Abstract
Atypical chemokine receptor CXCR7 (ACKR3) functions as a scavenger receptor for chemokine CXCL12, a molecule that promotes multiple steps in tumor growth and metastasis in breast cancer and multiple other malignancies. While normal vascular endothelium expresses low levels of CXCR7, marked upregulation of CXCR7 occurs in tumor vasculature in breast cancer and other tumors. To investigate effects of endothelial CXCR7 in breast cancer, we conditionally deleted this receptor from vascular endothelium of adult mice, generating CXCR7ΔEND/ΔEND animals. CXCR7ΔEND/ΔEND mice appeared phenotypically normal, although these animals exhibited a modest 35 ± 3% increase in plasma CXCL12 as compared with control. Using two different syngeneic, orthotopic tumor implant models of breast cancer, we discovered that CXCR7ΔEND/ΔEND mice had significantly greater local recurrence of cancer following resection, elevated numbers of circulating tumor cells, and more spontaneous metastases. CXCR7ΔEND/ΔEND mice also showed greater experimental metastases following intracardiac injection of cancer cells. These results establish that endothelial CXCR7 limits breast cancer metastasis at multiple steps in the metastatic cascade, advancing understanding of CXCL12 pathways in tumor environments and informing ongoing drug development targeting CXCR7 in cancer.
Keywords: chemokine, bioluminescence, imaging, animal model
Introduction
Chemokines, a family of peptides that regulate physiologic trafficking of cells, have emerged as key regulators of metastasis (1, 2). Compared with normal tissue, malignant and/or stromal cells upregulate multiple chemokines in primary and metastatic tumors. Chemokine CXCL12 is one of the most prominent regulators of tumor growth and metastasis in breast cancer and greater than 20 other human cancers (3). CXCL12 secreted by carcinoma-associated fibroblasts in primary breast cancers increases proliferation and invasion of malignant cells and recruits endothelial progenitor cells for tumor angiogenesis (4). These processes collectively contribute to vascular intravasation of cancer cells. CXCL12 in primary breast cancers selects for malignant cells that can home, survive, and proliferate in CXCL12-rich metastatic sites such as bone marrow, lung, and brain (5, 6). CXCL12 also confers drug resistance in multiple cancers, limiting eradication of cancer cells and further reducing survival (3, 7, 8).
CXCL12 binds to chemokine receptors CXCR4 and CXCR7 (recently designated ACKR3), both of which promote progression of cancer to metastatic disease. While CXCR4 typically correlates with greater metastases and reduced survival in cancers including breast, lung, and prostate, outcomes associated with CXCR7 have been less consistently associated with poor prognosis (9–11). Expression of CXCR7 in cancer cells promotes tumor growth and metastasis in mouse models of breast and prostate cancer (12, 13). In patients, increased levels of CXCR7 correlate with worse survival in malignancies of the gallbladder and kidney (14, 15). However, CXCR7 did not affect outcomes in a study of patients with esophageal cancer, and CXCR7 corresponded with better survival in neuroblastoma and rectal cancer (16–18). These discrepancies among effects of CXCR7 on survival suggest context-dependent effects of this receptor on cancer progression, including specific types of cells that express CXCR7.
In addition to malignant cells, CXCR7 is expressed on vascular endothelium under normal and pathologic conditions. CXCR7 critically regulates development of the cardiovascular system in two different animal models. Knocking down CXCR7 in zebrafish embryos prevents formation of intersegmental blood vessels, and mice lacking CXCR7 in all tissues frequently suffer early post-natal mortality due to myocardial degeneration and/or heart valve defects (12) (19, 20). CXCR7 is expressed highly in vasculature of human cancers, including breast, glioma, and renal (12, 21, 22). However, effects of tumor endothelial CXCR7 on tumor progression remain unknown.
We investigated effects of CXCR7 on tumor vasculature in breast cancer, a disease in which this receptor is expressed highly on vascular endothelium in mouse models and primary human breast tumors (12). Adult mice lacking endothelial CXCR7 appear phenotypically normal, although they have elevated levels of plasma CXCL12. Using two different syngeneic breast cancer cell lines, we discovered that mice lacking CXCR7 on vascular endothelium have greater spontaneous metastases from orthotopic tumor implants and experimentally-induced breast cancer metastases. Collectively, these results reveal that CXCR7 on vascular endothelium suppresses progression of breast cancer to metastatic disease.
Results
Deletion of endothelial CXCR7 elevates systemic levels of CXCL12
Since embryonic deletion of CXCR7 from vascular endothelium may cause early post-natal death, we deleted CXCR7 conditionally from endothelium of adult mice using the 5′ endothelial enhancer element from the stem cell leukemia (Scl) locus to drive tamoxifen-inducible Cre (CreERT). A previous study established this transgene drives expression of CreERT selectively in vascular endothelium (23). Treatment with tamoxifen deleted CXCR7 as determined by PCR of tail samples, a well-vascularized tissue used previously to screen for Cre-mediated excision of genes from vascular endothelium (24, 25) (Fig S1A). These mice also had reduced expression of CXCR7 on endothelium of the liver, an organ with low level expression of this receptor on blood vessels sinusoidal under basal conditions (26) (Fig S1B). We used this tamoxifen protocol to delete CXCR7 from vascular endothelium prior to all tumor studies, generating CXCR7ΔEND/ΔEND mice. Adult CXCR7ΔEND/ΔEND mice appeared phenotypically normal.
CXCR7 is a scavenger receptor for CXCL12, removing this chemokine from the extracellular space and degrading it (27, 28). Deleting endothelial CXCR7 significantly elevated basal levels of CXCL12 in plasma (p < 0.05) (Fig S2). We could not assess effects of CXCR7 on plasma levels of its second chemokine ligand, CXCL11, because a genetic mutation eliminates expression of this gene in C57BL/6 mice (19). Overall, these data show that while endothelial CXCR7 is not essential for survival of adult animals, this receptor does regulate circulating levels of CXCL12.
CXCR7ΔEND/ΔEND mice have greater spontaneous metastases from orthotopic breast tumors
We previously identified CXCR7 on tumor blood vessels in mouse xenograft models of breast cancer and primary human breast tumors, while this receptor was absent from vasculature in normal breast tissue (12). To investigate effects of endothelial CXCR7 on growth and metastasis in breast cancer, we used AT-3 breast cancer cells, a C57BL/6 mouse breast cancer cell line that expresses modest levels of cell surface CXCR4 and low amounts of CXCR7 (Fig S3). We implanted AT-3 cells expressing firefly luciferase (AT-3-FL) orthotopically into 4th inguinal mammary fat pads of control and CXCR7ΔEND/ΔEND mice. We also implanted mouse mammary fibroblasts stably expressing CXCL12-α to reproduce secretion of this chemokine by stromal cells in primary human breast cancer (4).
Bioluminescence imaging showed significantly greater growth of AT-3-FL breast cancer cells in CXCR7ΔEND/ΔEND mice on days 7–16 after implantation (p < 0.05) (Fig 1A). We resected orthotopic tumors ~2 weeks after implantation to simulate treatment of patients with breast cancer and allow time for disseminated tumor cells to form detectable metastases. Resected tumors from control and CXCR7ΔEND/ΔEND mice appeared similar by standard H&E staining and did not differ in relative vascularity assessed by CD31 (Fig 1B). Tumor blood vessels in control mice demonstrated extensive co-localization of CD31 with CXCR7, consistent with prior studies showing increased expression of CXCR7 on that vascular endothelium in breast cancer and other malignancies (Fig 1B) (12). By comparison, endothelial expression of CXCR7 and co-localization with CD31 were markedly diminished in tumors from CXCR7ΔEND/ΔEND mice. Immunofluorescence also confirmed expression of CXCR7 in AT-3-FL cells in the tumor.
Figure 1. Orthotopic AT-3-FL tumor implants in CXCR7ΔEND/ΔEND mice have more viable tumor cells.
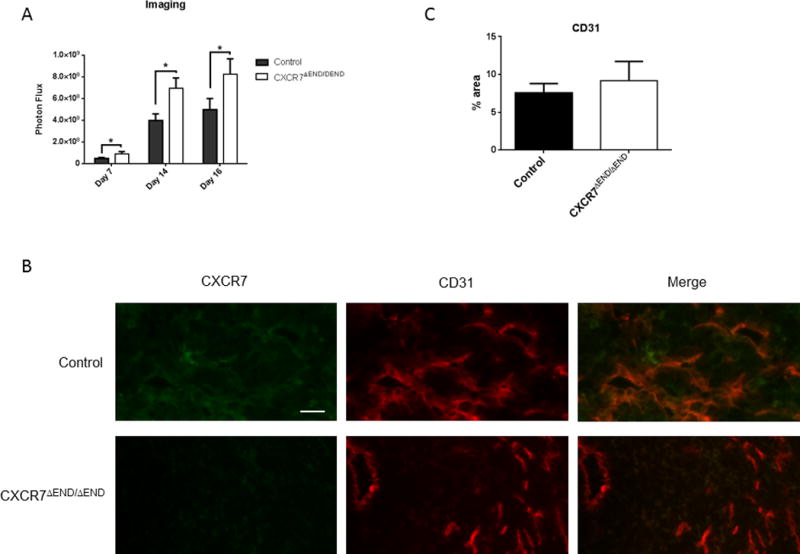
A) Growth of AT-3-FL breast cancer cells implanted orthotopically into CXCR7ΔEND/ΔEND and control mice monitored by bioluminescence imaging at indicated days (n = 8 mice per group). Graph depicts mean values + SEM for photon flux. *, p < 0.05. B) Immunofluorescence of excised orthotopic AT-3-FL tumors for CD31-tumor vasculature (green) and CXCR7 (red). White arrows show co-localization of CD31 and CXCR7 in merged images. AT-3-FL cells express CXCR7, accounting for staining throughout both tumors. Scale bar designates 50 μm. C. Graph shows mean values + SEM for area occupied by CD31+ blood vessels in tumors from each group of mice.
After resecting orthotopic tumors, we monitored mice for three more weeks for local recurrence and metastases before euthanizing mice on the same day. CXCR7ΔEND/ΔEND mice had significantly larger recurrent tumors in the mammary fat pad and greater overall systemic metastases quantified by bioluminescence (Fig 2A–C). When we analyzed by distribution of metastases in various anatomic sites, CXCR7ΔEND/ΔEND mice had greater metastatic AT-3-FL cells in all locations, although differences between groups of mice only were significant for the total abdomen (Fig 2D). Fluorescence microscopy of lungs ex vivo showed small clusters of metastatic AT-3-FL cells in CXCR7ΔEND/ΔEND mice, while control mice had frequent solitary metastatic breast cancer cells as well as cell clusters (Fig S4). CXCR7ΔEND/ΔEND mice also had more viable, circulating AT-3-FL cells than control animals (Fig S5). Since bone and bone marrow represent the most common sites of disseminated tumor cells and breast cancer metastases (29), we also recovered bone marrow from lower extremities of mice. We categorized mice as positive for tumor cells in bone marrow based on ex vivo bioluminescence above background levels in five independent experiments. Greater than 30% of CXCR7ΔEND/ΔEND mice had viable disseminated AT-3-FL breast cancer cells in bone marrow as compared with less than 15% of controls (Fig 2E). Overall, these data demonstrate that loss of CXCR7 vascular endothelium results in markedly greater spontaneous breast cancer metastases to multiple sites.
Figure 2. Increased local tumor recurrence and metastases in CXCR7ΔEND/ΔEND mice.
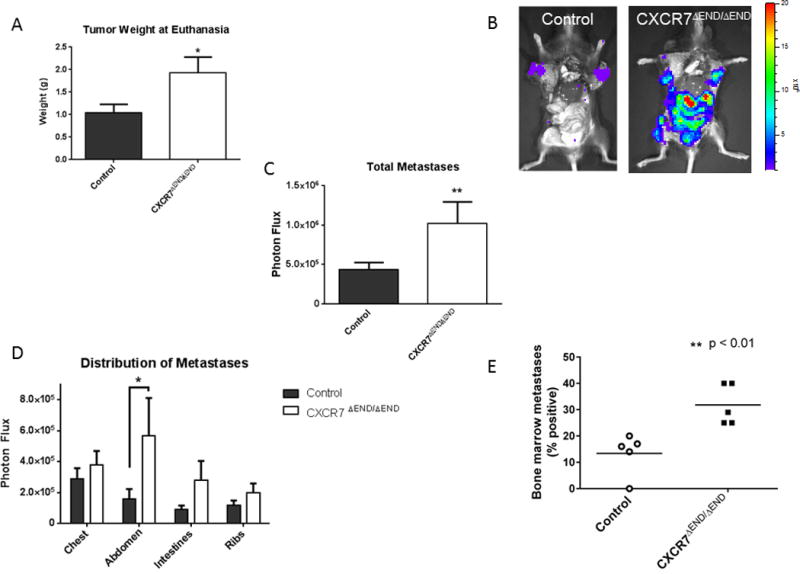
A) We resected recurrent tumors from mammary fat pads of CXCR7ΔEND/ΔEND and control mice at the time of euthanization. Weights of recurrent tumors are shown as mean values + SEM (n = 8 mice per group). B) Representative bioluminescence images of metastases in CXCR7ΔEND/ΔEND and control mice immediately after euthanization. C) We quantified total photon flux from AT-3-FL metastases in both groups of mice and graphed data as mean values + SEM. D) Graph shows mean values + SEM for bioluminescent metastases quantified at defined anatomic sites. E) We measured disseminated AT-3-FL cells recovered from bone marrrow of CXCR7ΔEND/ΔEND and control mice by ex vivo bioluminescence imaging and determined the percentage of mice in each group with detectable signal above background. Each data point shows percent of mice per group with disseminated tumor cells in bone marrow from five independent experiments (n = 8–10 mice per group per experiment). Horizontal line denotes the mean for each group. *, p < 0.05; **, p < 0.01.
To verify these results with a second breast cancer cell line, we used E0771 breast cancer cells, an estrogen receptor positive C57BL/6 medullary adenocarcinoma (30). E0771 cells express low levels of cell surface CXCR7 and no detectable CXCR4 by flow cytometry (Fig S3). We co-implanted E0771-FL cells and mouse mammary fibroblasts secreting CXCL12 into mammary fat pads of control and CXCR7ΔEND/ΔEND mice. Unlike AT-3-FL tumors, imaging data showed no difference in growth of E0771 orthotopic tumors through the time we resected tumors 21 days after implantation (Fig 3A). Immediately after resecting tumors, both cohorts had comparable bioluminescence, but imaging showed significantly greater regrowth of E0771-FL cells in CXCR7ΔEND/ΔEND animals by day 34 (p < 0.05) (Fig 3A). CXCR7ΔEND/ΔEND mice also had significantly shorter survival than control mice when we monitored animals until euthanized for humane end points or defined as cancer-free based on lack of bioluminescent signal and palpable tumor at 50 days (p < 0.01) (Fig 3B).
Figure 3. Reduced survival of CXCR7ΔEND/ΔEND mice with orthotopic tumor implants of E0771-FL breast cancer cells.
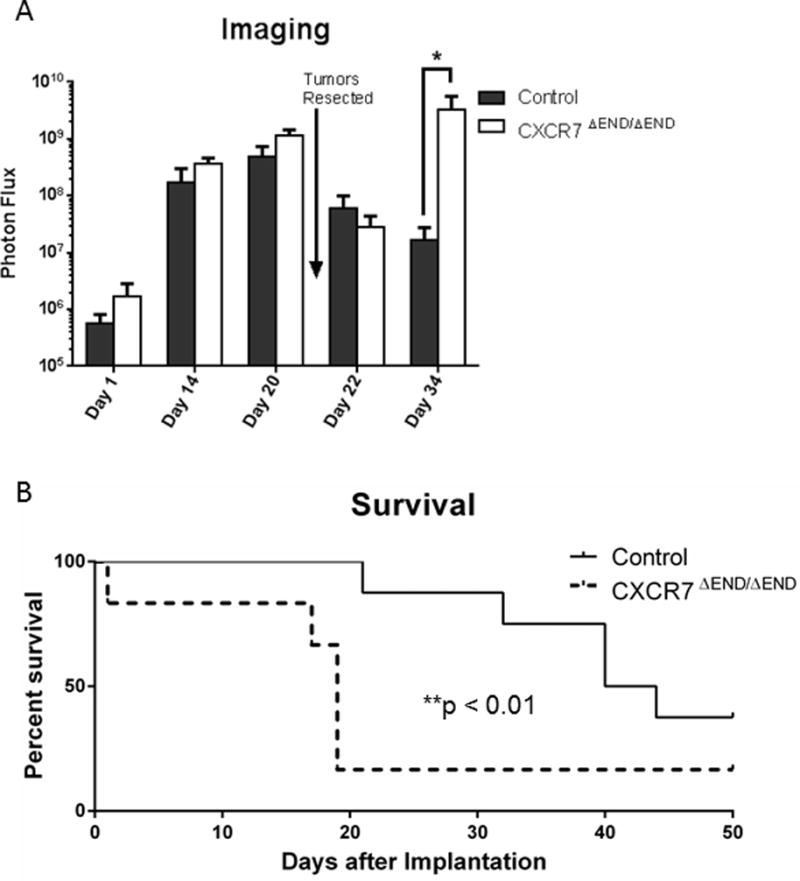
A) We measured tumor growth in mice with orthotopic implants of E0771-FL cells by bioluminescence imaging at indicated days. Graph shows mean values + SEM for photon flux in CXCR7ΔEND/ΔEND and control mice (n = 8 mice per group). Arrow designates when tumor implants were resected on day 21. B) Following resection of orthotopic tumor implants, we monitored CXCR7ΔEND/ΔEND and control mice until each animal had to be euthanized for humane end points. Mice surviving through 50 days had no detectable tumor as assessed by palpation of mammary fat pads and bioluminescence imaging. *, p < 0.05.
In an independent experiment with orthotopic E0771 tumors, we resected tumor implants and then euthanized all mice on the same day to quantify spontaneous metastasis. CXCR7ΔEND/ΔEND mice had significantly greater overall spontaneous metastases (p < 0.01) (Fig 4A, B). When analyzed by anatomic location, CXCR7ΔEND/ΔEND mice had more metastases in all body sites, although only differences in abdominal metastases were significant (p < 0.05) (Fig 4C). As described for AT-3-FL implants, we recovered blood and bone marrow from extremities and quantified circulating and disseminated tumor cells by ex vivo bioluminescence. Combining data from four independent experiments, 57 ± 8% of CXCR7ΔEND/ΔEND mice had circulating E0771-FL cells, while no control mice had any detectable viable cells. We also identified a significantly higher percentage of CXCR7ΔEND/ΔEND mice with disseminated tumor cells in bone marrow (p < 0.05); control mice only had E0771-FL cells in bone marrow in one of four experiments (Fig 4D). Therefore, we established that CXCR7ΔEND/ΔEND mice have greater spontaneous metastases using two different breast cancer cell lines.
Figure 4. CXCR7ΔEND/ΔEND mice with orthotopic E0771-FL tumors have greater spontaneous metastases.
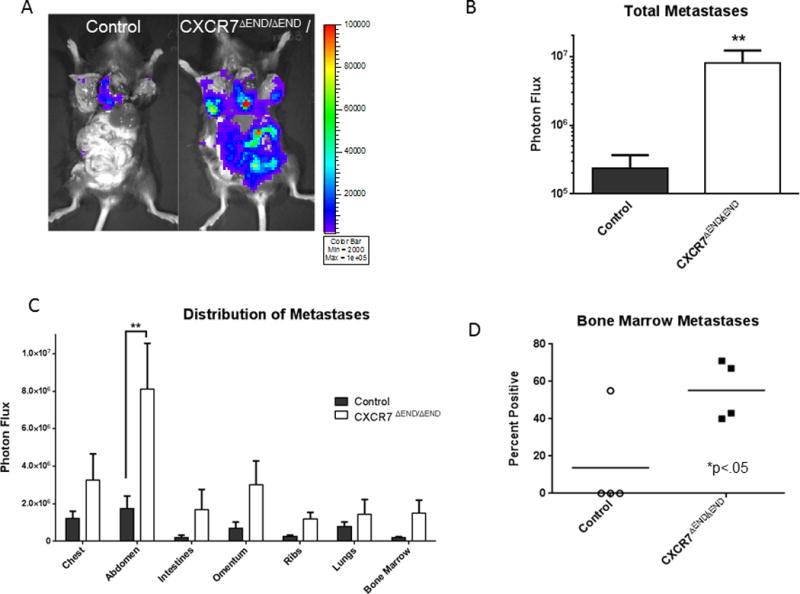
A) Representative bioluminescence images of metastatic E0771-FL cells in euthanized CXCR7ΔEND/ΔEND and control mice. B) Graph of total metastases measured in euthanized mice presented as mean values + SEM (n = 8 per group). C) Anatomic distribution of metastasis quantified by bioluminescence imaging of mice immediately after euthanization. Data are presented as mean values + SEM. D) We quantified disseminated E0771-FL cells in bone marrow by ex vivo bioluminescence imaging. Each point shows the percentage of CXCR7ΔEND/ΔEND and control mice with detectable breast cancer cells in bone marrow in a given experiment (n = 8–10 mice per group per experiment). Horizontal line shows mean values for percent mice with disseminated tumor cells in four independent experiments. **, p < 0.01.
CXCR7ΔEND/ΔEND mice have greater experimentally-induced metastases
CXCR7ΔEND/ΔEND mice had greater numbers of circulating tumor cells than control animals using orthotopic implants of either AT-3-FL or E0771-FL cells, respectively. Potentially, greater metastasis in CXCR7ΔEND/ΔEND mice could be due to enhanced ability of malignant cells to move from a mammary tumor into the vasculature and/or capability of cells to survive and proliferate in secondary organs.
To eliminate steps in metastasis through vascular intravasation from a localized tumor, we injected breast cancer cells directly into the left ventricle to produce experimental metastases. After intracardiac injection of AT-3-FL cells, we euthanized all animals 15 days later to quantify metastases. Bioluminescence imaging immediately after euthanization showed significantly greater total burden of AT-3-FL cells in CXCR7ΔEND/ΔEND mice with widespread metastases throughout these animals (Fig 5A, B). We observed significantly greater AT-3-FL cells in multiple organs and tissues, including abdomen, omentum, and liver (p < 0.05) (Fig 5C). In a separate experiment, we followed mice for survival after intracardiac injection of AT-3-FL cells instead of euthanizing animals at the same time. CXCR7ΔEND/ΔEND mice had significantly shorter survival with 50% of animals reaching humane endpoints by 18 days (p < 0.05) (Fig 5D). By comparison, 50% of control mice survived until day 26. CXCR7ΔEND/ΔEND mice also had significantly greater percentages of mice with disseminated AT-3-FL cells in bone marrow (Fig S6). CXCR7ΔEND/ΔEND mice with intracardiac injection of E0771 cells also showed a trend toward shorter survival than control mice, although differences between groups were not significant (Fig S7). These studies demonstrate that greater metastasis in CXCR7ΔEND/ΔEND mice is not solely due to more intravasation of breast cancer cells from mammary tumors, particularly for AT-3-FL cells that express both CXCR4 and CXCR7.
Figure 5. AT-3-FL cells produce greater experimental metastases in CXCR7ΔEND/ΔEND mice.
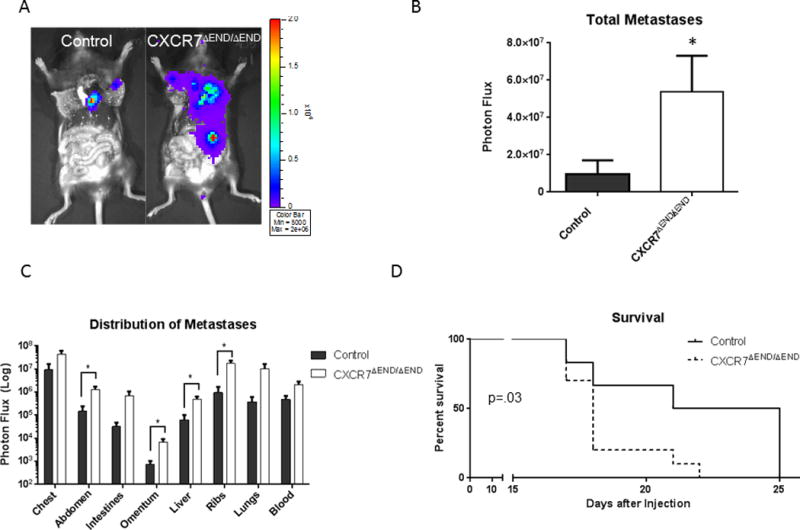
A) We injected AT-3-FL cells systemically via injection into left ventricles of CXCR7ΔEND/ΔEND and control mice. Presented images are representative of relative amounts of metastatic AT-3-FL cells at the time of euthanization. B) We quantified total AT-3-FL metastases by bioluminescence imaging and graphed data as mean values + SEM for each group (n = 12 each). C) We determined site-specific localization of metastases by bioluminescence imaging and quantified tumor burden as mean values + SEM for photon flux in each site. D) Survival curves for a separate experiment in which AT-3-FL cells were injected into CXCR7ΔEND/ΔEND and control mice and monitored until animals were euthanized for humane endpoints (n = 12 per group). * p < 0.05.
Discussion
Atypical chemokine receptor CXCR7 (ACKR3) is a prominent component of tumor microenvironments in multiple human cancers. Increased expression of CXCR7 commonly occurs on malignant cells in several different human cancers, including breast, renal, colon, and esophagus (14–16, 31). CXCR7 also is expressed by endothelial cells in tumor-associated blood vessels found in primary and metastatic sites (12, 21, 32). Studies in animal models typically demonstrate that CXCR7 in cancer cells promotes tumor growth and metastasis (12, 33, 34). While small molecule inhibitors of CXCR7 reduce tumor growth and metastasis in animal models, these systemic agents affect the receptor on both cancer and stromal cells (35–37). Therefore, effects of endothelial CXCR7 on cancer progression remain unknown.
To investigate endothelial CXCR7 in cancer, we used a genetically-engineered mouse model to conditionally delete this receptor from vascular endothelium. Cre-mediated excision of endothelial CXCR7 in adult mice had no gross phenotype, unlike peri-natal lethality caused by deleting this receptor from endothelium in utero (19). Using two different syngeneic breast cancer cell lines, we discovered that mammary tumor implants spontaneously metastasize more extensively in CXCR7ΔEND/ΔEND mice. These mice also had greater metastases following systemic injection of breast cancer cells, which bypasses the vascular intravasation step in metastasis. Effects on metastasis occurred without altering overall CD31+ vasculature in mammary tumors, indicating that endothelial CXCR7 is not required for tumor angiogenesis in this model.
Similar to other members of the newly designated family of atypical chemokine receptors, CXCR7 functions in part as a scavenger receptor for CXCL12. In cell-based assays, CXCR7 efficiently removes CXCL12 from the extracellular space and degrades it (27, 28). Prior studies demonstrate that CXCR7 also controls amounts of CXCL12 in living organisms (31, 38). Plasma CXCL12 increases by approximately 4-6-fold in mice with complete knockout of this receptor from all tissues or treated systemically with an inhibitor of CXCR7. In the current study, selective genetic deletion of CXCR7 only from vascular endothelium elevates plasma CXCL12 by ~35% in mice without breast tumors. This result confirms a recent study by Berahovich et al showing immunodetectable CXCR7 on endothelium of venules in organs including spleen, lung, and kidney, suggesting that endothelial CXCR7 could control systemic levels of CXCL12 (38).
Local and systemic increases in CXCL12 produce different outcomes, depending on amounts of CXCL12 and context. In some experimental systems, loss of CXCR7 and associated elevation of CXCL12 cause phenotypes due to reduced levels of cell surface CXCR4. CXCR7 scavenging of CXCL12 establishes chemotactic gradients necessary for migration of lateral line primordial cells in zebrafish and neurons during brain development in mice (27, 39–42). For both of these systems, loss of CXCR7 produces persistent, relatively high levels of CXCL12 that cause internalization and degradation of CXCR4 on migrating cells, resulting in loss of chemotaxis. However, we did not observe reduced levels of CXCR4 on natural killer cells in bone marrow of CXCR7ΔEND/ΔEND mice (data not shown), suggesting that modest elevations of CXCL12 in these mice did not affect cell surface expression of CXCR4. We note that a requirement for CXCR7 scavenging in CXCL12-CXCR4 chemotaxis depends on overall levels of this chemokine. Using a microfluidic model of chemotaxis, high levels of CXCL12 produced loss of cell surface CXCR4 and cell migration in the absence of cells with CXCR7, but cells maintained cell surface CXCR4 and migrated toward low levels of CXCL12 even without CXCR7 scavenging (43). Reinforcing integrated effects of CXCL12 levels and CXCR7, CXCR7ΔEND/ΔEND mice did not have greater spontaneous breast cancer metastasis without co-implanted fibroblasts to reproduce known secretion of CXCL12 by carcinoma-associated fibroblasts in primary human breast cancer (data not shown) (4, 44).
Modest elevations of plasma CXCL12 in CXCR7ΔEND/ΔEND mice may be more consistent with conditions that maintain CXCR4 signaling to promote tumor progression and metastasis. The ~35% increase in CXCL12 in CXCR7ΔEND/ΔEND mice is similar to the 14% increase in systemic CXCL12 reported for patients with breast cancer (45). In this clinical study, progressive increases in CXCL12 correlated with established markers of poor prognosis, including high grade tumors and basal subtype, suggesting that our mouse model may reproduce effects of moderately elevated CXCL12 on patient outcomes.
CXCL12 regulates multiple steps in tumor growth and metastasis, many of which increased in CXCR7ΔEND/ΔEND mice. In both AT-3 and E0771 orthotopic mammary tumor models, deletion of endothelial CXCR7 increased circulating tumor cells, suggesting greater vascular intravasation of breast cancer cells and/or survival in the circulation. CXCR4 and CXCR7 promote cell migration through tissues and resistance to anoikis, processes that would increase circulating tumor cells (6, 46–48). CXCR7ΔEND/ΔEND mice had greater local recurrence and spontaneous metastasis to multiple organs including bone marrow, one of the most common sites of metastatic breast cancer cells in patients. Greater spontaneous metastases in CXCR7ΔEND/ΔEND mice indicates that elevated CXCL12 may promote trafficking of circulating tumor cells to CXCL12-rich organs, enhance survival of disseminated tumor cells in secondary sites, and/or increase proliferation to produce overt metastases. Increases in growth of breast cancer cells after implantation into mammary fat pads were more pronounced with AT-3 cells. We note that AT-3 cells express cell surface CXCR4 and CXCR7, while flow cytometry showed only CXCR7 in E0771. The difference in initial growth of breast cancer cells in mammary fat pads suggests that CXCL12 signaling through CXCR4 is more important for this step in tumor progression. Similarly, when we bypassed the vascular intravasation step by injecting breast cancer cells systemically, we also determined that only CXCR7ΔEND/ΔEND mice with AT-3 cells had significantly shorter survival. This result suggests that CXCL12-CXCR7 interactions on cancer cells may promote intravasation in the mammary tumor environment, which is consistent with a prior cell culture study demonstrating that CXCR7 functions in transendothelial migration of cells (46).
To nominate other potential mechanisms through which endothelial CXCR7 may regulate affect breast cancer, we performed a bioinformatics analysis of the TCGA breast cancer data base. While we recognize that the TCGA does not segregate expression data by cell type, endothelial cells in almost all breast cancers express CXCR7. GSEA revealed that CXCR7 correlates negatively with proliferation in comparison with 4 of 5 data sets, suggesting that loss of endothelial CXCR7 in tumor environments may increase proliferation of breast cancer cells in primary tumors and/or metastases (Fig S8). In some contexts, endothelial CXCR7 may increase influx of immune cells into organs and tissues, suggesting potential effects on tumor immunity and regulation of metastasis by immune cells (49, 50).
While CXCR7 has been reported to increase proliferation of endothelial cells in vitro, this receptor was not essential for tumor angiogenesis in our mouse model as assessed by CD31 staining of mammary tumors (51) (52). This discordance likely reflects the large number of cytokines and chemokines that control angiogenesis in living systems, providing redundancy that compensates for loss of a single molecule. Such redundancy has been evident in resistance to therapies targeting a single regulator of tumor angiogenesis (53). Previous studies demonstrate that other tumor endothelial markers do not necessarily decrease tumor angiogenesis, showing our results are not unique to CXCR7 in tumor vasculature (54, 55). Further studies are planned to establish functions of endothelial CXCR7 in other types of cancer and different diseases.
Overall, we determined that CXCR7ΔEND/ΔEND mice have modestly elevated systemic levels of CXCL12, showing that endothelial CXCR7 contributes to overall regulation of this chemokine in vivo. Deletion of endothelial CXCR7 results in greater spontaneous and experimentally-induced breast cancer metastasis, indicating that vascular CXCR7 suppresses processes including tumor growth, vascular intravasation, and survival and proliferation of breast cancer cells in multiple organs and tissues. These data advance our understanding of CXCR7 and emphasize tissue context-dependent functions of this receptor in cancer. We note that in some experimental systems, pharmacologic inhibition of a molecular target may produce different outcomes than genetic deletion in adult animals (56, 57). However, our results suggest that targeting small molecule inhibitors of CXCR7 selectively to cancer cells, such as with nanoparticles, may improve therapeutic benefits by blocking oncogenic functions of this receptor on malignant cells while maintaining effects of endothelial CXCR7 to limit tumor progression.
Materials and Methods
Cells
C57BL/6 mouse breast cancer cell lines AT-3 and E0771 (gifts of Dr. Abrams and Dr. Mihich, Roswell Park Cancer Institute) were cultured in DMEM with 10% fetal bovine serum and 1% glutamine/penicillin/streptomycin (Life Technologies, Carlsbad, CA). We also cultured immortalized mammary fibroblasts from C57BL/6 mice (gift of Dr. Moses, Vanderbilt University) in the same medium. We cultured all cells at 37°C with 5% CO2.
Lentiviruses
We stably transduced mouse mammary fibroblasts with CXCL12-α fused to fluorescent protein mCherry (MMF-CXCL12) and breast cancer cells with firefly luciferase (AT-3-FL and E0771-FL, respectively) as described (58, 59).
Mice
The University of Michigan Committee for Care and Use of Animals approved all mouse procedures. We crossed mice with CXCR7 flanked by LoxP sites (gift of ChemoCentryx) with transgenic mice carrying tamoxifen-inducible Cre-ERT recombinase under control of the stem cell leukemia (Scl) stem cell enhancer (gift of Dr. Begley and the University of Western Australia) for endothelial-specific expression (23, 26). We backcrossed mice 12 generations on a C57BL/6 background.
To activate Cre, we treated mice every other day for seven days with 5 mg/kg taxmoxifen (Sigma, St. Louis, MO) in corn oil by intraperitoneal injection. We treated control mice with vehicle only. We also performed selected experiments with Cre-negative mice treated with tamoxifen rather than vehicle only and obtained similar results. We verified deletion of CXCR7 by PCR of tail biopsies. Since mouse tail tissue is well-vascularized, this strategy has been used previously to verify Cre-mediated deletion of a target gene from endothelium (24, 25). PCR primers for genotyping are shown in Table 1.
Table 1.
PCR primers for analyzing mice.
| Cre | forward | TCGATGCAACGAGTGATGAG |
| reverse | TTCGGCTATACGTAACAGGG | |
| LoxP flanked CXCR7 | forward | CTACAGCTTCATCAACCGCAAC |
| reverse | GTCCTGGRGCTGGCTTTGAT | |
| CXCR7 deletion | forward | GCAAGTTTGGGGTACAGTCC |
| reverse | GAGAGACAGGAAGCATGGTCAC |
Mouse tumor model
We established orthotopic tumors by implanting 2 × 105 breast cancer cells (E0771-FL or AT-3-FL) with 1 × 105 MMF-CXCL12 cells in 4th inguinal mammary fat pads of age-matched, 5–8 week old female CXCR7ΔEND/ΔEND or littermate control mice (n = 8–12 per group with specific numbers for each experiment listed in figure legends). These samples sizes are based on prior experience with imaging and disease progression studies in mice. Mice were assigned to groups based on genotype, and investigators knew the genotype of experimental groups. To generate systemic metastases, we injected 1 × 105 breast cancer cells directly into the left ventricle of the heart (n = 8–10 per group as listed in figure legends). We used bioluminescence imaging (IVIS Spectrum, Perkin-Elmer, Waltham, MA) to measure tumor burden and metastases as described (60).
When experiments were terminated for humane reasons, we injected luciferin i.p. prior to euthanizing an animal and then imaged metastases immediately after death (31). We quantified metastases based on region-of-interest analysis of bioluminescence for total body, chest, and abdomen. We subdivided the chest region into ribs and lungs, while we defined sub-regions of the abdomen as omentum, liver, and intestines with associated mesenteric lymph nodes. In selected experiments, we collected blood for circulating tumor cells via a 28 gauge needle inserted into the right side of the heart of anesthetized mice immediately before euthanization. We cultured blood samples overnight in DMEM growth medium; washed cells with PBS to remove red blood cells; and then measured firefly luciferase bioluminescence in cancer cells after one week. To detect tumor cells in bone marrow, we flushed each femur and tibia with PBS (1 ml per bone); dissociated bone marrow plugs by repeated pipetting; and then cultured cells in DMEM growth medium for one week. We quantified relative numbers of viable tumor cells in bone marrow by bioluminescence.
ELISA
We measured levels of CXCL12 in plasma by ELISA according to the manufacturer’s directions (R&D Systems, Minneapolis, MN).
Immunostaining
We fixed tumors in 4% buffered formalin overnight and then embedded tissues in paraffin for sectioning. To stain CXCR7 in tumor vasculature, we snap froze tumors or livers (Tissue-TEK®, VWR International, Radnor, PA) for subsequent cryosectioning of samples at 8 μm thickness. We stained paraffin sections with hematoxylin and eosin for tumor histology. We identified tumor blood vessels with an antibody to CD31 (eBioscience, San Diego, CA). In selected experiments, we also stained sections with CXCR7 (11G8, gift of ChemoCentryx, Mountain View, CA) to co-localize this receptor to tumor blood vessels (12). For immunostaining of CXCR7 with antibody 11G8, we blocked tumor sections with 0.1 mg/ml Fab fragment goat anti-mouse IgG (H+L) (Jackson Immunoresearch, West Grove, PA) for one hour to prevent background from mouse-on-mouse staining prior to adding primary antibodies. For CD31 staining, we quantified the percent area stained with antibody in four randomly selected 20X microscopic fields using ImageJ (61).
Flow cytometry
We analyzed cell surface expression of CXCR4 or CXCR7 on breast cancer cells with fluorescent-conjugated antibodies 12G5 and 11G8, respectively (R&D Systems) (62).
GSEA analysis
We performed RNAseq data retrieval and normalization as described previously and then correlated all individual genes to CXCR7 expression in the TCGA breast samples to generate a pre-ranked list based on Spearman’s Rho (63). To perform Gene Set Enrichment Analysis (GSEA), we used the MsigDB collections C2, C5, and C6 (64). We presented findings for gene sets related to proliferation and also performed an unbiased review of the top positively and negatively enriched gene sets to nominate potential mechanisms of action.
Statistical analysis
We performed animal studies 2–5 times with independent cohorts of mice. Graphs present mean values with standard error of the mean (SEM). We analyzed data using one-way ANOVA (GraphPad Prism, San Diego, CA) with p ≤ 0.05 defining statistically significant differences.
Supplementary Material
Acknowledgments
The authors thank Dr. Mark Penfold and ChemoCentryx for CXCR7 loxP mice and antibody 11G8. Dr. Jun-Lin Guan provided SCL CreERT through courteous permission of Dr. Glenn Begley. We thank Dr. Aaron Robida from the University of Michigan Flow Cytometry Core for assistance. Research was supported by United States National Institute of Health grants R21CA182333, R01CA136553, R01CA142750, R01CA170198, and P50CA093990.
Footnotes
Author Contributions
ACS, JF, SPC, KS, and GDL performed experiments and analyzed data. KEL contributed new reagents. SZ and SLC performed GSEA analyses. KEL and GDL conceptualized and directed the research. ACS, JF, SPC, and GDL wrote the manuscript.
Competing Interests
The authors have no competing financial interests.
References
- 1.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4(7):540–50. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 2.Ali S, Lazennec G. Chemokines: novel targets for breast cancer metastasis. Cancer Metastasis Rev. 2007;26(3–4):401–20. doi: 10.1007/s10555-007-9073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teicher B, Fricker S. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 2010;16(11):2927–31. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 4.Orimo A, Gupta P, Sgroi D, Arenzana-Seisdedos F, Delaunay T, Naeem R, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121(3):335–48. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Jin X, Malladi S, Zou Y, Wen Y, Brogi E, et al. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell. 2013;154(5):1060–73. doi: 10.1016/j.cell.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanon M, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–6. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 7.Singh S, Srivastava S, Bhardwaj A, Owen L, Singh A. CXCL12-CXCR4 signalling axis confers gemcitabine resistance to pancreatic cancer cells: a novel target for therapy. Br J Cancer. 2010;103(11):1671–9. doi: 10.1038/sj.bjc.6605968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoellenriegel J, Zboralski D, Maasch C, Rosin N, Wierda W, Keating M, et al. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood. 2014;123(7):1032–9. doi: 10.1182/blood-2013-03-493924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akashi T, Koizumi K, Tsuneyama K, Saiki I, Takano Y, Fuse H. Chemokine receptor CXCR4 expression and prognosis in patients with metastatic prostate cancer. Cancer Sci. 2008;99(3):539–42. doi: 10.1111/j.1349-7006.2007.00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu T, Shen H, Liu L, Shu Y. The impact of chemokine receptor CXCR4 on breast cancer prognosis: a meta-analysis. Cancer Epidemiol. 2013;37(5):725–31. doi: 10.1016/j.canep.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 11.Wald O, Shapira O, Izhar U. CXCR4/CXCL12 axis in non small cell lung cancer (NSCLC) pathologic roles and therapeutic potential. Theranostics. 2013;3(1):26–33. doi: 10.7150/thno.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miao Z, Luker K, Summers B, Berahovich R, Bhojani M, Rehemtulla A, et al. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proc Natl Acad Sci U S A. 2007;104(40):15735–40. doi: 10.1073/pnas.0610444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Shiozawa Y, Wang J, Wang Y, Jung Y, Pienta K, et al. The role of CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate cancer. J Biol Chem. 2008;283(7):4283–94. doi: 10.1074/jbc.M707465200. [DOI] [PubMed] [Google Scholar]
- 14.Yao X, Zhou L, Han S, Chen Y. High expression of CXCR4 and CXCR7 predicts poor survival in gallbladder cancer. J Int Med Res. 2011;39(4):1253–64. doi: 10.1177/147323001103900413. [DOI] [PubMed] [Google Scholar]
- 15.D’Alterio C, Consales C, Polimeno M, Franco R, Cindolo L, Portella L, et al. Concomitant CXCR4 and CXCR7 expression predicts poor prognosis in renal cancer. Curr Cancer Drug Targets. 2010;10(7):772–81. doi: 10.2174/156800910793605839. [DOI] [PubMed] [Google Scholar]
- 16.Tachezy M, Zander H, Gebauer F, von Loga K, Pantel K, Izbicki J, et al. CXCR7 expression in esophageal cancer. J Transl Med. 2013;11:238. doi: 10.1186/1479-5876-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberman J, Sartelet H, Flahaut M, Muhlethaler-Mottet A, Coulon A, Nyalendo C, et al. Involvement of the CXCR7/CXCR4/CXCL12 axis in the malignant progression of human neuroblastoma. PLoS One. 2012;7(8):e43665. doi: 10.1371/journal.pone.0043665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Alterio C, Avallone A, Tatangelo F, Delrio P, Pecori B, Cella L, et al. A prognostic model comprising pT stage, N status and the chemokine receptors CXCR4 and CXCR7 powerfully predicts outcome in neo-adjuvant resistant rectal cancer patients. Int J Cancer. 2013;135(2):379–90. doi: 10.1002/ijc.28689. [DOI] [PubMed] [Google Scholar]
- 19.Sierro F, Biben C, Martinez-Munoz L, Mellado M, Ransohoff R, Li M, et al. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci U S A. 2007;104(37):14759–64. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerrits H, van Ingen Schenau D, Bakker N, van Disseldorp A, Strik A, Hermens L, et al. Early postnatal lethality and cardiovascular defects in CXCR7-deficient mice. Genesis. 2008;46(5):235–45. doi: 10.1002/dvg.20387. [DOI] [PubMed] [Google Scholar]
- 21.Madden S, Cook B, Nacht M, Weber W, Callahan M, Jiang Y, et al. Vascular gene expression in nonneoplastic and malignant brain. Am J Pathol. 2004;165(2):601–8. doi: 10.1016/s0002-9440(10)63324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maishi N, Ohga N, Hida Y, Akiyama K, Kitayama K, Osawa T, et al. CXCR7: a novel tumor endothelial marker in renal cell carcinoma. Pathol Int. 2012;62(5):309–17. doi: 10.1111/j.1440-1827.2012.02792.x. [DOI] [PubMed] [Google Scholar]
- 23.Gőthert J, Gustin S, Hall M, Green A, Gőttgens B, Izon D, et al. In vivo fate-tracing studies using the Scl stem cell enhancer: embryonic hematopoietic stem cells significantly contribute to adult hematopoiesis. Blood. 2005;105(7):2724–32. doi: 10.1182/blood-2004-08-3037. [DOI] [PubMed] [Google Scholar]
- 24.Liao Y, Day K, Damon D, Duling B. Endothelial cell-specific knockout of connexin 43 causes hypotension and bradycardia in mice. Proc Natl Acad Sci USA. 2001;98(17):9989–94. doi: 10.1073/pnas.171305298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suárez Y, Fernández-Hernando C, Yu J, Gerber S, Harrison K, Pober J, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci USA. 2008;105(37):14082–7. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding B-S, Cao Z, Lis R, Nolan D, Guo P, Simons M, et al. Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature. 2013;505(7481):97–102. doi: 10.1038/nature12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boldajipour B, Mahabaleshwar S, Kardash E, Reichman-Fried M, Blaser H, Minina S, et al. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132(3):463–73. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 28.Luker K, Steele J, Mihalko L, Luker G. Constitutive and chemokine-dependent internalization and recycling of CXCR7 in breast cancer cells to degrade chemokine ligands. Oncogene. 2010;29:4599–610. doi: 10.1038/onc.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coleman R. Clinical features of metastatic bone disease and risk of skeletal morbidity. Clin Cancer Res. 2006;12(20 Pt 2):6243s–9s. doi: 10.1158/1078-0432.CCR-06-0931. [DOI] [PubMed] [Google Scholar]
- 30.Ewens A, Mihich E, Ehrke M. Distant metastasis from subcutaneously grown E0771 medullary breast adenocarcinoma. Anticancer Res. 2005;25(6B):3905–15. [PubMed] [Google Scholar]
- 31.Luker K, Lewin S, Mihalko L, Schmidt B, Winkler J, Coggins N, et al. Scavenging of CXCL12 by CXCR7 regulates tumor growth and metastasis of CXCR4-positive breast cancer cells. Oncogene. 2012;31(45):4570–8. doi: 10.1038/onc.2011.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guillemot E, Karimdjee-Soilhi B, Pradelli E, Benchetrit M, Goquet-Surmenian E, Millet M, et al. CXCR7 receptors facilitate the progression of colon carcinoma within lung not within liver. Br J Cancer. 2012;107(12):1944–9. doi: 10.1038/bjc.2012.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang S, Peng Y, Ye L, Wang Y, Qian Z, Chen Y, et al. Stimulation of TLR4 by LMW-HA induces metastasis in human papillary thyroid carcinoma through CXCR7. Clin Dev Immunol. 2013;2013:712561. doi: 10.1155/2013/712561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue T, Chen R, Han D, Chen J, Xue Q, Gao D, et al. Down-regulation of CXCR7 inhibits the growth and lung metastasis of human hepatocellular carcinoma cells with highly metastatic potential. Exp Ther Med. 2012;3(1):117–23. doi: 10.3892/etm.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burns J, Summers B, Wang Y, Melikian A, Berahovich R, Miao Z, et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203(9):2201–13. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goquet-Surmenian E, Richard-Fiardo P, Guillemot E, Benchetrit M, Gomez-Brouchet A, Buzzo P, et al. CXCR7-mediated progression of osteosarcoma in the lungs. Br J Cancer. 2013;109(6):1579–85. doi: 10.1038/bjc.2013.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walters M, Ebsworth K, Berahovich R, Penfold M, Liu S, Al Omran R, et al. Inhibition of CXCR7 extends survival following irradiation of brain tumours in mice and rats. Br J Cancer. 2014;110(5):1179–88. doi: 10.1038/bjc.2013.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berahovich R, Zabel B, Lewen S, Walters M, Ebsworth K, Wang Y, et al. Endothelial expression of CXCR7 and the regulation of systemic CXCL12 levels. Immunology. 2014;141(1):111–22. doi: 10.1111/imm.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Li G, Stanco A, Long J, Crawford D, Potter G, et al. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69(1):61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez-Alcaniz J, Haege S, Mueller W, Pla R, Mackay F, Schulz S, et al. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 2011;69(1):77–90. doi: 10.1016/j.neuron.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 41.Dona E, Barry J, Valentin G, Quirin C, Khmelinskii A, Kunze A, et al. Directional tissue migration through a self-generated chemokine gradient. Nature. 2013;503(7475):285–9. doi: 10.1038/nature12635. [DOI] [PubMed] [Google Scholar]
- 42.Venkiteswaran G, Lewellis S, Wang J, Reynolds E, Nicholson C, Knaut H. Generation and Dynamics of an Endogenous, Self-Generated Signaling Gradient across a Migrating Tissue. Cell. 2013;155(3):674–87. doi: 10.1016/j.cell.2013.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cavnar S, Ray P, Moudgil P, Chang S, Luker K, Linderman J, et al. Microfluidic source-sink model reveals effects of biophysically distinct CXCL12-isoforms in breast cancer chemotaxis. Integr Biol. 2014;6(5):564–76. doi: 10.1039/c4ib00015c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allinen M, R B, Cai L, Brennan C, Lahti-Domerci J, Huang H, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6(1):17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 45.Potter S, Dwyer R, Curran C, Hennesy E, Harrington K, Griffin D, et al. Systemic chemokine levels in breast cancer patients and their relationship with circulating menstrual hormones. Breast Cancer Res Treat. 2009;115(2):279–87. doi: 10.1007/s10549-008-0078-2. [DOI] [PubMed] [Google Scholar]
- 46.Zabel B, Wang Y, Lewen S, Berahovich R, Penfold M, Zhang P, et al. Elucidation of CXCR7-mediated signaling events and inhibition of CXCR4-mediated tumor cell transendothelial migration by CXCR7 ligands. J Immunol. 2009;183(5):3204–11. doi: 10.4049/jimmunol.0900269. [DOI] [PubMed] [Google Scholar]
- 47.Kochetkova M, Kumar S, McColl S. Chemokine receptors CXCR4 and CCR7 promote metastasis by preventing anoikis in cancer cells. Cell Death Differ. 2009;16(5):664–73. doi: 10.1038/cdd.2008.190. [DOI] [PubMed] [Google Scholar]
- 48.Sun Y, Mao X, Fan C, Liu C, Guo A, Guan S, et al. CXCL12-CXCR4 axis promotes the natural selection of breast cancer cell metastasis. Tumour Biol. 2014;35(8):7765–73. doi: 10.1007/s13277-014-1816-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cruz-Orengo L, Holman D, Dorsey D, Zhou L, Zhang P, Wright M, et al. CXCR7 influences leukocyte entry into the CNS parenchyma by controlling abluminal CXCL12 abundance during autoimmunity. J Exp Med. 2011;208(2):327–39. doi: 10.1084/jem.20102010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kitamura T, Qian B-Z, Pollard J. Immune cell promotion of metastasis. Nat Rev Immunol. 2015;15:73–86. doi: 10.1038/nri3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raggo C, Ruhl R, McAllister S, Koon H, Dezube B, Fruh K, et al. Novel cellular genes essential for transformation of endothelial cells by Kaposi’s sarcoma-associated herpesvirus. Cancer Res. 2005;65(12):5084–95. doi: 10.1158/0008-5472.CAN-04-2822. [DOI] [PubMed] [Google Scholar]
- 52.Totonchy J, Osborn J, Botto S, Clepper L, Moses A. Aberrant proliferation in CXCR7+ endothelial cells via degradation of the retinoblastoma protein. PLoS One. 2013;8(7):e69828. doi: 10.1371/journal.pone.0069828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casanovas O, Hicklin D, Bergers G, Hanahan D. Drug resistance by evasion of antiangiogenic targeting of VEGF signaling in late-stage pancreatic islet tumors. Cancer Cell. 2005;8(4):299–309. doi: 10.1016/j.ccr.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 54.Nanda A, Karim B, Peng Z, Liu G, Qiu W, Gan C, et al. Tumor endothelial marker 1 (Tem1) functions in the growth and progression of abdominal tumors. Proc Natl Acad Sci USA. 2006;103(9):3351–6. doi: 10.1073/pnas.0511306103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cullen M, Seaman S, Chaudhary A, Yang M, Hilton M, Logsdon D, et al. Host-derived tumor endothelial marker 8 promotes the growth of melanoma. Cancer Res. 2009;69(15):6021–6. doi: 10.1158/0008-5472.CAN-09-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yokoyama K, Ishikawa N, Igarahsi S, Kawano N, Hattori K, Miyazaki T, et al. Discovery of potent CCR4 antagonists: Synthesis and structure-activity relationship study of 2,4-diaminoquinazolines. Bioorg Med Chem. 2008;16(14):7021–31. doi: 10.1016/j.bmc.2008.05.036. [DOI] [PubMed] [Google Scholar]
- 57.Walters M, Ebsworth K, Sullivan T, Zhang P, Powers J, Jaen J, et al. CCR9 inhibition does not interfere with the development of immune tolerance to oral antigens. Immunol Lett. 2013;151(1–2):44–7. doi: 10.1016/j.imlet.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 58.Smith M, Luker K, Garbow J, Prior J, Jackson E, Piwnica-Worms D, et al. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64(23):8604–12. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- 59.Ray P, Mihalko L, Coggins N, Moudgil P, Ehrlich A, Luker K, et al. Carboxy-terminus of CXCR7 regulates receptor localization and function. Int J Biochem Cell Biol. 2012;44(4):669–78. doi: 10.1016/j.biocel.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luker G, Pica C, Song J, Luker K, Piwnica-Worms D. Imaging 26S proteasome activity and inhibition in living mice. Nat Med. 2003;9(7):969–73. doi: 10.1038/nm894. [DOI] [PubMed] [Google Scholar]
- 61.Fenner J, Stacer A, Winterroth F, Johnson T, Luker K, Luker G. Macroscopic stiffness of breast tumors predicts metastasis. Sci Rep. 2014;4:5512. doi: 10.1038/srep05512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salomonnson E, Stacer A, Ehrlich A, Luker K, Luker G. Imaging CXCL12-CXCR4 signaling in ovarian cancer therapy. PLoS One. 2013;8(1):e51500. doi: 10.1371/journal.pone.0051500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao S, Chang S, Linderman J, Feng F, Luker G. A comprehensive analysis of CXCL12 isoforms in breast cancer. Transl Onc. 2014 doi: 10.1016/j.tranon.2014.04.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subramanian A, Tamayo P, Mootha V, Mukherjee S, Ebert B, Gillette M, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


