Abstract
The tumor suppressor serine/threonine kinase 11 (STK11 or LKB1) is mutated in 20–30% of non-small cell lung cancer (NSCLC) patient tumors. Loss of LKB1-AMPK signaling confers sensitivity to metabolic inhibition or stress-induced mitochondrial insults. We tested the hypothesis that loss of LKB1 sensitizes NSCLC cells to energetic stress induced by treatment with erlotinib. LKB1-deficient cells exhibited enhanced sensitivity to erlotinib in vitro and in vivo that was associated with alterations in energy metabolism and mitochondrial dysfunction. Loss of LKB1 expression altered the cellular response to erlotinib treatment, resulting in impaired ATP homeostasis and an increase in reactive oxygen species (ROS). Furthermore, erlotinib selectively blocked mTOR signaling, inhibited cell growth, and activated apoptosis in LKB1-deficient cells. Erlotinib treatment also induced AMPK activation despite loss of LKB1 expression, which was partially reduced by the application of a CaMKKβ inhibitor (STO-609) or calcium chelator (BAPTA-AM). These findings may have significant implications for the design of novel NSCLC treatments that target dysregulated metabolic and signaling pathways in LKB1-deficient tumors.
INTRODUCTION
Lung cancer is the leading cause of cancer mortality worldwide and is linked to 28% of all cancer deaths in the United States 1. Despite advances in traditional therapeutic strategies involving surgery, ionizing radiation therapy, and chemotherapy, the 5-year survival remains less than 20% 2. In recent years, it has become clear that non-small cell lung cancer has a high frequency of somatically acquired genetic alterations that define critical subsets of tumors with distinct behaviors 3. An improved understanding of potential vulnerabilities of lung cancer subsets has led to the development of effective targeted therapies for tumors with certain activated oncogenes 4, 5, but little is known about specific susceptibilities that may derive from the loss of classical tumor suppressor genes, such as LKB1.
LKB1 is a tumor suppressive serine/threonine kinase that activates diverse downstream kinases, thus regulating a variety of cellular phenotypes including metabolism, invasion, proliferation, and polarity 6. LKB1 inactivation occurs in 20–30 percent of lung adenocarcinomas 7, and LKB1 deficiency in combination with KRAS mutation leads to an aggressive tumor phenotype at high prevalence in mouse models, surpassing that of KRAS mutation alone 8. Many of the metabolic regulatory functions of LKB1 are mediated by its interaction with adenosine monophosphate-activated protein kinase (AMPK). LKB1 phosphorylates and activates AMPK 6, which functions to regulate cellular energy metabolism under conditions of low ATP 9. AMPK also contributes to inactivation of mTOR when ATP levels fall, which leads to inhibition of protein synthesis and cell growth 10. Therefore, loss of LKB1 leads to dysregulation of cellular metabolism and cell growth under conditions of energy stress 11, resulting in enhanced sensitivity to drug treatments that target bioenergetic pathways 12.
Most lung cancers express the epidermal growth factor receptor (EGFR), and this signaling pathway is the major target of several drugs in the clinic. EGFR tyrosine kinase inhibitors including gefitinib and erlotinib have been shown to suppress oncogenic signaling through downstream pathways such as PI3K-Akt-mTOR and Mek-Erk 13. NSCLC tumors with certain activating mutations in EGFR show enhanced sensitivity to these compounds 14. However, the majority of NSCLC patient tumors possess wild-type EGFR, with only 10–30% harboring a mutant EGFR allele 15. Although erlotinib has clear therapeutic efficacy in some NSCLC tumors bearing wild-type EGFR 16, 17 and is approved for patients with wild-type EGFR tumors 18, it is unclear how to best identify which of these patients may benefit from treatment with EGFR-targeted inhibitors. Furthermore, the mechanism by which erlotinib induces selective cell death in wild-type EGFR tumors is not completely known.
In EGFR mutant NSCLC cells, erlotinib causes apoptosis through activation of intrinsic pathways mediated by the induction of BH3-only BIM protein or activation of caspase 3 19, 20. In these studies, erlotinib treatment was associated with loss of mitochondrial potential, which resulted in mitochondrial-mediated apoptosis. Interestingly, recent studies suggest that LKB1 deficiency causes an accumulation of defective mitochondria and loss of mitochondrial membrane potential, resulting in depletion of hematopoietic stem cells through disruption of mitophagy and mitochondrial homeostasis21. Furthermore, the mitochondrial complex I inhibitor phenformin enhanced apoptosis of LKB1-deficient tumor cells by depletion of mitochondrial membrane potential compared to wild-type LKB1-reconstituted cells 12. Therefore, we hypothesized that erlotinib would be more effective at inducing apoptosis in LKB1-deficient NSCLC cells due to disruption of normal mitochondrial function, even in the presence of wild-type EGFR.
In the present study, we tested whether LKB1-deficient cells were more prone to apoptosis in response to erlotinib treatment, both in vitro and in vivo, in the absence of mutationally activated EGFR. Our data demonstrate enhanced antitumor effects of erlotinib in LKB1-deficient cells that are associated with loss of mitochondrial function and blockage of mTOR signaling. Overall, our findings suggest that the LKB1-deficient molecular phenotype may serve as a predictive marker for erlotinib sensitivity and, if substantiated by further studies, may provide a rationale for the therapeutic use of erlotinib to treat LKB1-deficient NSCLC. This is significant because LKB1 mutations are preferentially found in NSCLC tumors with wild-type EGFR 22, 23, and thus LKB1 loss may define a novel subset of tumors with enhanced erlotinib sensitivity that is largely distinct from the classical subset possessing targetable EGFR mutations.
RESULTS
LKB1-deficient cells are more sensitive to inhibition of EGFR-PI3K-mTOR signaling
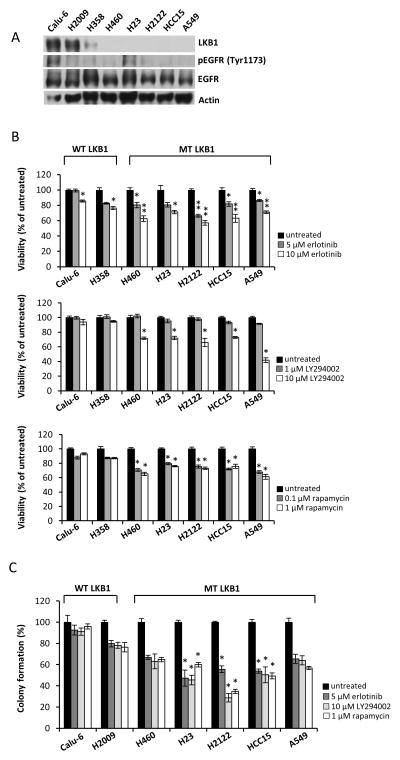
We sought to determine the relative inhibitory effects of erlotinib, as well as other compounds that target the EGFR-PI3K-mTOR signaling pathway, across a panel of NSCLC cell lines that express either wild-type or mutant LKB1. The expressions of LKB1 and EGFR were assessed by Western blotting in NSCLC cells (Fig. 1A). The NSCLC cells tested have been reported to express wild-type EGFR 24, 25, and the expression level of phosphorylated EGFR is shown in Fig 1A. Calu-6, H2009, and H358 cells were previously reported to express wild-type LKB1 26–28, and we confirmed LKB1 protein expression in these three lines. Next, we asked whether erlotinib could selectively inhibit the viability of LKB1 mutant cells harboring KRAS mutations but with wild-type EGFR. We found that LKB1 mutant cells were more sensitive to erlotinib on average (Fig. 1B). Furthermore, 10 μM of the PI3K inhibitor LY294002 did not reduce viability of LKB1 wild-type NSCLC cells, whereas 30–50% inhibition was observed in LKB1 mutant cells. Sensitivity to rapamycin was also exacerbated in LKB1 mutant cells. We further assessed the survival of NSCLC cells using a colony-forming assay where cells were pretreated with inhibitor for 72 h and then grown in inhibitor-free media for two weeks. The colony-forming assay was more capable of detecting differences in viability at low inhibitor concentrations and confirmed the finding that LKB1 mutant cells were more sensitive to inhibition of EGFR-PI3K-mTOR signaling (Fig. 1C). These results suggest that LKB1 loss confers enhanced sensitivity to inhibition of the EGFR-PI3K-mTOR signaling pathway in NSCLC cells harboring wild-type EGFR.
Figure 1. Erlotinib and inhibitors of PI3K-mTOR signaling selectively inhibit growth of LKB1-deficient NSCLC cells.
(a) Levels of LKB1 expression and EGFR phosphorylation in our panel of NSCLC cells. Actin was used as a loading control. (b) Effect of treatment with erlotinib, LY294002, or rapamycin on viability of NSCLC cells. Cells were seeded in 96-well plates. After 24 h, cells were treated with indicated concentrations of erlotinib, LY294002, or rapamycin for 48 h. Cell viability was determined using MTT assay. *; p<0.005, **; p<0.00001, untreated/erlotinib, *; p<0.0001, untreated/LY294002 or untreated/rapamycin. Data are mean ± SEM (n=6). (c) Effects of treatment with erlotinib, LY294002, or rapamycin on colony-forming ability of NSCLC cells. After 72 h treatment with erlotinib, LY294002, or rapamycin, cells were re-plated in 12-well plates at low density. Colonies were counted after 2 weeks of growth. *; p<0.0001, untreated/erlotinib, untreated/LY294002, or untreated/rapamycin. Data are mean ± SEM (n=3).
LKB1-deficient transgenic cells exhibit increased sensitivity to erlotinib treatment
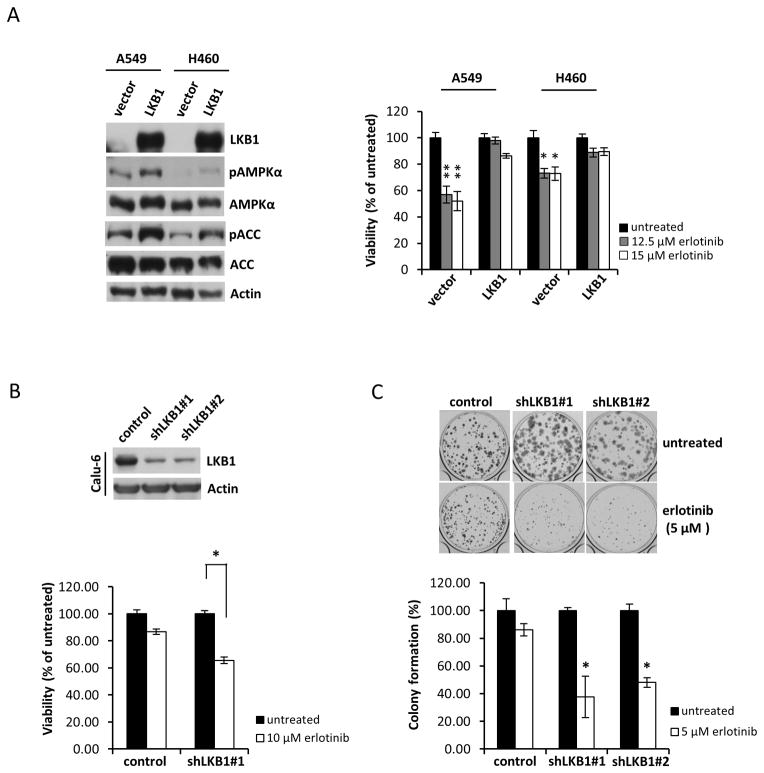
A549 and H460 cells (both wild-type EGFR and mutant LKB1) were transduced with retroviral constructs to stably express wild-type LKB1 or an empty vector control. Expression of LKB1 and related downstream signaling proteins were verified in these stable transgenic cells (Fig. 2A, left). LKB1 phosphorylates Thr172 in the activation loop of AMPKα 29, 30 and activates AMPK under elevated AMP levels 31. Activated AMPK subsequently phosphorylates and inhibits acetyl-CoA carboxylase (ACC). Overexpression of LKB1 in A549 and H460 resulted in increased basal phosphorylation of AMPKα and its downstream target ACC. Erlotinib inhibited growth of LKB1 non-expressing A549 (p<0.0001) and H460 cells (p<0.005) more than LKB1 overexpressing cells (Fig. 2A, right). Conversely, in stable LKB1-knockdown lines generated from LKB1 wild-type Calu-6 cells, erlotinib induced a more substantial reduction in growth than the vector control (Fig. 2B, lower panel). Additionally, erlotinib reduced the colony-forming ability of LKB1-knockdown cells more significantly than the vector control (Fig. 2C). These results suggest that LKB1 deficiency enhances sensitivity to erlotinib by suppressing viability and tumorigenic potential of NSCLC cells.
Figure 2. Erlotinib selectively inhibits growth of LKB1 mutant cells and LKB1 knockdown cells.
(a) Left panel: Levels of LKB1 expression and LKB1-related downstream proteins in A549 and H460 cells stably expressing empty pBABE vector (vector) or wild-type LKB1 (LKB1). Right panel: Cell viability determined by MTT assay. Cells were treated with indicated concentrations of erlotinib for 48 h. *; p<0.005, **; p<0.0001, untreated/erlotinib. Data are mean ± SEM (n=6). (b) Upper panel: Western blots of whole-cell lysates from LKB1 stable knockdown and non-targeted shRNA control Calu-6 cells after puromycin selection: control, pLKO.1 control vector; shLKB1#1 and shLKB1#2, LKB1 shRNA vectors. Lower panel: Cell viability determined by MTT assay. *; p<0.0001, untreated/erlotinib. Data are mean ± SEM (n=6). (c) Colony-forming ability of LKB1 stable knockdown Calu-6 cells. After 72 h treatment with 5 μM erlotinib, cells were replated in 12-well plates at low density. Colonies were counted after 2 weeks of growth. *; p<0.001, untreated/erlotinib. Data are mean ± SEM (n=3).
Erlotinib treatment selectively inhibits oxidative metabolism and disrupts energy homeostasis in LKB1-deficient cells
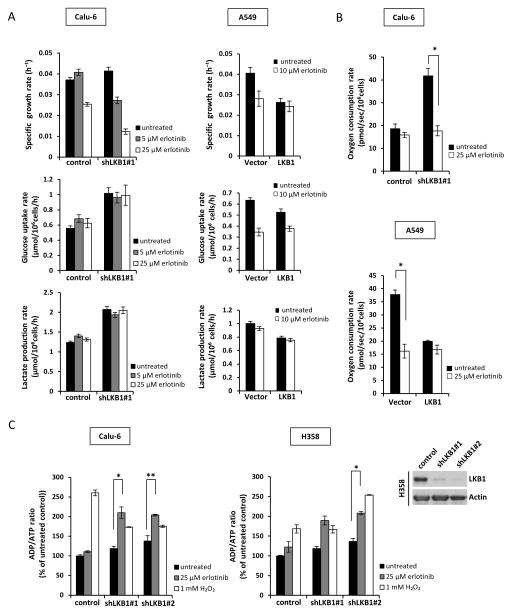
We next determined whether erlotinib treatment alters cellular metabolism through analysis of glucose, lactate, and oxygen exchange rates. Knockdown of LKB1 in Calu-6 cells caused basal glucose consumption and lactate production to increase significantly compared to vector control cells (Fig. 3A, left), consistent with previously published results 32. Measurement of oxygen consumption rate (OCR) also revealed a two-fold increase in basal oxidative metabolism by shLKB1 Calu-6 cells (Fig. 3B, upper), which was further confirmed in shLKB1 H358 cells (Supplementary Fig. S1A). Erlotinib treatment reduced specific growth rates of both vector control and shLKB1 Calu-6 cells in a dose-dependent manner without affecting glucose uptake or lactate production (Fig. 3A, left). However, the OCR of shLKB1-expressing Calu-6 and H358 cells was halved by erlotinib treatment (Fig. 3B and Supplementary Fig. S1A). Similar results were observed after re-expressing LKB1 in A549 and H460 cells. LKB1 mutant A549 and H460 cells exhibited increased glucose consumption and lactate production compared to LKB1 overexpressing cells (Fig. 3A, right and Supplementary Fig. 2). Furthermore, OCR was significantly elevated in LKB1 mutant cells but was reduced >50% by erlotinib treatment (Fig. 3B and Supplementary Fig. S1B). In contrast to its effects on glycolytic metabolism, the effect of erlotinib to reduce OCR was significantly enhanced in the LKB1 mutant lines.
Figure 3. Erlotinib reduces growth rate and selectively inhibits OXPHOS in LKB1-deficient cells.
(a) Cell growth rate, glucose uptake rate, and lactate production rate of Calu-6 and A549 cells. Cells were grown for 72 h in the presence of treatments. (b) Oxygen consumption rate (OCR) of Calu-6 and A549 cells. Cells were treated with erlotinib for 24 h. Data are mean ± SEM (n=4). *; p<0.005, untreated/erlotinib. (c) Left panel: ADP/ATP ratio of Calu-6 and H358 cells. Cells were treated with 25 μM erlotinib for 24 h or 1 mM H2O2 (positive control) for 1 h prior to measurement. Right panel: Western blot analysis of LKB1 expression in H358 cells. Data are mean ± SEM. *; p<0.05, **; p<0.005, untreated/erlotinib.
The reduction in OCR by erlotinib treatment was associated with a significant increase in the ADP/ATP ratio in shLKB1-expressing Calu-6 and H358 cells (Fig. 3C). In contrast, the OCR and ADP/ATP ratio of control Calu-6 and H358 cells were not significantly affected by erlotinib. These results indicate that LKB1-deficient cells were not able to maintain their elevated rate of oxidative phosphorylation (OXPHOS) in the presence of erlotinib treatment, thus leading to ATP depletion. Several recent articles have shown that loss of LKB1 function leads to increased expression of mitochondria-associated genes 23 but also induces defective mitophagy and accumulation of dysfunctional mitochondria 12, 21. Therefore, we hypothesized that mitochondrial impairments may play a role in the response of LKB1-deficient cells to erlotinib treatment.
Erlotinib treatment unmasks mitochondrial defects in LKB1-deficient cells
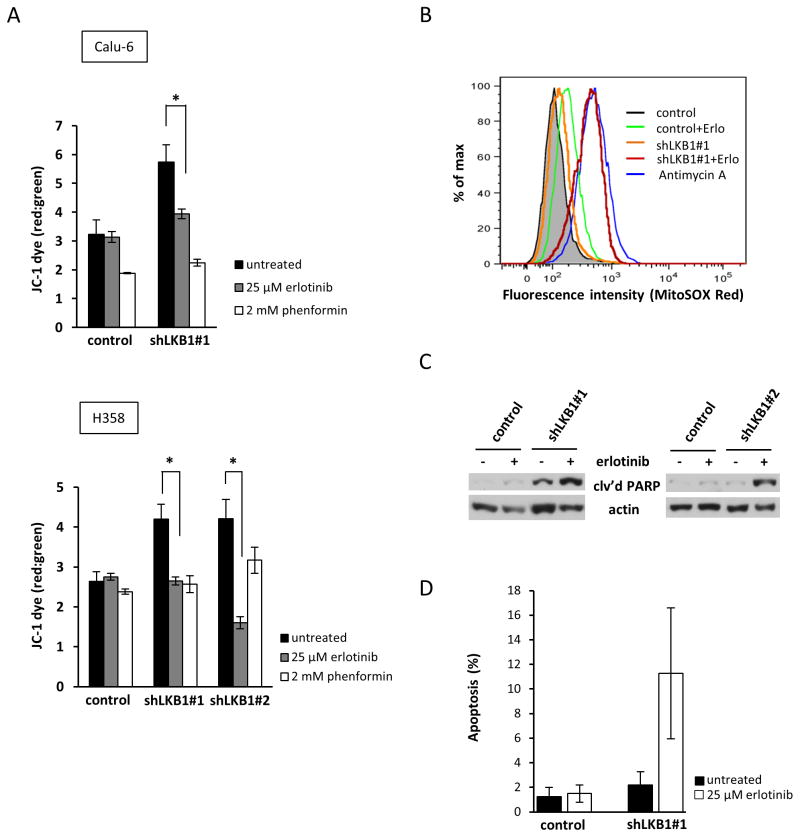
Erlotinib-induced apoptosis is associated with loss of mitochondrial membrane potential in EGFR mutant NSCLC cells 20. Therefore, we asked whether EGFR wild-type NSCLC cells bearing loss-of-function mutations in LKB1 would show similar mitochondrial defects following erlotinib treatment. We measured the effect of erlotinib treatment on mitochondrial potential by using the cationic dye JC-1. Phenformin, a mitochondrial complex I antagonist, was applied as a positive control to impair mitochondria. As shown in Fig. 4A, following treatment with erlotinib or phenformin, Calu-6 and H358 cells expressing shLKB1 exhibited enhanced dissipation of Δψm compared to control cells (red to green ratio of JC-1), reflecting impaired mitochondrial membrane integrity. Significantly, the Δψm of untreated shLKB1 cells was higher than vector controls, which is indicative of increased basal mitochondrial activity and is consistent with the elevated OCR of untreated shLKB1 cells (Fig. 3B). Calu-6 cells expressing shLKB1 also exhibited increased mitochondrial production of reactive oxygen species (ROS) in response to erlotinib treatment (Fig. 4B). We hypothesized that ATP depletion, loss of mitochondrial membrane potential, and ROS accumulation might by associated with increased apoptosis in LKB1-deficient cells. As shown in Fig. 4C and 4D, treatment of shLKB1 Calu-6 cells with erlotinib induced cleavage of PARP and an accumulation of apoptotic cells, suggesting that erlotinib enhances mitochondrial-mediated apoptosis in LKB1-deficient cells. These data indicate that LKB1-deficient cells rely more heavily on mitochondria for energy metabolism under basal conditions, but they exhibit profound mitochondrial dysfunction in response to erlotinib treatment as evidenced by Δψm collapse, ATP depletion, ROS accumulation, and apoptosis activation.
Figure 4. Erlotinib induces mitochondrial defects and enhanced apoptosis in stable LKB1 knockdown NSCLC cells.
(a) Mitochondrial potential was measured using JC-1 fluorescence in both Calu-6 and H358 cells. Cells were treated with 25 μM erlotinib for 24 h. Data are mean ± SEM. *; p<0.05, untreated/erlotinib. (b) Mitochondrial ROS accumulation by Calu-6 cells. Cells were treated with 25 μM erlotinib for 24 h or 20 μM antimycin A (positive control) for 40 min prior to staining with MitoSOX Red and analysis by flow cytometry. (c) Induction of apoptosis by erlotinib in Calu-6 cells. Control and shLKB1 cells were treated with 25 μM erlotinib for 24 h and cell lysates were then subjected to Western blot analysis using antibody to cleaved PARP. (d) Apoptosis was detected with Annexin V staining in control and shLKB1 Calu-6 cells following 48 h treatment with erlotinib.
Erlotinib activates AMPKα and blocks mTOR signaling in LKB1-deficient cells
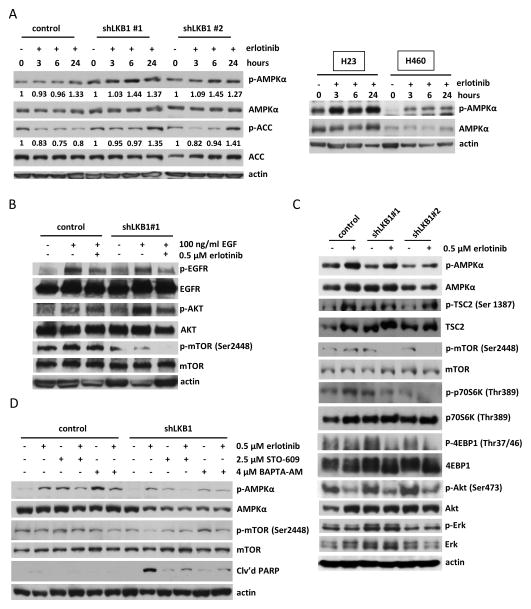
Next, we evaluated the effect of erlotinib on phosphorylation of AMPKα at 3, 6, and 24 h post-treatment. Interestingly, erlotinib induced phosphorylation of AMPKα after 6 h treatment in shLKB1 Calu-6 cells (Fig. 5A, left panel), with increased phosphorylation of acetyl-CoA carboxylase (ACC) apparent by 24 h. A similar increase in ACC phosphorylation was observed after 24 h treatment in shLKB1 H358 cells (Supplementary Fig. S3). This effect was not observed in the control cells. In non-transgenic LKB1 mutant NSCLC cells, erlotinib also induced phosphorylated AMPKα in both H23 and H460 cells after 3 h treatment (Fig. 5A, right panel). These results indicate that AMPKα remains sensitive to changes in ATP levels induced by erlotinib treatments in spite of reduced or abolished LKB1 activity. To confirm the effect of erlotinib to suppress EGFR activation and signaling through downstream pathways, we performed Western blot analysis of EGFR, Akt, and mTOR phosphorylation in the presence of exogenous EGF (Fig. 5B). EGF treatment stimulated phosphorylation of EGFR and Akt in both control and shLKB1 cells. EGFR phosphorylation was effectively reduced by erlotinib treatment in both cell types, but only shLKB1 Calu-6 cells exhibited reduced phosphorylation of mTOR in response to erlotinib. Therefore, we hypothesized that activation of AMPKα in LKB1-deficient cells might confer sensitivity to erlotinib through inhibition of mTOR signaling. As shown in Fig. 5C, erlotinib induced phosphorylation of AMPKα in both control and shLKB1 Calu-6 cells after 24 h, but shLKB1 cells exhibited complete disruption of mTOR pathway signaling following erlotinib treatment. In particular, AMPKα can directly phosphorylate TSC2 at Ser 1387, which is involved in inhibition of mTOR signaling. shLKB1 Calu-6 cells exhibited enhanced phosphorylation of TSC2 (Ser1387) by erlotinib treatment in comparison to control cells. Furthermore, erlotinib decreased phosphorylation of the 70-kDa ribosomal protein S6 kinase (p70S6K1) and 4EBP-1, both downstream effectors of mTOR signaling. Erlotinib treatment reduced phosphorylation of Akt in all cells, but phosphorylated Erk was not changed. These effects of erlotinib on AMPK and mTOR pathway signaling were further confirmed in H358, A549, and H460 cells. For example, shLKB1 H358 cells exhibited enhanced phosphorylation of AMPKα and inhibition of mTOR signaling in response to erlotinib treatment (Supplementary Fig. S4A), whereas LKB1 overexpressing A549 and H460 cells were less sensitive to mTOR inhibition by erlotinib in comparison to vector control cells (Supplementary Fig. S4B). These results indicate that erlotinib induces phosphorylation of AMPKα in both control and LKB1-deficient cells, but its effect to block mTOR pathway signaling is enhanced in LKB1-deficient cells, resulting in repression of protein synthesis and inhibition of cell growth.
Figure 5. Erlotinib-induced activation of AMPKα selectively inhibits mTOR signaling pathway in LKB1-deficient cells.
(a) Left panel: Erlotinib activated AMPKα and inhibited ACC in shLKB1 Calu-6 cells. Following treatment with 0.5 μM erlotinib for the indicated time, cell lysates were analyzed by Western blotting. The expression of phosphorylated AMPKα was calculated as the ratio of phosphorylated AMPKα to total AMPKα protein expression using densitometric analysis. The ratio of phosphorylated to total ACC was calculated similarly. All expression ratios were normalized to the untreated group. Right panel: Erlotinib also induced phosphorylated AMPKα in LKB1 mutant H23 and H460 cells. (b) Erlotinib inhibited EGF-stimulated EGFR phosphorylation and mTOR phosphorylation in shLKB1 Calu-6 cells. Cells were treated with 0.5 μM erlotinib for 72 h. After serum starvation for 24 h, cells were stimulated by 100 ng/ml EGF for 10 minutes and cell lysates were then analyzed by Western blotting. (c) Erlotinib blocked mTOR pathway signaling and activated AMPKα in shLKB1 Calu-6 cells. After 24 h of 0.5 μM erlotinib treatment, cell lysates were analyzed by Western blotting using specific antibodies for the indicated proteins. Blockade of mTOR activity following erlotinib treatment resulted in decreased phosphorylation of p70S6K and p-4EBP1 in shLKB1 Calu-6 cells. (d) Erlotinib-induced pAMPKα phosphorylation was reduced by addition of STO-609 (a CAMKKβ inhibitor) or BAPTA-AM (an intracellular calcium chelator) in shLKB1 Calu-6 cells. Cells were co-treated with 0.5 μM erlotinib and 2.5 μM STO-609 or 4 μM BAPTA-AM for 24 h and cell lysates were analyzed by Western blotting. The blots are representative of three independent experiments.
We then asked whether an alternative kinase other than LKB1 could be responsible for erlotinib-induced activation of AMPKα in LKB1-deficient cells. In a previous study, it was shown that AMPKα can become phosphorylated in response to energy stress, even in the absence of functional LKB1, through the action of calcium/calmodulin-dependent protein kinase kinase 2 (CaMKKβ) 33. To elucidate the mechanism by which erlotinib treatment can induce phosphorylation of AMPKα in LKB1-deficient cells, we used STO-609, a CaMKKβ inhibitor, and BAPTA-AM, an intracellular calcium chelator. After 24 h of treatment, both STO-609 and BAPTA-AM were able to partially inhibit erlotinib-induced AMPK phosphorylation in shLKB1 Calu-6 cells (Fig. 5D). This partially blocked erlotinib’s ability to inhibit mTOR and induce apoptosis in shLKB1 Calu-6 cells. These results indicate that AMPKα activation can be stimulated by erlotinib-induced changes in intracellular ADP/ATP levels and CAMKKβ activity, even in the absence of functional LKB1. This provides a mechanistic explanation for erlotinib’s ability to activate AMPKα and suppress mTOR signaling in LKB1-deficient cells.
Erlotinib reduces tumor volume in shLKB1 Calu-6 tumor xenografts
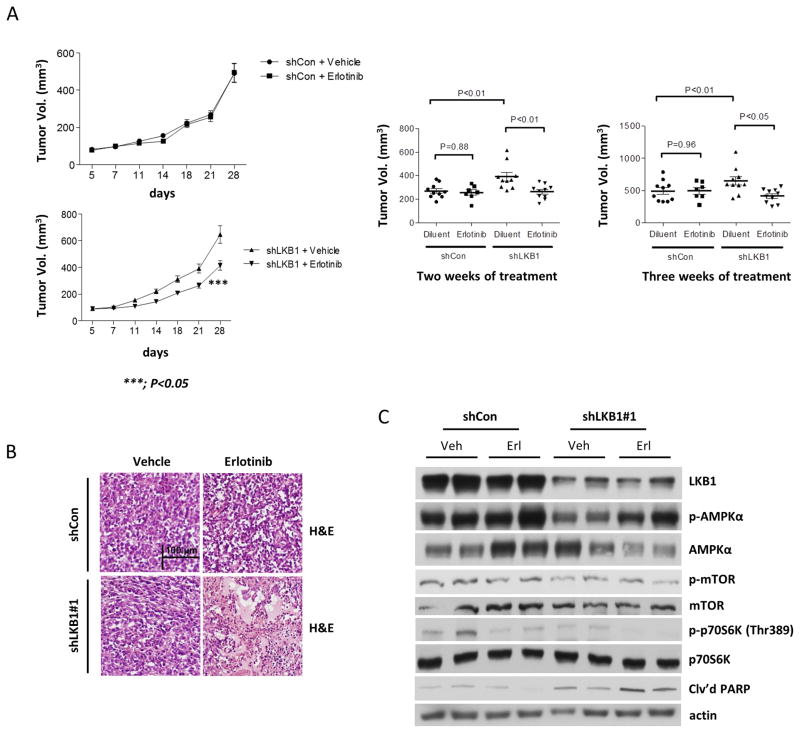
Finally, we investigated the in vivo antitumor activity of erlotinib in nude mice bearing control and shLKB1 Calu-6 subcutaneous tumors. As expected, mice bearing shLKB1 Calu-6 tumor xenografts showed the highest tumor volumes (Fig. 6A, left panel). Mean tumor volume in mice bearing shLKB1 tumors was significantly reduced at 2 and 3 weeks of erlotinib treatment, as compared with control-treated mice (Fig. 6A, right panel). No toxic effects (such as severe reduction in body weight) were observed in erlotinib-treated groups. We performed histological analyses of tumor samples by H&E immunohistochemical staining and found clear signs of large necrotic areas in shLKB1 tumor xenografts treated with erlotinib, but not in control xenografts (Fig. 6B). We also collected tissue samples from tumors to determine the in vivo effects of erlotinib treatment on activation of AMPKα, mTOR, p70S6K, and cleaved PARP (Fig. 6C). We first detected levels of LKB1 expression in control and shLKB1 Calu-6 tumor tissues from mice. As shown by Western blotting, levels of LKB1 protein were highly reduced in shLKB1 Calu-6 xenograft tumors as compared to control tumors. Consistent with in vitro results, erlotinib treatment enhanced AMPK phosphorylation, inhibited mTOR-related downstream signaling proteins, and increased PARP cleavage in shLKB1 Calu-6 xenograft tumors only. These results suggest that erlotinib treatment significantly reduced tumor growth in shLKB1 Calu-6 tumor xenografts through inhibition of mTOR signaling and induction of apoptosis.
Figure 6. Erlotinib induces a significant reduction of tumor volume in shLKB1 mice.
(a) Mean tumor volumes for each treatment group of the pLKO.1 control vector (dil, n=10, erlo, n=8) and shLKB1 vector (dil, n=10, erlo, n=10) mice after 1 week of treatment. Female nude mice were injected subcutaneously into the left/right flanks with 5 × 106 control or shLKB1 Calu-6 cells per mouse mixed 1:1 with 100% Matrigel. After 7 days, mice were treated with diluent (dil) or erlotinib (erlo) in sodium carboxymethylcellulose and Tween 80 solution o.p. at 100 mg/kg daily. Body weight was measured twice a week, and drug administration was adjusted accordingly. Tumors were measured twice a week with a caliper. Dots: individual tumor volumes (mm3). Horizontal lines: median and inter-quartile ranges. p-values are from linear regression analyses and Mann Whitney U test. (b) H&E immunohistochemical staining of sectioned tumor tissues. Original magnification, ×20. Scale bars, 100 μm. (c) Erlotinib inhibited mTOR signaling and enhanced cleaved PARP in shLKB1 Calu-6 tumor xenografts. The blots are representative of three independent experiments.
DISCUSSION
Although erlotinib exhibits potent anti-cancer activity against human NSCLC tumors with activating mutations in EGFR, it also has activity in some patients with wild-type EGFR. It has been unclear what molecular mechanisms drive this sensitivity and, furthermore, how to identify wild-type EGFR tumors that might be susceptible to erlotinib treatment. In the present study, we found that NSCLC cells harboring wild-type EGFR show varying sensitivity to erlotinib treatment that is dependent on LKB1 status. We demonstrated that erlotinib suppressed viability and enhanced apoptosis of LKB1-deficient NSCLC cells both in vitro and in vivo. Another study recently reported that glioblastoma cells with wild-type EGFR exhibited sensitivity to erlotinib treatment that was associated with inhibition of autophagy but was independent of caspase activity 34. Consistent with this report, our findings indicate that erlotinib can be beneficial even in tumors that are not EGFR mutated. However, we show that apoptosis is elevated in erlotinib-treated cells and that the inhibitory effects of erlotinib are enhanced in tumors that have dysregulated LKB1-AMPK pathway signaling. Because this signaling pathway normally serves to maintain mitochondrial function and bioenergetic homeostasis in response to metabolic stress, we hypothesized that LKB1-deficient cells may be compromised in their ability to cope with metabolic alterations induced by erlotinib treatment.
To further examine the mechanism of erlotinib-induced apoptosis in LKB1-deficient NSCLC cells, we analyzed changes in glycolytic metabolism and mitochondrial function resulting from LKB1 knockdown. Interestingly, we observed that glucose consumption and lactate production by Calu-6 cells increased by more than 50% in response to LKB1 knockdown, which correlated with increased growth and colony formation (Fig. 2C). We also observed substantial increases in OCR (Fig. 3B) and Δψm (Fig. 4A) after expression of shLKB1 in Calu-6 and H358 cells, consistent with another recent study that profiled gene expression of over 600 patient tumors and reported upregulation of OXPHOS and mitochondria-associated genes in samples classified as LKB1-deficient 23. The effect of LKB1 loss to activate glycolysis and oxidative metabolism was further confirmed by expressing wild-type LKB1 in mutant A549 and H460 cell lines (Figs. 2A, 2B, Supplementary Figs. S1, S2). Faubert et al. 32 previously reported simultaneous increases in both glucose uptake and lactate production in response to complete LKB1 knockout in transgenic mouse embryonic fibroblasts (MEFs). They also noted enhanced flux of both glucose and glutamine into the TCA cycle of LKB1 mutant A549 cells in comparison to cells expressing wild-type LKB1, based on isotope labeling experiments with 13C-glucose and 13C-glutamine tracers. These previous findings are consistent with our own results indicating increased dependence of LKB1-deficient cells on mitochondrial metabolism to support enhanced cell proliferation.
Despite elevated rates of oxidative metabolism, we observed only slight changes in basal ADP/ATP ratios and ROS levels in LKB1 knockdown cells (Fig. 3C and 4B). However, the effect of LKB1 depletion to alter mitochondrial function became readily apparent when shLKB1 cells were challenged with erlotinib. Substantial increases in ADP/ATP and ROS levels were observed, which were accompanied by depletion of Δψm (Fig. 4A). In contrast, control cells were able to maintain normal ADP/ATP, ROS, and Δψm levels in response to the same concentration of erlotinib (Fig. 3C, 4A, 4B). Taken together, these data provide a clear indication that LKB1 is required for maintenance of mitochondrial function in the presence of erlotinib treatment. As a result, LKB1-deficient cells cannot effectively restore ATP levels or prevent ROS accumulation in response to erlotinib, resulting in selective growth inhibition (Fig. 2) and apoptosis activation (Fig. 4C and 4D). In this context, we view the decoupling of ROS from OCR and Δψm measurements as an indicator of mitochondrial dysfunction following erlotinib treatment. These results are similar to those reported by Shackelford et al. 12, where significant increases in ROS and decreases in Δψm were simultaneously observed in response to phenformin treatment of LKB1-mutant A549 cells. Our results are also consistent with another previous study that showed AMPK-deficient cells are unable to maintain sufficient NADPH levels needed to neutralize mitochondrial ROS during metabolic stress 35, a mechanism which may further contribute to the enhanced erlotinib sensitivity we observed in LKB1-deficient cells.
Despite the previously described similarities, our observations of increased basal mitochondrial activity in LKB1-deficient cells are also partially at odds with the results of Shackelford et al. 12. Their study showed that re-expressing wild-type LKB1 in A549 cells led to enhancements of basal OCR and Δψm, implying reduced mitochondrial activity in the parental, LKB1-deficient cells 12. In contrast, a more recent study by Faubert et al. 32 showed no change in OCR after re-expressing LKB1 in A549 cells and a reduction in OCR after re-expressing LKB1 in A427 cells, which are also LKB1 mutant-type. Therefore, there is some variability in the prior literature concerning the effects that manipulating LKB1 expression has on markers of mitochondrial activity. It is possible that these differences could be attributable to adaptation of LKB1 mutant cells to the re-expression of LKB1 over many generations. Since our studies were completed within ten passages following antibiotic selection, our results likely capture the immediate response to LKB1 re-expression in the absence of long-term adaptation.
Another unexpected outcome of our study was that LKB1-deficient cells still exhibited a robust ability to activate AMPK in response to increases in ADP/ATP ratio following erlotinib treatment. AMPK activation by adenine nucleotides involves at least three independent mechanisms: (1) increased AMP-dependent phosphorylation by LKB1, (2) inhibitory effects of AMP on its dephosphorylation, and (3) allosteric activation by AMP and ADP 36. The latter two mechanisms do not require LKB1. We also found that the ability of erlotinib to induce phosphorylation of AMPKα in LKB1-deficient cells was partially inhibited by co-treatment with either STO-609 or BAPTA-AM (Fig. 5D), indicating involvement of CaMKKβ as an alternative kinase that contributes to erlotinib-induced AMPK phosphorylation. Two prior articles have reported that AMPK phosphorylation by CaMKKβ is dependent on AMP and ADP levels 37, 38. The large increase in ADP/ATP ratio we observed following erlotinib treatment could therefore be sufficient to activate AMPK even when LKB1 was depleted, possibly by a CaMKKβ-dependent mechanism. We cannot rule out, however, that changes in Ca2+ levels may synergize with changes in energy charge to stimulate AMPK activation in response to erlotinib treatment. Consistent with this hypothesis, gefitinib has been reported to increase cytosolic and mitochondrial calcium levels of Bcl-2-overexpressing cells 39.
Erlotinib-mediated AMPKα activation led to inhibition of mTOR phosphorylation in LKB1-deficient cells, which correlated with inhibition of phosphorylated p70S6K and 4EBP-1 (Fig. 5C) as major downstream effectors of the mTOR signaling pathway. These data are consistent with the known role of AMPK to suppress mTOR signaling in response to low ATP levels, leading to inhibition of protein synthesis and proliferation of cancer cells 10, 40. Previous studies suggested that loss of LKB1 induced increased cell size and mass due to failure of AMPK to inhibit mTOR signaling (31, 32). In contrast, we found that only LKB1-deficient cells effectively suppressed mTOR signaling and induced PARP cleavage in response to erlotinib treatments, both in vitro and in vivo. This led to a significant decrease in growth of LKB1-deficient xenograft tumors. It is likely that mitochondrial dysfunction associated with LKB1 deficiency sensitizes cells to erlotinib treatment by enhancing changes in ADP/ATP and ROS levels, which subsequently activate AMPK and suppress mTOR signaling.
Inhibition of Erk signaling has also been shown to induce pyruvate dehydrogenase kinase 4 (PDK4) expression and redirect cellular metabolism in mammary epithilia 41. However, we do not expect that changes in Mek/Erk signaling or PDK4 expression were directly involved in regulating the metabolic response to erlotinib treatment in our studies. First, no significant change in Erk phosphorylation was observed in response to erlotinib treatment of Calu-6 cells (Fig. 5C, Supplementary Fig. S4). Second, if PDK4 expression was induced by erlotinib treatment, we would expect to see increased lactate production due to inhibition of pyruvate dehydrogenase flux. However, this was not observed in any of the cell lines tested. Finally, to confirm these observations, we performed Western blots with an anti-PDK4 antibody in LKB1 overexpressing and stable knockdown cell lines (Supplementary Fig S5). No significant changes in PDK4 expression were observed after 24 hours of erlotinib treatment. Taken together, we conclude that erlotinib treatment reduces OCR in LKB1-deficient cells independently of Erk and PDK4.
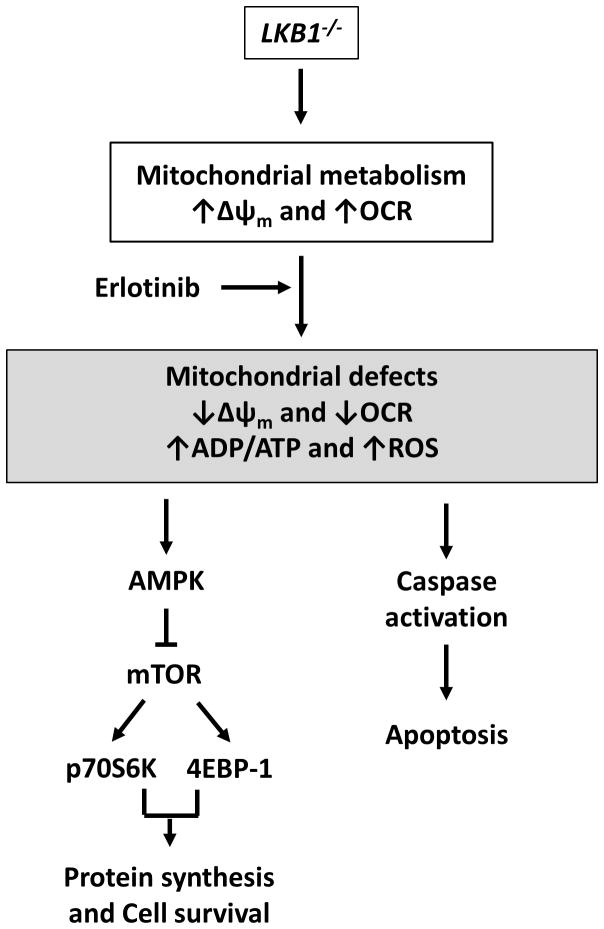
In conclusion, we demonstrate for the first time that erlotinib induces apoptosis in EGFR wild-type NSCLC cells by a mechanism that is enhanced by LKB1 loss of function (Fig. 7). Although LKB1-deficient cells relied more heavily on mitochondrial OXPHOS for energy production under basal conditions, they could not properly respond to metabolic stress induced by erlotinib treatment in order to maintain energetic homeostasis. These metabolic alterations were associated with more pronounced AMPK activation and inhibition of mTOR signaling in LKB1-deficient cells, resulting in increased sensitivity to erlotinib treatment. Taken together, our findings provide a rationale for erlotinib treatment of LKB1-deficient NSCLC tumors, even in the absence of EGFR activating mutations. These tumors may constitute a distinct NSCLC molecular subtype with enhanced erlotinib sensitivity, which motivates the development of novel diagnostic approaches (e.g., mRNA expression profiling 23) capable of identifying LKB1-deficient patient tumors that are potentially susceptible to erlotinib treatment.
Figure 7. Hypothetical schema of erlotinib-induced cell death in LKB1-deficient cells.
LKB1-deficient cells exhibit enhanced mitochondrial metabolism as indicated by increased mitochondrial potential and OCR. Erlotinib treatment selectively induces mitochondrial dysfunction in LKB1-deficient cells, decreasing mitochondrial potential and OCR while increasing ADP/ATP and ROS. This has the dual effect of inhibiting growth (via activation of AMPK and suppression of mTOR signaling) and activating apoptosis. This mechanism is based on composite data from our in vitro studies on multiple LKB1 wild-type (Calu-6, H358) and LKB1 mutant (A549, H460) cell lines, as well as in vivo data from Calu-6 mouse xenografts.
MATERIALS AND METHODS
Cell culture and reagents
Human NSCLC Calu-6, H358, H2009, H460, H23, H2122, HCC15, and A549 cell lines were generously shared with us by John Minna and Luc Girard (University of Texas, Southwestern). Cells were tested to ensure that they were mycoplasma negative. The human NSCLC cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS; GIBCO-BRL, Gaithersburg, MD) in a humidified atmosphere with 5% CO2. A549 and H460 cells stably expressing either an empty pBABE vector (vector) or wild-type LKB1 (LKB1) were generated by retroviral transduction. The empty pBABE viral plasmid and pBABE-LKB1 plasmid were obtained from Addgene (Cambridge, MA). Phoenix cells were transfected with viral plasmids and retroviral particles were harvested from media supernatant 48 hours after transfection. Viruses were added to target cells with polybrene, and selection with 1μg/ml puromycin was begun 48–72 hours after infection. Cells were selected under puromycin for one to two weeks before performing subsequent experiments, with experiments being completed within two months. Erlotinib (an inhibitor of EGFR), LY294002 (an inhibitor of PI3K), and rapamycin (an inhibitor of mTOR) were purchased from LC Laboratories (Woburn, MA).
Cell viability analysis, colony formation assays, and growth rate measurements
Cells were plated in 96-well plates in complete medium and treated with various concentrations of erlotinib, LY294002, or rapamycin. Cell viability was analyzed after 2 days of treatment using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay according to the manufacturer’s instructions (Sigma, St. Louis, MO). Cell proliferation was analyzed every 8 or 16 h for 3 days after erlotinib treatment using the CyQUANT® Direct Cell Proliferation Assay according to the manufacturer’s protocol (Invitrogen, Frederick, MD). The fluorescence intensity was measured with a fluorescence microplate reader (Bio-Tek Instruments, Winooski, VT). Conditioned medium samples were collected from each well prior to CyQUANT analysis, frozen, and stored for metabolite analysis. Colony formation assays were performed following treatment with 5 μM erlotinib for 3 days, at which time the cells were replated in six-well plates at low density (1 × 103 cells per well) in complete medium for 2 weeks. Colonies were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich, St. Louis, MO). Colony numbers were assessed visually and colonies measuring at least 50 μm were counted.
Metabolite analysis and oxygen consumption rate
Conditioned medium samples collected during cell growth experiments were subjected to glucose and lactate analysis using a YSI 2300 biochemical analyzer (YSI Life Science, Yellow Springs, OH). The ETA software package was applied to calculate specific rates of glucose consumption and lactate production using an exponential growth model 42. Oxygen consumption rate (OCR) was measured using the Oxygraph-2K (Oroboros, Innsbruck, Austria), which contains two chambers with separate oxygen probes to monitor on-line changes in oxygen concentration. The instrument was set to a temperature of 37 °C, and the stirring speed for each chamber was 750 rpm. After treatment with erlotinib for 24 h, cells were then trypsinized, resuspended in the same culture medium at a concentration of 1–2 × 106 cells per mL, and injected into the Oxygraph instrument.
LKB1 stable knockdown using lentiviral short-hairpin RNA
For generation of stable knockdown cells, lentiviral LKB1 short-hairpin RNA (shRNA) constructs and a non-targeting shRNA control were purchased from Open Biosystems (Huntsville, AL). These constructs were cloned into a pLKO.1 vector. HEK293T cells (2 × 106 cells) were plated on 100 mm dishes and one day later were co-transfected with 5 μg shRNA constructs, 3 μg pCMV-dR8.2 dvpr (Addgene) and 2 μg pCMV-VSV-G (Addgene) using a calcium phosphate transfection kit (Invitrogen). After 6 h, the medium was exchanged with fresh medium, and then the viral stock was collected for 1–2 days and filtered to remove non-adherent HEK293T cells. To obtain stable shRNA expressing clones, Calu-6 or H358 cells were plated on six-well plates, and one day later infected with 1 ml of lentiviral suspension plus 4 μg/ml polybrene. Puromycin selection (0.5–2 μg/ml) was initiated 2 days after lentiviral infection.
Western blotting
Total cell lysates were resolved using sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, and then transferred onto polyvinylidene fluoride (PVDF) membranes (EMD Millipore Corporation, Billerica, MA). Western blotting was performed as described previously 43. The following antibodies were purchased from Cell Signaling (Danvers, MA): rabbit polyclonal antibodies against LKB1, phosphorylated AMPKα, phosphorylated acetyl-CoA carboxylase (ACC), phosphorylated Tuberin/TSC2 (Ser1387), phosphorylated mTOR (Ser2448), phosphorylated 4EBP1 (Thr37/46), phosphorylated Akt (Ser473), phosphorylated Erk (Thr202/Tyr204), poly(ADP-ribose) polymerase (PARP), and mouse monoclonal antibodies against phosphorylated p70S6K (Thr389). A goat polyclonal anti-actin antibody was purchased from Santa Cruz (Santa Cruz, CA). All experiments were performed in triplicate.
Flow cytometry
Cells were seeded in six-well plates containing complete medium and treated with 5 μM or 10 μM erlotinib. After 72 h, cells were harvested, pooled, and fixed with 1% paraformaldehyde and 70% ethanol. Apoptosis was assessed with flow cytometry using the Annexin V-FITC Apoptosis kit (Clontech, Mountain View, CA). To measure mitochondrial ROS after 24 h of erlotinib treatment, cells were stained with MitoSOX Red (Molecular Probes, Eugene, OR) for 40 min at 37 °C in the dark. Annexin V and MitoSOX Red were analyzed using a LSRII flow cytometer (Becton Dickinson, San Jose, CA).
Assessment of ADP/ATP ratio and mitochondrial membrane potential
Cells were seeded in 96-well plates and treated with 25 μM erlotinib for 24 h. Intracellular ADP/ATP ratio was measured using a bioluminescent ADP/ATP Ratio Assay Kit according to the manufacturer’s instructions (Abcam, Cambridge, MA). Mitochondrial membrane depolarization was determined using the JC-1 fluorescence probe according to the manufacturer’s instructions (Molecular Probes). Cells were labeled with 2 μM JC-1 for 30 min at 37 °C in the dark and then analyzed at 488 nm excitation with 530/30 or 585/42 nm bypass emission filters using a fluorescence microplate reader (Bio-TeK Instruments).
Animal experiments
The research protocol for animal studies was approved by and carried out in accordance with the Vanderbilt Institutional Animal Care & Use Committee. Four- to five-week old female Foxn1nu/nu nude mice were purchased from Harlan Laboratories (Indianapolis, IN). Both non-targeting shRNA (shCon, LKB1 wild-type) and LKB1 shRNA (shLKB1, LKB1 knockdown) Calu-6 cells were injected s.c. into the left /right flanks at 5 × 106 cells per mouse mixed 1:1 with 100% growth factor-reduced Matrigel (BD Bioscience, San Jose, CA). The mice were randomly assigned to shCon + vehicle solution (in sodium carboxymethylcellulose and Tween 80), shCon + erlotinib, shLKB1+ vehicle solution, and shLKB1 + erlotinib, with each group containing eight to ten mice. After 1 week, mice were treated with vehicle or erlotinib (100 mg/kg) daily by oral gavage for 3 weeks. Tumor size was calculated by measuring the tumors in three dimensions using calipers twice a week 44, while body weight was measured twice weekly. On day 21, mice were euthanized and tumor tissues were harvested for pathologic examination (immunohistochemical analysis) and analyzed by Western blotting.
Statistical analyses
The data acquired with MTT, colony formation, and ADP/ATP assays were analyzed using the Student’s t-test. The Mann-Whitney U test was used to compare differences in tumor weight. In all of the statistical analyses, two-sided p-values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by NIH P50 CA090949, DOD W81XWH-12-1-0383, NSF CBET-1105991, and an award from Uniting Against Lung Cancer. Dr. Joshua P. Fessel was also supported by NIH K08 HL121174. The authors thank Dr. Changki Lee for his assistance with the animal experiments.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA: A Cancer Journal for Clinicians. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Rapp E, Pater JL, Willan A, Cormier Y, Murray N, Evans WK, et al. Chemotherapy can prolong survival in patients with advanced non-small-cell lung cancer--report of a Canadian multicenter randomized trial. 1988. [DOI] [PubMed] [Google Scholar]
- 3.West L, Vidwans SJ, Campbell NP, Shrager J, Simon GR, Bueno R, et al. A novel classification of lung cancer into molecular subtypes. PLoS One. 2012;7:e31906. doi: 10.1371/journal.pone.0031906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. The New England journal of medicine. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. The New England journal of medicine. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 6.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, et al. Inactivation of LKB1/STK11 Is a Common Event in Adenocarcinomas of the Lung. 2002. [PubMed] [Google Scholar]
- 8.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 9.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carretero J, Medina PP, Blanco R, Smit L, Tang M, Roncador G, et al. Dysfunctional AMPK activity, signalling through mTOR and survival in response to energetic stress in LKB1-deficient lung cancer. Oncogene. 2007;26:1616–1625. doi: 10.1038/sj.onc.1209951. [DOI] [PubMed] [Google Scholar]
- 12.Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23:143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faber AC, Li D, Song Y, Liang M-C, Yeap BY, Bronson RT, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proceedings of the National Academy of Sciences. 2009;106:19503–19508. doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 16.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, et al. Erlotinib in Previously Treated Non–Small-Cell Lung Cancer. New England Journal of Medicine. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 17.Cappuzzo F, Ciuleanu T, Stelmakh L, Cicenas S, Szczésna A, Juhász E, et al. Erlotinib as maintenance treatment in advanced non-small-cell lung cancer: a multicentre, randomised, placebo-controlled phase 3 study. The Lancet Oncology. 2010;11:521–529. doi: 10.1016/S1470-2045(10)70112-1. [DOI] [PubMed] [Google Scholar]
- 18.Garassino MC, Marsoni S, Floriani I. Testing Epidermal Growth Factor Receptor Mutations in Patients With Non–Small-Cell Lung Cancer to Choose Chemotherapy: The Other Side of the Coin. 2011. [DOI] [PubMed] [Google Scholar]
- 19.Deng J, Shimamura T, Perera S, Carlson NE, Cai D, Shapiro GI, et al. Proapoptotic BH3-Only BCL-2 Family Protein BIM Connects Death Signaling from Epidermal Growth Factor Receptor Inhibition to the Mitochondrion. 2007. [DOI] [PubMed] [Google Scholar]
- 20.Ling Y-H, Lin R, Perez-Soler R. Erlotinib Induces Mitochondrial-Mediated Apoptosis in Human H3255 Non-Small-Cell Lung Cancer Cells with Epidermal Growth Factor ReceptorL858R Mutation through Mitochondrial Oxidative Phosphorylation-Dependent Activation of BAX and BAK. 2008. [DOI] [PubMed] [Google Scholar]
- 21.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumoto S, Iwakawa R, Takahashi K, Kohno T, Nakanishi Y, Matsuno Y, et al. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene. 2007;26:5911–5918. doi: 10.1038/sj.onc.1210418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufman JM, Amann JM, Park K, Arasada RR, Li H, Shyr Y, et al. LKB1 Loss Induces Characteristic Patterns of Gene Expression in Human Tumors Associated with NRF2 Activation and Attenuation of PI3K-AKT. Journal of Thoracic Oncology. 2014:9. doi: 10.1097/JTO.0000000000000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Schaeybroeck S, Kyula J, Kelly DM, Karaiskou-McCaul A, Stokesberry SA, Van Cutsem E, et al. Chemotherapy-induced epidermal growth factor receptor activation determines response to combined gefitinib/chemotherapy treatment in non–small cell lung cancer cells. Molecular Cancer Therapeutics. 2006;5:1154–1165. doi: 10.1158/1535-7163.MCT-05-0446. [DOI] [PubMed] [Google Scholar]
- 25.Tracy S, Mukohara T, Hansen M, Meyerson M, Johnson BE, Jänne PA. Gefitinib Induces Apoptosis in the EGFRL858R Non–Small-Cell Lung Cancer Cell Line H3255. Cancer Research. 2004;64:7241–7244. doi: 10.1158/0008-5472.CAN-04-1905. [DOI] [PubMed] [Google Scholar]
- 26.Onozato R, Kosaka T, Achiwa H, Kuwano H, Takahashi T, Yatabe Y, et al. LKB1 gene mutations in Japanese lung cancer patients. Cancer Science. 2007;98:1747–1751. doi: 10.1111/j.1349-7006.2007.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Marks K, Cowley GS, Carretero J, Liu Q, Nieland TJF, et al. Metabolic and Functional Genomic Studies Identify Deoxythymidylate Kinase as a Target in LKB1-Mutant Lung Cancer. Cancer Discovery. 2013;3:870–879. doi: 10.1158/2159-8290.CD-13-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahoney CL, Choudhury B, Davies H, Edkins S, Greenman C, Haaften G, et al. LKB1/KRAS mutant lung cancers constitute a genetic subset of NSCLC with increased sensitivity to MAPK and mTOR signalling inhibition. Br J Cancer. 2009;100:370–375. doi: 10.1038/sj.bjc.6604886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LGD, Neumann D, et al. LKB1 Is the Upstream Kinase in the AMP-Activated Protein Kinase Cascade. Current Biology. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 30.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MS, PG, BH, MS, DC . Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faubert B, Vincent EE, Griss T, Samborska B, Izreig S, Svensson RU, et al. Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1alpha. Proc Natl Acad Sci U S A. 2014;111:2554–2559. doi: 10.1073/pnas.1312570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA. The Ca2+/Calmodulin-dependent Protein Kinase Kinases Are AMP-activated Protein Kinase Kinases. Journal of Biological Chemistry. 2005;280:29060–29066. doi: 10.1074/jbc.M503824200. [DOI] [PubMed] [Google Scholar]
- 34.Eimer S, Belaud-Rotureau M-A, Airiau K, Jeanneteau M, Laharanne E, Véron N, et al. Autophagy inhibition cooperates with erlotinib to induce glioblastoma cell death. Cancer Biology & Therapy. 2011;11:1017–1027. doi: 10.4161/cbt.11.12.15693. [DOI] [PubMed] [Google Scholar]
- 35.Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gowans GJ, Hawley SA, Ross FA, Hardie DG. AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metab. 2013;18:556–566. doi: 10.1016/j.cmet.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oakhill JS, Steel R, Chen ZP, Scott JW, Ling N, Tam S, et al. AMPK is a direct adenylate charge-regulated protein kinase. Science. 2011;332:1433–1435. doi: 10.1126/science.1200094. [DOI] [PubMed] [Google Scholar]
- 38.Oakhill JS, Chen ZP, Scott JW, Steel R, Castelli LA, Ling N, et al. beta-Subunit myristoylation is the gatekeeper for initiating metabolic stress sensing by AMP-activated protein kinase (AMPK) Proc Natl Acad Sci U S A. 2010;107:19237–19241. doi: 10.1073/pnas.1009705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zannetti A, Iommelli F, Fonti R, Papaccioli A, Sommella J, Lettieri A, et al. Gefitinib Induction of In vivo Detectable Signals by Bcl-2/Bcl-xL Modulation of Inositol Trisphosphate Receptor Type 3. Clinical Cancer Research. 2008;14:5209–5219. doi: 10.1158/1078-0432.CCR-08-0374. [DOI] [PubMed] [Google Scholar]
- 40.Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 41.Grassian AR, Metallo CM, Coloff JL, Stephanopoulos G, Brugge JS. Erk regulation of pyruvate dehydrogenase flux through PDK4 modulates cell proliferation. Genes Dev. 2011;25:1716–1733. doi: 10.1101/gad.16771811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy TA, Young JD. ETA: Robust software for determination of cell specific rates from extracellular time courses. Biotechnology and Bioengineering. 2013;110:1748–1758. doi: 10.1002/bit.24836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whang YM, Kim YH, Kim JS, Yoo YD. RASSF1A Suppresses the c-Jun-NH2-Kinase Pathway and Inhibits Cell Cycle Progression. 2005. [DOI] [PubMed] [Google Scholar]
- 44.Park SI, Lee C, Sadler WD, Koh AJ, Jones J, Seo JW, et al. Parathyroid Hormone–Related Protein Drives a CD11b+Gr1+ Cell–Mediated Positive Feedback Loop to Support Prostate Cancer Growth. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.