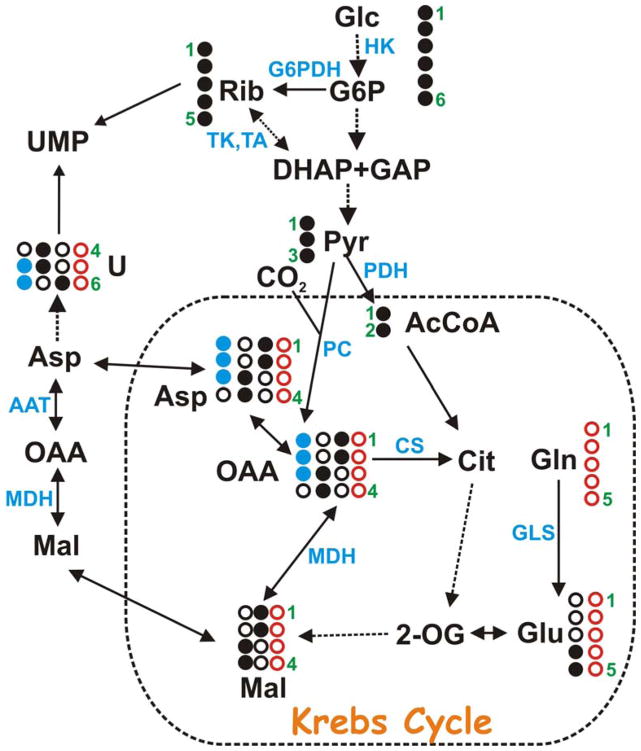
Glutamine is converted to glutamate via glutaminase, and amidotransferase reactions. The glutamate product can be incorporated directly into GSH, or enter the Krebs cycle at 2-oxoglutarate (Figure 2) via the transamination or oxidative deamination reactions. OAA formed in the Krebs cycle is subsequently transaminated to form aspartate and removed from the Krebs cycle for pyrimidine biosynthesis (Figure 2). Aspartate provides three of the four carbon atoms of uracil (Figure 2) and cytosine (i.e. C4, C5 and C6). As expected for a direct precursor-product relationship, the labeling pattern in Asp and the pyrimidine rings was very similar (Table 1). The Gln to Asp route is more direct than the glucose to Asp path, which involves both glycolysis and the Krebs cycle (Figure 2). This supports the above conclusion that glutamine is the preferred carbon source for pyrimidine ring synthesis, even in the presence of excess glucose. In addition, the presence of the scrambled 13C pattern in both aspartate and the pyrimidine rings when cells were grown on 13C-Gln + 12C-Glc indicates that a significant mixing of carbon sources via the Krebs cycle occurred under these conditions.
Figure 2. Metabolic pathways involving nucleotide biosynthesis.
The ribose subunit of nucleotides (Rib) is synthesized from [U-13C]-glucose (filled black circles) via hexokinase (HK) and glucose-6-phosphate dehydrogenase (G6PDH) (the oxidative branch of the PPP) or via transaldolase (TA) and transketolase (TK) (the non-oxidative branch of the PPP). The pyrimidine nucleobases are synthesized using one carbon from CO2 and three carbons from Asp, which is readily produced by transamination of oxalacetate (OAA) derived from the Krebs cycle by aspartate aminotransferase (AAT). The 13C labeling pattern of the pyrimidine ring thus reflects [U-13C]-glucose metabolism via glycolysis to pyruvate, entry of pyruvate into the Krebs cycle via acetyl CoA (AcCoA) formation catalyzed by pyruvate dehydrogenase (PDH), leading to respective 13C labeling of Asp at C1/C2 or C3/C4 in the first cycle, which are in turn incorporated into C6 or C4/C5 of uracil (U) base, respectively. Likewise, entry of 13C-glucose carbons into the Krebs cycle via oxalacetate (OAA) production catalyzed by pyruvate carboxylase (PC) leads to 13C label in Asp at C1/C2/C3(filled blue circles), which are in turn incorporated into C5/C6 of the uracil base. [U-13C,15N]-Glutamine (open red circles) can also serve as the precursor for Asp and therefore pyrimidine bases by replenishing 2-oxoglutarate (2-OG) via glutaminase (GLS) plus transaminase reactions, and the latter half of the Krebs cycle sequence. The glucose and Gln pathways give different 13C labeling patterns in both Asp and pyrimidine rings. Open black circles represents 12C from unlabeled sources. The shuttling in and out of the mitochondrion (dashed box) also involves isoforms of malate dehydrogenase (MDH). CS: citrate synthase.
The 13C labeling pattern of the ribose component of the free nucleotides had an opposite trend to that of the pyrimidine rings. With [U-13C]-glucose as tracer, the ribose rings of the free nucleotides were >92% 13C labeled whereas little 13C label was detected in ribose with 13C-Gln as the source (Figs. 1E, 1F; Table 1). There was also no detectable scrambling of ribosyl label from [U-13C]-glucose, indicating that the ribose rings were synthesized directly from glucose via the pentose phosphate pathway. The lack of incorporation of 13C into ribose from 13C-Gln implies that there was no significant gluconeogenesis under these conditions.
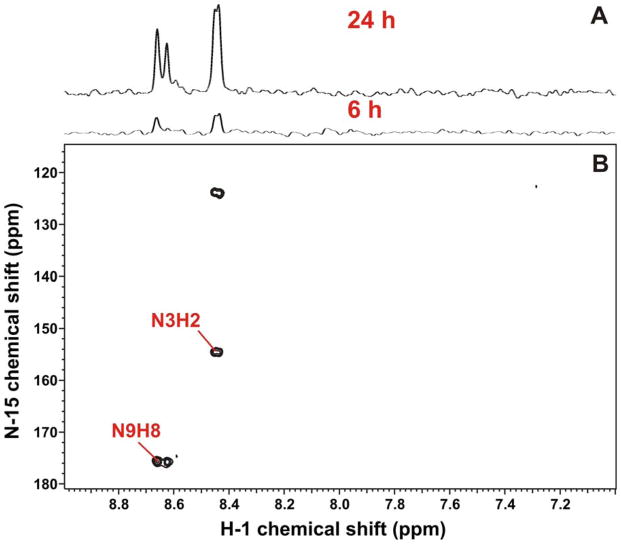
Although glutamine is not expected to provide carbon directly to purine biosynthesis, the amido nitrogen of Gln is incorporated into N3 and N9 of the purine ring (Murray et al. 2009). 15N at these positions can be readily detected by 2-bond coupling to H2 and H8 of the adenine rings, respectively. Figure 3 shows 1-D and 2-D 1H-{15N}-HSQC spectra of the A549 extracts grown in [U-15N,13C]-Gln, where H2 and H8 signals were evident as a result of the coupling to 15N at N3 and N9. The degree of labeling at 24 h was calculated from the 1-D proton spectrum (not shown) by comparing the intensity of the 15N satellites with that of the central or unlabeled H8 peak of the adenine ring. The adenine nucleobases showed 70% of 15N enrichment, which is consistent with de novo synthesis over a period of more than one cell doubling.
Figure 3. 1H{15N}-HSQC spectra of A549 cell extracts labeled with [U-13C,15N]-Gln.
(A) 1-D 1H-{15N} HSQC spectra were recorded as described in the Methods. The H8(N9) is at 8.65 ppm and the H2(N3) is at 8.44 ppm.
(B) The 2D 1H-{15N} HSQC spectrum. The 15N3 and 15N9 endocyclic nitrogens of purine rings (derived from the amido N of Gln) were observed indirectly via the coupled protons. A significant incorporation was observed even at 6 h of labeling, implying rapid de novo synthesis of the purine nucleotides, similar to that of the pyrimidine nucleotides.
The free nucleotide pool comprises all four common nucleotides plus NAD+. Because the intracellular concentration of the nucleotides was maintained essentially constant during the growth, the sizable presence of de novo synthesized ribonucleotides indicates that the pre-existing nucleotides were incorporated into a pool that was not extracted by aqueous acetonitrile. The most likely product is polymeric RNA, which led us to examine the incorporation of 13C from [U-13C]-glucose into the nucleotide components of RNA.
Incorporation of newly synthesized nucleotides into RNA
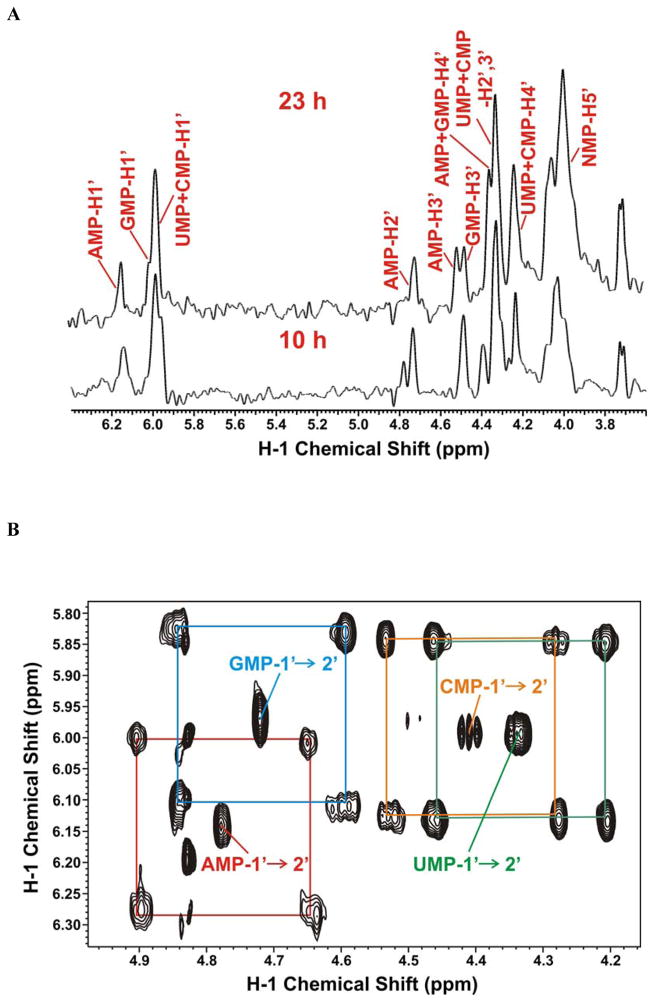
Total RNA was extracted from A549 cells at different times after growth in [U-13C]-glucose and digested with P1 nuclease as described in the Methods. To monitor the hydrolysis of RNA to its component nucleoside monophosphates, we followed the absorbance at 260 nm and verified the production of free mononucleotides by 1-D 1H NMR. As we have demonstrated for cellular extracts, it was unnecessary to separate the nucleotides by chromatography, as each nucleotide was sufficiently resolved by NMR. Furthermore, the RNA isolation step was very effective in removing contaminating non-nucleic acid components, resulting in simple NMR spectra, as shown in Figure 4. Figure 4A shows 1-D 1H-{13C}-HSQC spectra at 10 h and 23 h post labeling with 13C-glucose. These spectra show that the incorporation of labeled glucose carbon into the ribose moieties of both purine and pyrimidine nucleotides increased as the cells proliferated. Furthermore, all five ribosyl carbons were labeled, as expected from using [U-13C]-glucose as the source (see above).
Figure 4. RNA labeling in A549 cells grown in [U-13C]-Glc.
A549 cells were grown in the presence of [U-13C]-glucose and the RNA was extracted and digested for NMR analysis as described in the Methods. NMR spectra were recorded at 20 °C and 14.1 T.
A. 1-D 1H-{13C}-HSQC of the ribose region of the nucleotides at 10 and 23 h post labeling. Resonance assignments of the four nucleoside monophosphates (NMP) under these buffer conditions are denoted on the figure.
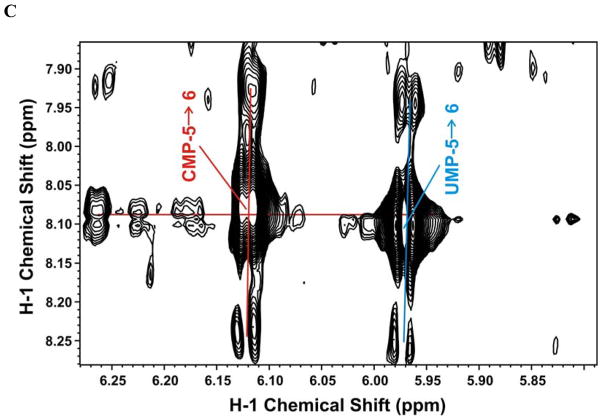
B. 2-D 1H TOCSY spectrum of the nucleotides showing the 13C C1′-C2′ satellites of the ribose units of AMP (red), GMP (cyan), UMP (green), and CMP (orange)
C. 2-D 1H TOCSY spectrum of the nucleotide riboses showing the 13C C5-C6 satellites of CMP (red) and UMP (cyan) nucleotides.
The fractional 13C incorporation into the pyrimidine rings and the ribosyl units of nucleotides hydrolyzed from RNA at 23 h were determined from the TOCSY spectrum shown in Figure 4B and 4C respectively. The isotopomer quantifications are summarized in Table 2. The pyrimidine rings showed incorporation of 13C from labeled glucose at either C5 or C6, which together accounted for about 10% of the total pool; very little was doubly labeled (Fig 4C, Table 2) compared with approximately 20% in the free nucleotide pool (Table 1). In contrast, the ribose moieties of the RNA nucleotides were much more heavily labeled from 13C-glucose (Fig. 4B) (ca. 50%, Table 2) as observed for the free nucleotide pool (see above). This is again roughly half the level observed in the free nucleotide pool (>95%, Table 1) after 23 h labeling with 13C-glucose. On the other hand, the cells doubled only once in this period, which is consistent with half of the isolated RNA coming from pre-existing unlabeled material in the cells prior to adding 13C glucose. The low extent of labeling in the pyrimidine rings of RNA nucleotides may reflect not only pre-existing unlabeled RNA but also the preference for carbon sources other than glucose (e.g. glutamine; see above) for de novo pyrimidine ring biosynthesis.
Table 2. 13C incorporation into nucleotides derived from cellular RNA.
Cellular RNA was extracted and hydrolyzed to the free nucleotides as described in the methods. Incorporation was quantified from TOCSY recorded at 20°C at 14.1 T after 23 h labeling with [U-13C]-glucose, as in Table 1. Enrichments have been corrected for differential relaxation of protons attached to 13C or 12C (Lane and Fan 2007).
| Isotopomer species | % Fraction |
|---|---|
| Uracil in UXP | |
| 12C512C6 | 90±3 |
| 12C513C6 | 7±2 |
| 13C512C6 | 3±2 |
| 13C513C6 | <1 |
| Cytosine in CXP | |
| 12C512C6 | 90±3 |
| 12C513C6 | 7±2 |
| 13C512C6 | 3±2 |
| 13C512C6 | <1 |
| Ribose in AXP | |
| 12C1′12C2′ | 52±5 |
| 13C1′13C2′ | 48±5 |
| Ribose in GXP | |
| 12C1′12C2′ | 49±5 |
| 13C1′13C2′ | 51±5 |
| Ribose in UXP + CXP | |
| 12C1′12C2′ | 58±5 |
| 13C113C2 | 42±5 |
| 12C112C3 | 47±5 |
| 13C113C3 | 53±5 |
It should be noted that these analyses were performed with only 30 μg of total 100 μg RNA extracted from cells grown on a single 10 cm plate, leaving sufficient material also for a typical gene microarray experiment.
Effect of MSA on the biosynthesis of free nucleotides and RNA
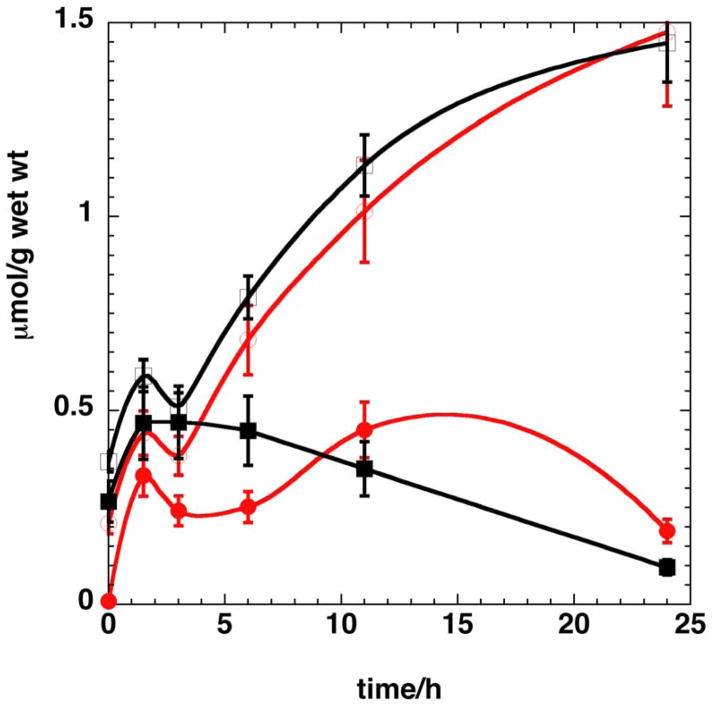
MSA is a potent inhibitor of cancer cell proliferation (IC50 = 5 μM for A549 cells) and induces numerous metabolic and morphological changes (T. W-M. Fan et al., unpublished data). Central metabolism including nucleotide metabolism is expected to decrease as growth is inhibited. Here we focused on nucleotide biosynthesis via the pentose phosphate pathway and pyrimidine ring synthesis, and subsequent incorporation into RNA. As Table 1 indicates, the incorporation of 13C from labeled glucose into the ribose moieties of the free nucleotides and uracil/cytosine base rings was substantially lower at 23 h in the presence of 5 μM MSA. Also attenuated was the degree of labeling in the Krebs cycle markers Glu and Asp. The reduced labeling of the precursor Asp presumably led to the reduced synthesis of the pyrimidine ring products (Table 1). Figure 5 shows a representative time course of 13C incorporation from [U-13C]-glucose into the C1′ of free nucleotide riboses, in the absence (open symbols) and presence (filled symbols) of 5 μM MSA. The reduced rate of synthesis in the presence of MSA was pronounced for both the purine and pyrimidine nucleotides, and was accompanied by a substantial decrease in proliferation. This is consistent with an inhibitory effect on 13C incorporation into RNA (Supplementary materials). The reduced incorporation rate into both the ribose moiety and the pyrimidine rings suggest a close link between inhibition of cell proliferation and nucleotide synthesis.
Figure 5. Effect of MSA on the time course of nucleotide biosynthesis.
A549 Cells were labeled with [U-13C]-glucose in the absence or presence of 5 μM MSA for different periods and the nucleotides were analyzed as described in the methods. The amount of each nucleotide was normalized to the cell wet weight. The time courses shown are representative of two independent experiments.
Open symbols control, filled symbols + MSA. Red is AXP, black is UXP. The lines are used to guide the eye. The control and MSA treatment at 11 and 24 h were significantly different for both nucleotide classes (p<0.005, unpaired t-test).
4. Concluding Remarks
We have shown that nucleotide synthesis in the free pools and their incorporation into the major cellular sink, RNA, can be readily followed by 1-D and 2-D NMR using stable isotope tracers such as [U-13C]-glucose or [U-13C,15N]-glutamine with less than 2 mg of dry cell materials. The NMR analysis enabled the determination of fractional enrichment and atomic position of label incorporated into both ribose and nucleobase units of individual intact nucleotides, as well as in precursors and other metabolites in the same experiment. This provides many more crucial parameters for pathway modeling than existing mass spectrometry-based methods. We found that [U-13C]-glucose was preferentially incorporated into the ribose moiety of all nucleotides, whereas 13C,15N-glutamine was the preferred carbon source for the synthesis of the pyrimidine rings, in addition to providing nitrogen for the N9 and N3 positions of the purine rings. This in part accounts for the requirement of glutamine for cancer cell proliferation (Yuneva 2008). We have observed similar differences in label incorporation into the nucleotide pools of several other cancer cells in culture (Fan et al. 2008; Fan and Lane 2011) (and A.N. Lane, T.W-M. Fan, unpublished data), suggesting that the present observations in A549 cells may reflect a generally distinct role of glucose and glutamine in nucleotide biosynthesis. As nucleotides are used in a wide variety of cellular processes in addition to RNA and DNA biosynthesis (Morrish et al. 2009; Murray et al. 2009; Moseley et al. 2011), a simple method to determine nucleotide biosynthesis and utilization is necessary for understanding the regulation of cell proliferation. The direct observation of labeling patterns in nucleotides enables such a conclusion, which might otherwise be ambiguous based on the labeling patterns of metabolically remote metabolites (DeBerardinis et al. 2007; Hiller et al. 2010) or from total pool analysis without tracers (Lu et al. 2010). Furthermore, the importance of nucleotide and RNA biosynthesis in modulating cancer cell proliferation can be inferred from the parallel inhibition of both processes by MSA, which was presumably mediated through the reduced production of the Asp precursor. This interference with nucleotide metabolism by MSA may be an important factor underlying its anti-cancer action.
Supplementary Material
Acknowledgments
Financial Support
This work was supported in part by National Science Foundation EPSCoR grant # EPS-0447479; NIH NCRR Grant 5P20RR018733, 1R01CA118434-01A2 (to TWMF), 1RO1 CA101199 (to TWMF), R21CA133668-01 (to ANL) from the National Cancer Institute; the Kentucky Challenge for Excellence, and the Brown Foundation.
We thank Dr. S. Arumugam for assistance in the NMR measurements.
Abbreviations used
- DSS
2,2′dimethylsilapentane-5-sulfonate
- GSH
reduced glutathione
- HSQC
Heteronuclear single quantum coherence
- MSA
Methyl Seleninic Acid
- NSCLC
non small cell lung cancer
- SIRM
Stable Isotope Resolved Metabolomics
- TOCSY
Total Correlation Spectroscopy
References
- Bak LK, Waagepetersen HS, Melo TM, Schousboe A, Sonnewald U. Complex glutamate labeling from [U-C-13]glucose or [U-C-13]lactate in co-cultures of cerebellar neurons and astrocytes. Neurochemical Research. 2007;32:671–680. doi: 10.1007/s11064-006-9161-4. [DOI] [PubMed] [Google Scholar]
- Beger RD, Hansen DK, Schnackenberg LK, Cross BM, Fatollahi JJ, Lagunero FT, Sarnyai Z, Boros LG. Single valproic acid treatment inhibits glycogen and RNA ribose turnover while disrupting glucose-derived cholesterol synthesis in liver as revealed by the [U-13C6]-D-glucose tracer in mice. Metabolomics. 2009;5:336–345. doi: 10.1007/s11306-009-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boren J, Cascante M, Marin S, Comin-Anduix B, Centelles JJ, Lim S, Bassilian S, Ahmed S, Lee WNP, Boros LG. Gleevec (ST1571) influences metabolic enzyme activities and glucose carbon flow toward nucleic acid and fatty acid synthesis in myeloid tumor cells. Journal of Biological Chemistry. 2001;276:37747–37753. doi: 10.1074/jbc.M105796200. [DOI] [PubMed] [Google Scholar]
- Centelles JJ, Ramos-Montoya A, Lim S, Bassilian S, Boros LG, Marin S, Cascante M, Lee WNP. Metabolic profile and quantification of deoxyribose synthesis pathways in HepG2 cells. Metabolomics. 2007;3:105–111. [Google Scholar]
- Clark LC, Combs GF, Turnbull BW, Slate EH, Chalker DK, Chow J, Davis LS, Glover RA, Graham GF, Gross EG, Krongrad A, Lesher JL, Park HK, Sanders BB, Smith CL, Taylor JR. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin a randomized controlled trial - A randomized controlled trial. Jama-Journal of the American Medical Association. 1996;276:1957–1963. [PubMed] [Google Scholar]
- Combs GF. Status of selenium in prostate cancer prevention. British Journal of Cancer. 2004;91:195–199. doi: 10.1038/sj.bjc.6601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado TC, Castro MM, Geraldes CF, Jones JG. Quantitation of erythrocyte pentose pathway flux with [2-(13)]Glucose and H-1 NMR analysis of the lactate methyl signal. Magnetic Resonance in Medicine. 2004;51:1283–1286. doi: 10.1002/mrm.20096. [DOI] [PubMed] [Google Scholar]
- Fan TWM, Bandura L, Higashi RM, Lane AN. Metabolomics-edited transcriptomics analysis of Se anticancer action in human lung cancer cells. Metabolomics. 2005;1:1–15. [Google Scholar]
- Fan TW-M, Kucia M, Jankowski K, Higashi RM, Rataczjak MZ, Rataczjak J, Lane AN. Proliferating Rhabdomyosarcoma cells shows an energy producing anabolic metabolic phenotype compared with Primary Myocytes. Molecular Cancer. 2008;7:79. doi: 10.1186/1476-4598-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TWM, Lane AN. Structure-based profiling of Metabolites and Isotopomers by NMR. Progress in NMR Spectroscopy. 2008;52:69–117. [Google Scholar]
- Fan TW-M, Lane AN. Assignment strategies for NMR resonances in metabolomics research. In: Lutz N, Sweedler JV, Wever RA, editors. Methodologies for Metabolomics: Experimental Strategies and Techniques. New York: Cambridge University Press; 2011. in press. [Google Scholar]
- Fan TW-M, Lane AN. NMR-based Stable Isotope Resolved Metabolomics in Systems Biochemistry. J Biomolec NMR. 2011;267–280:267–280. doi: 10.1007/s10858-011-9484-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TW-M, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM. Altered Regulation of Metabolic Pathways in Human Lung Cancer Discerned by 13C Stable Isotope-Resolved Metabolomics (SIRM)) Molecular Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TWM, Yuan P, Lane AN, Higashi RM, Wang Y, Hamidi A, Zhou R, Guitart-Navarro X, Chen G, Manji HK, Kaddurah-Daouk R. Stable Isotope Resolved Metabolomic Analysis of Lithium Effects on Glial-Neuronal Interactions. Metabolomics. 2010;6:165–179. doi: 10.1007/s11306-010-0208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TWM, Higashi RM, Lane AN. Integrating metabolomics and transcriptomics for probing Se anticancer mechanisms. Drug Metabolism Reviews. 2006;38:707–732. doi: 10.1080/03602530600959599. [DOI] [PubMed] [Google Scholar]
- Fan TWM, Higashi RM, Lane AN, Jardetzky O. Combined use of proton NMR and gas chromatography-mass spectra for metabolite monitoring and in vivo proton NMR assignments. Biochimica et Biophysica Acta. 1986;882:154–167. doi: 10.1016/0304-4165(86)90150-9. [DOI] [PubMed] [Google Scholar]
- Ganther HE. Selenium metabolism, selenoproteins and mechanisms of cancer prevention: Complexities with thioredoxin reductase. Carcinogenesis (Oxford) 1999;20:1657–1666. doi: 10.1093/carcin/20.9.1657. [DOI] [PubMed] [Google Scholar]
- Hiller K, Metallo CM, Kelleher JK, Stephanopoulos G. Nontargeted Elucidation of Metabolic Pathways Using Stable-Isotope Tracers and Mass Spectrometry. Analytical Chemistry. 2010;82:6621–6628. doi: 10.1021/ac1011574. [DOI] [PubMed] [Google Scholar]
- Hu VW, Black GE, Torres-Duarte A, Abramson FP. H-3-thymidine is a defective tool with which to measure rates of DNA synthesis. Faseb Journal. 2002;16:1456. doi: 10.1096/fj.02-0142fje. [DOI] [PubMed] [Google Scholar]
- Ip C, Dong Y, Ganther HE. New concepts in selenium chemoprevention. Cancer and Metastasis Reviews. 2002;21:281–289. doi: 10.1023/a:1021263027659. [DOI] [PubMed] [Google Scholar]
- Ippel J, Wijmenga S, de Jong B, Heus H, Hilbers C, Vroom E, van de Marel C, van Boom J. Heteronuclear scalar couplings in the bases and sugar rings of nucleic acids: their determination and application in assignment and conformational analysis. Magnetic Resonance in Chemistry. 1996;34:S156–S176. [Google Scholar]
- Lane AN, Fan TWM. Quantification and identification of isotopomer distributions of metabolites in crude cell extracts using 1H TOCSY. Metabolomics. 2007;3:79–86. [Google Scholar]
- Lane AN, Fan TWM, Higashi RM. Isotopomer-based metabolomic analysis by NMR and mass spectrometry. Biophysical Tools for Biologists. 2008;84:541–588. doi: 10.1016/S0091-679X(07)84018-0. [DOI] [PubMed] [Google Scholar]
- Lane AN, Fan TWM, Higashi RM, Tan J, Bousamra M, Miller DM. Prospects for clinical cancer metabolomics using stable isotope tracers. J Exp Molec Pathol. 2009;86:165–173. doi: 10.1016/j.yexmp.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, Ford LG, Parnes HL, Minasian LM, Gaziano JM, Hartline JA, Parsons JK, Bearden JD, Crawford ED, Goodman GE, Claudio J, Winquist E, Cook ED, Karp DD, Walther P, Lieber MM, Kristal AR, Darke AK, Arnold KB, Ganz PA, Santella RM, Albanes D, Taylor PR, Probstfield JL, Jagpal TJ, Crowley JJ, Meyskens FL, Baker LH, Coltman CA. Effect of Selenium and Vitamin E on Risk of Prostate Cancer and Other Cancers The Selenium and Vitamin E Cancer Prevention Trial (SELECT) Jama-Journal of the American Medical Association. 2009;301:39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Bennet B, Mu E, Rabinowitz J, Kang Y. Metabolomic Changes Accompanying Transformation and Acquisition of Metastatic Potential in a Syngeneic Mouse Mammary Tumor Model. J Biol Chem. 2010;285:9317–9321. doi: 10.1074/jbc.C110.104448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso A, Zhu AZ, Beardsley NJ, Glickson JD, Wehrli S, Pickup S. Artificial tumor model suitable for monitoring P-31 and C-13 NMR spectroscopic changes during chemotherapy-induced apoptosis in human glioma cells. Magnetic Resonance in Medicine. 2005;54:67–78. doi: 10.1002/mrm.20545. [DOI] [PubMed] [Google Scholar]
- Mason GF, Petersen KF, de Graaf RA, Shulman GI, Rothman DL. Measurements of the anaplerotic rate in the human cerebral cortex using C-13 magnetic resonance spectroscopy and [1-C-13] and [2-C-13] glucose. Journal of Neurochemistry. 2007;100:73–86. doi: 10.1111/j.1471-4159.2006.04200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes AC, Caldeira MM, Silva C, Burgess SC, Merritt ME, Gomes F, Barosa C, Delgado TC, Franco F, Monteiro P, Providencia L, Jones JG. Hepatic UDP-glucose C-13 isotopomers from [U-C-13]glucose: A simple analysis by C-13 NMR of urinary menthol glucuronide. Magnetic Resonance in Medicine. 2006;56:1121–1125. doi: 10.1002/mrm.21057. [DOI] [PubMed] [Google Scholar]
- Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene. 2009;28:2485–2491. doi: 10.1038/onc.2009.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley HNB, Lane AN, Belshoff AC, Higashi RM, Fan WWM. Non-Steady State Modeling of UDP-GlcNAc Biosynthesis is Enabled by Stable Isotope Resolved Metabolomics (SIRM) BMC Biology. 2011;9:37. [Google Scholar]
- Murray RK, Bender DA, Botham KM, Kennelly PJ, Rodwell VW, Weil PA. Harper’s Illustrated Biochemistry. New York: McGraw-Hill; 2009. [Google Scholar]
- Nelson DL, Cox MM. Lehninger Principles of Biochemistry. New York: W.H. Freeman and Company; 2005. [Google Scholar]
- Telang S, Lane AN, Nelson KK, Arumugam S, Chesney JA. The oncoprotein H-RasV12 increases mitochondrial metabolism. Molec Cancer. 2007;6:77. doi: 10.1186/1476-4598-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizan P, Alcarraz-Vizan G, Diaz-Moralli S, Rodriguez-Prados JC, Zanuy M, Centelles JJ, Jauregui O, Cascante M. Quantification of intracellular phosphorylated carbohydrates in HT29 human colon adenocarcinoma cell line using liquid chromatography-electrospray ionization tandem mass spectrometry. Analytical Chemistry. 2007;79:5000–5005. doi: 10.1021/ac070170v. [DOI] [PubMed] [Google Scholar]
- Vizan P, Boros LG, Figueras A, Capella G, Mangues R, Bassilian S, Lim S, Lee WNP, Cascante M. K-ras Codon-Specific Mutations Produce Distinctive Metabolic Phenotypes in Human Fibroblasts. Cancer Res. 2005;65:5512–5515. doi: 10.1158/0008-5472.CAN-05-0074. [DOI] [PubMed] [Google Scholar]
- Yuneva M. Finding an “Achilles’ heel” of cancer: the role of glucose and glutamine metabolism in the survival of transformed cells. Cell Cycle. 2008;7:2083–2089. doi: 10.4161/cc.7.14.6256. [DOI] [PubMed] [Google Scholar]
- Zwingmann C, Leibfritz D. Regulation of glial metabolism studied by C-13-NMR. Nmr in Biomedicine. 2003;16:370–399. doi: 10.1002/nbm.850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.