Abstract
Nanoparticle-mediated delivery of therapeutics holds great potential for the diagnosis and treatment of a wide range of diseases. Significant advances have been made in the design of new polymeric nanoparticle carriers through modulation of their physical and chemical structures and biophysical properties. Nanoparticle shape has been increasingly proposed as an important attribute dictating their transport properties in biological milieu. In this review, we highlight three major methods for preparing polymeric nanoparticles that allow for exquisite control of particle shape. Special attention is given to various approaches to controlling nanoparticle shape by tuning copolymer structural parameters and assembly conditions. This review also provides comparisons of these methods in terms of their unique capabilities, materials choices, and specific delivery cargos, and summarizes the biological effects of nanoparticle shape on transport properties at the tissue and cellular levels.
Keywords: polymeric nanoparticles, shape control, drug delivery, gene therapy
Introduction
Therapeutic delivery using polymeric nanoparticles has received considerable attention for a wide range of biological applications, including drug and gene delivery, tissue engineering and regenerative medicine, and the detection of biomarkers and diagnosis of disease states.1–4 The potential cargo delivered by nanoparticles is vast: some of the most common include small molecular weight compounds, chemotherapeutics, proteins, nucleic acids, and imaging and diagnostic agents.5–9 While new systems continue to be developed, success of nanoparticle delivery in vivo is often limited, particularly following systemic administration where less than 5% of the total dose successfully reaches the target site.10, 11 Improving the delivery efficiency of nanoparticles is paramount to fully harness their potential as both a research tool and as a potential pharmaceutical agent. For these reasons, renewed focus has been given to engineering nanoparticles through modulation of a specific physical characteristic, nanoparticle shape. Natural pathogens, such as viruses, often display unique shapes, ranging from spherical Heptatitis A virons to micron-sized, worm-shaped Ebola virons.12, 13 While shape is not the only factor that guides the tissue tropism of the virus, applying it to nanoparticle engineering, particularly for polymeric nanoparticles, holds great potential for therapeutic delivery applications. In this review, we highlight the key methods and materials used to generate polymeric nanoparticles with controlled shapes. Using these tools, studies have begun to reveal the biological responses to nanoparticles with controlled shapes, leading to significant differences at the cellular level, tissue level, and systemic level.
Methods of Preparing Polymeric Nanoparticles with Controlled Shapes
Lithography-based Methods
Particle Replication in Nonwetting Template (PRINT) Method
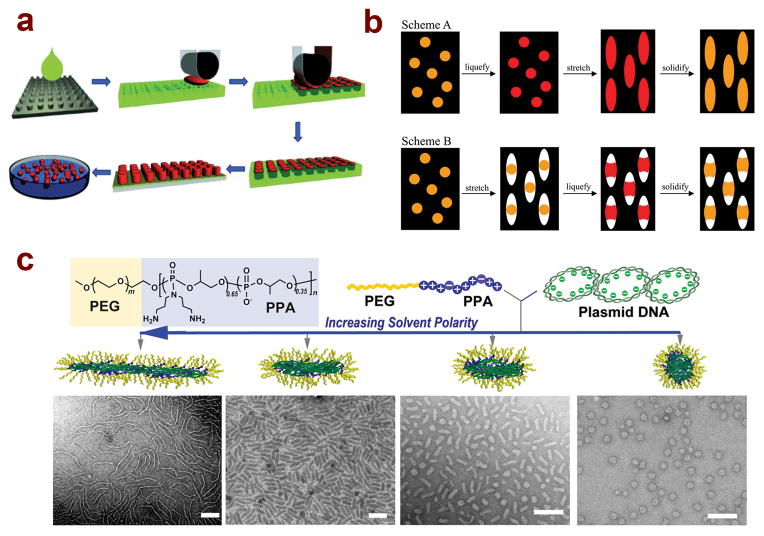
PRINT is the most popular among several photolithography techniques that have been developed to prepare polymeric nanoparticles with controlled sizes. It is a top-down approach that utilizes highly fluorinated surfaces, which are nonwetting to the materials being used to generate nanoparticles.14 Because of this, it is much easier to shape and harvest particles using PRINT, particularly in the nanometer scale, whereas other lithography techniques develop a residual film between the desired object and the mold due to interactions at the interface15 (Figure 1a). This method allows for the formation of polymeric nanoparticles in the micro- and nano-scale size ranges with a high degree of uniformity.15, 16 The range of shapes that can be produced through PRINT fabrication is also very wide; particles can range from cubic/cylindrical with an aspect ratio of 1, to worm-like with an aspect ratio as high as 60.17–19 A variety of materials can also be used to formulate these particles, including hydrogels comprised of crosslinked poly(ethylene glycol) (PEG),15, 18–20 solid particles formulated with poly(lactic acid) (PLA) and poly(lactic acid-co-glycolic acid) PLGA,17, 21, 22 as well as biopolymer-based particles formulated with serum albumin and insulin.23, 24 Incorporation of various cargos for therapeutics and diagnostics have been achieved, including chemotherapy drugs,17, 22 siRNA,20, 21 RNA replicons,24 and contrast agents.25 Recent advances in PRINT technology also allow for the fabrication of Janus particles and end-labeled particles.26 A study by Morton et al. demonstrated a spray method to incorporate layer-by-layer (LbL) coatings onto the surface of PRINT fabricated nanoparticles, providing a method for particle synthesis with precise control of nanoparticle size, shape, and surface characteristics.27 Importantly, the PRINT process can be performed in small batches as well as a continuous, automated roll-to-roll system, providing a large-scale production system for potential clinical applications.14
Figure 1.
Several methods have been developed to control the shape of polymeric nanoparticles for therapeutic delivery applications. (A) PRINT technology allows for the generation of particles with controlled shapes and surface chemistries through harvesting from polymer molds with low surface energy;14 (reprinted with permission, ©2013 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim) (B) Two distinct methods for generating nanoparticles with nonspherical shapes through stretching and liquefication of precursor films, leading to the formation of rod shaped and barrel shaped particles;32 (©2007 National Academy of Sciences, USA) (C) Self-assembly of PEG-polycation/DNA nanoparticles in solutions with varying solvent polarity leads to the formation of different shapes, including spheres, rod-like, and worm-like particles.125 (reprinted with permission, ©2013 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.)
Step and flash imprint lithography (S-FIL)
S-FIL method has also been used to generate polymeric nanoparticles with controlled sizes.28–31 S-FIL is a commercially available lithography technique that utilizes a patterned quartz template to mold photocrosslinkable solutions into defined patterns on a silicon surface.28 Compared to the PRINT method, S-FIL offers advantages in ease of particle harvesting through the use of a water-soluble PVA release layer,28, 30 whereas PRINT requires surgical blades or shear forces to release the particles.28 S-FIL still allows for precise control of particle size and shape, with feature sizes down to 50 nm. S-FIL has primarily been used to generate PEG hydrogel-based nanoparticles, which have been loaded with biomolecules including antibodies, nucleic acids, and anticancer drugs.29 In addition, this method has been used to incorporate stimuli-sensitive materials into the nanoparticle matrix, allowing for the release of cargo under physiological conditions through enzyme-mediated cleavage of the carrier materials.28
Membrane Stretching Methods
Another method for generating polymeric nanoparticles with different shapes involves the stretching of spherical particles into complex shapes (Figure 1b).32–38 Spherical particles, typically polystyrene (PS) with diameters ranging from 60 nm to 10 μm, are suspended in a solution of poly(vinyl alcohol) (PVA) and generated into films, liquefied in solvent or heated above the glass transition temperature, and stretched in one or two dimensions to generate aspherical shapes, typically rods or elliptical disks, with aspect ratios ranging from 2 to 15. 32 A second method involves stretching the PVA film first, followed by liquefying the spherical particles to fill the void left in the PVA film. This method can be used to generate barrel and lens-shaped objects.32 In addition to PS, PLGA has also been used to generate stretched particles.33, 34 Conjugation of antibodies for tissue-specific targeting, including anti-ICAM-1 and anti-transferrin for lung and brain delivery, respectively, has been successfully demonstrated.38
Self-Assembly Methods
Self-assembly of amphiphilic copolymers
Self-assembly of amphiphilic copolymers in aqueous or organic media has been widely used to generate nanoparticles. This bottom-up approach relies on the spontaneous assembly of single or multiple block copolymers to generate micellar nanoparticles with well-defined shapes. These amphiphilic copolymers are composed of two or more blocks with distinct polarities, and when placed in selected solvents can form supramolecular assemblies as a result of the thermodynamic incompatibility between the different blocks.39–42 The final aggregate morphology is dependent on the hydrodynamic volume fraction of one block with respect to the other; the interfacial tension between the aggregate and the solvent mixture, and the kinetics of phase separation, which is dictated by the method used to trigger the self-assembly of copolymers. The typical structures formed by these amphiphilic macromolecules comprise spherical micelles, cylindrical or worm-like micelles and bilayer structures (i.e. polymersomes). In addition, more complex aggregate morphologies including toroidal, disc-like, and compartmentalized particles have been engineered by fine-tuning the block lengths or molecular weights, polymer architecture, and chemical composition.43–45 Aside from the molecular characteristics of block copolymers, the method used to trigger the self-assembly of copolymers is crucial to the generated micelle morphology.
Conventional methods for synthesizing block copolymer nanoparticles involve the slow addition of a block copolymer dissolved in water-miscible organic solvent into a larger quantity of water, resulting in the formation of aggregates at near thermodynamically equilibrated state. This method has frequently been used to study the kinetics of copolymer nanoparticle formation and to isolate aggregates of unique morphologies.44, 45 Although nanoparticles with various sizes and shapes have been prepared using this method as drug delivery carriers and diagnostic devices,46, 47 the slow mixing kinetics (occurs typically at a time scale of seconds) in comparison with the aggregation events, and the highly heterogeneous nature of the mixing conditions yield nanoparticle preparations with poorly controlled size and with broad distribution, poor colloidal stability, and low drug encapsulation efficiency.
To address these challenges, methods to improve the uniformity of mixing and phase-separation kinetics and to accelerate the mixing rates have been developed recently. For example, microfluidic mixing devices have been developed to yield more uniform mixing; and preparations based on confined impinging jets and multi-inlet vortex mixers rely on rapid mixing in a time scale of milliseconds. Compared to conventional methods, these new techniques allow for the formation of smaller and uniform aggregates in a continuous process, which is amenable for easier scale-up. They also offer a higher degree of versatility and control over particle size and distribution, higher drug encapsulation efficiency, and improved colloidal stability.48, 49
These methods have predominantly been adopted for preparing spherical-like nanoparticles from block copolymers. Only recently, the Moffitt’s group has established conditions to tune aggregate shape using microfluidic devices.50–53 They employed a gas-liquid multiphase microfluidic reactor, in which argon gas is introduced into the merged liquid streams, compartmentalizing the colaminar flow into segmented liquid plugs. The chaotic advection observed within the liquid plugs enables fast mixing (>1 s) between water and polystyrene-b-poly(acrylic acid) copolymer solution triggering the formation of aggregates. The aggregates are subsequently exposed to strong and localized shear forces through the processing channel resulting in the formation of a myriad of aggregate structures including Y-Junctions, cylinders, vesicles and networks. The interplay between the chemical conditions (copolymer concentration, solvent polarity, and ionic strength) and flow kinetics enables the control over the size and the shape of the final aggregate.50–52 Another interesting example by Förster et al. shows that one can also prepare size-controlled unilamellar copolymer vesicles in a fast, continuous and reproducible fashion using perpendicular hydrodynamic flow focusing in a microfluidic device.54
Another approach to improve micelle stability and drug release kinetics is to directly conjugate hydrophobic drugs to the polymer block forming the core of the micelles.55–63 The ability to create multiple aggregate morphologies from a single copolymer is particularly attractive for investigating the effect of shape on aggregate stability, cellular internalization, trafficking, and drug delivery. In a recent example, Hu et al. synthesized polyprodrug amphiphiles consisting of polyethylene glycol (PEG) and a reduction-responsive camptothecin prodrug block to prepare particles of different shape including spheres, smooth disks, flower-like large compound vesicles (LCVs), and staggered lamellae with spiked periphery.64 The self-assembled nanostructures were prepared under highly controlled mixing rate and concentrations; and particles with various shapes were prepared depending on the water addition rate and the organic solvent polarity. The study has shown that particle shape plays a critical role in particle-cell interaction and in vivo biodistribution. The staggered lamellae nanoparticles exhibited the fastest cellular uptake and highest stability in biological media. In addition, the blood circulation half-life of these nanoparticles was nearly 15-fold higher than their spherical analogue. In another study, Geng et al. found that self-assembly of PEG-b-polycaprolactone (PCL) copolymers in water led to the formation of long, filamentous particles, which they termed filomicelles.65 The filomicelles in aqueous medium displayed lengths ranging from 2 microns up to 20 microns, and filomicelles loaded with paclitaxel were also successfully prepared for anticancer applications.
Conventional copolymer micelles as drug delivery carriers, however, may exhibit limited stability following the extreme dilution upon systemic delivery or as a result of serum protein-copolymer interaction and blood flow stress.66 Additionally, the rapid dissociation of drug-loaded copolymer micelles in the bloodstream leads to deficient drug accumulation in the tumor and undesired side effects. To suppress these limitations, several research groups have introduced crosslinkable copolymer micelles that present both prolonged circulation in vivo and a better control over their drug-release properties. 67–69 Traditionally, crosslinkable copolymer micelles can be prepared through the addition of crosslinking agents that will react with functional domains present in the block copolymer micelle. Three main strategies have been pursued: (i) core-crosslinked stabilized copolymer micelles, where crosslinking occurs either at the hydrophobic chain-end or along the hydrophobic chain; (ii) core-shell interface stabilized copolymer micelles; and (iii) shell-crosslinked stabilized micelles. The location at which the crosslinking occurs as well as the extent of crosslinking significantly impaacts the physicochemical properties of the final aggregate structures. Furthermore, stimuli-responsive crosslinked copolymer micelles can be prepare by introducing bifunctional crosslinking agents that respond to external stimuli such as temperature, light, pH and redox agents.67, 70–75 The Wooley group has developed several shell crosslinked copolymer micelles for the delivery of therapeutics or as theranostic devices.71, 76, 77 In an elegant example, they prepared crosslinked copolymer micelles of different shape, spheres and cylinders with different aspect ratios, to which a cell penetrating peptide (HIV Tat PTD) was conjugated. Both the aggregate shape and the amount of peptide conjugated were found to play a role in the endocytosis and exocytosis of the aggregates evaluated.78
Unimolecular micelles prepared from molecular amphiphilic brush-like polymers (MABPs) are an attractive alternative since their covalent nature ensures micellar stability without compromising the drug release profile or biodegradability.79–81 MABPs are macromolecules comprised of a linear polymer backbone and densely grafted polymer side chains.82, 83 Trigger-sensitive unimolecular micelles can be prepared to achieve on-demand release of encapsulated drugs. For example, aggregates resembling unimolecular micelles of size ranging from 6 to 50 nm containing 10 – 400 doxorubicin or camptothecin molecules per brush have been prepared. In this micelle system, drug molecules were fully protected by the dense PEG chains and released in its free form upon irradiation with UV light.84, 85 Incorporation of pH- and redox-cleavable linkers in a similar manner can expand the utility of MABPs for the delivery of anticancer drugs.86 Johnson et al. have synthesized nanoparticles with stimuli-responsive dual MRI and NIR optical fluorescence imaging capabilities.81 The brush polymers contained nitroxide radicals that in the native state partially quench the fluorescence intensity of the NIR dye. Upon exposure to ascorbate or ascorbate/glutathione in cancer cells, nitroxide radicals are reduced, resulting in a significant decrease of the MRI contrast and concomitant increase of the fluorescence intensity. The high in vivo stability, long circulation time, and the easy to conjugate cellular targeting moieties to these MABP micelles make them appealing as theranostic devices for tumor imaging and treatment.
Negatively charge polymers, such as polyacrylate and nucleic acid, can also be used as the hydrophilic block instead of PEG to construct amphiphilic micelles. For example, poly(acrylic acid)-b-polystyrene (PAA-b-PS) block copolymers can self assemble into micelles with different shapes either by changing the block lengths of PAA and PS, or by varying solvent polarity. Furthermore, the micelle shape can be reversibly transformed from spheres to rods to vesicles through modulation of solvent polarity even after micelles are formed.87, 88 More relevant to biomedical application, self-assembly of nucleic acid-based amphiphiles is highlighted in a recent review by Kwak and Herrmann.89 In these assemblies, the oligonucleotide chain, mostly DNA, is conjugated to a hydrophobic polymer chain, forming either linear block copolymer or brush-like graft copolymer. These DNA-based amphiphiles self-assemble into micellar nanoparticles with different shapes, including spherical particles, vesicles, rod-like particles, and long cylinders.90, 91 Shape control is achieved based on the same assembly principle as discussed above via tuning copolymer composition and architecture of the block copolymers, and solvent conditions. The length of the oligonucleotides used in these DNA amphiphiles typically is on the order of tens to hundreds of base pairs. Incorporation of longer chains, including functional DNA sequences or plasmid DNA, has not been reported. Use of this strategy for the delivery of siRNA or antisense oligonucleotides, however, may be of interest for the field.
Self-assembly through complimentary base-pairing
Recent studies have also reported the generation of complex nanostructures through complementary base-pairing of DNA molecules and other short stranded oligonucleotides.92 Because of the self-recognition of complementary DNA strands, a multitude of complex two- and three-dimensional assemblies can be rationally designed, ranging from dendritic structures, cubes, polyhedrons, and nanotubes.92–97 Concerns exist over the stability of DNA nanostructures against nuclease degradation, although studies have shown that the nanostructures exhibit less degradation than nucleotides themselves.98–100 These DNA nanoparticles have also been used for therapeutic delivery applications, including small molecule drugs, antibodies, nucleic acids, and vaccine adjuvants.92, 101–105 Lee et al. have developed a DNA tetrahedron, generated through complementary base-pairing of single stranded oligonucleotides, capable of packaging and delivering small interfering RNA (siRNA) to silence genes of interest in tumors.106 One unique advantage of this method is the ability to precisely control the density and the spatial orientation of ligand placement of targeting ligands independently in the context of improving transfection efficiency. This assembly approach has also been extended to RNA.107–109 Recently, Afonin et al. have employed an in silico design strategy to synthesize functional RNA nanoparticles in the form of nanocubes and nanorings.110 While these assembled structures are capable of packaging multiple siRNA sequences, their utility was hindered by similar factors that affect naked RNA delivery in vivo, such as degradation by nucleases, lack of cell uptake, and rapid clearance. To overcome these shortcomings, cationic amphiphiles were used as carriers to effectively deliver the RNA payload. In this approach, the RNA molecules designed to have unique shapes were further condensed into spherically shaped micelles when complexed with an amphiphile.111
An alternative strategy to improve the RNA stability is to generate oligomerized RNA sequences. Shopsowitz et al. have reported a method to synthesize a RNA microsponges (~500 nm) densely loaded with RNA generated from a circular DNA template.112 These RNAi microsponges were condensed into complexes and successfully mediated gene knockdown when used with Lipofectamine and linear polyethylenimine (lPEI) as the transfection agents in vitro and in vivo in a mouse tumor model.113 While shape control using DNA and RNA as building blocks has been possible, controlling the shapes of nanoparticles following condensation of these species with polycations remains a challenge that continues to be addressed.
Self-assembly of polyelectrolytes
Self-assembly of oppositely charged polyelectrolytes defines a class of nanoparticles called complex core micelles, also referred to as polyion complex micelles.114 These micelles exhibit a unique core-shell structure, with the polyelectolyte complex forming a stable core surrounded by a neutral, hydrophilic corona. Complex core micelle formation has been reviewed in several papers,114–116 although shape control of such assemblies has not received significant attention until recently. DNA as a unique polyanion itself has been used as a building block to generate a variety of complex core micelles with different shapes.117–119 Using PEG-b-poly(L-lysine) (PEG-b-PLL) copolymers to package the plasmid DNA, Osada et al. found that the length of the PLL segment significantly influenced the shape of the micelle.120 Short PLL blocks led to the formation of rod and toroid-like particles, whereas longer PLL blocks led to spherical shapes. Further tuning of the length of the PLL segment could be used to control the length of the rod-like shapes, with short PLL segments (degree of polymerization of 19) forming rod lengths greater than 200 nm, and longer PLL segments (degree of polymerization of 70) generating rods with lengths of 50–100 nm.121, 122 It appears that the PEG chain crowdedness (i.e. PEG chain density on micelle core surface) as a key factor in controlling micelle rod length.122, 123 For shorter PLL segments, a greater amount of polymer is needed to neutralize the DNA charge, resulting in a higher density of PEG on micelle surface. Due to steric repulsion of the PEG chains, the micelles tend to elongate to provide additional surface area for the PEG corona. In a separate study using N-(2-hydroxypropyl) methacrylamide (HPMA) as the hydrophilic block, Shi et al. showed that the molecular weight of PLL in a HPMA-b-PLL copolymer also effectively changed the shape of complex core micelles formed with plasmid DNA.124 Nanoparticles formed with copolymers containing longer PLL block exhibited higher aspect ratios than that with shorter PLL blocks, which exhibited more spherical shapes.
Interestingly, solvent quality and polarity has been shown to significantly influence the shape control in this complex core assembly. A recent study by Jiang et al. has demonstrated the feasibility of shape variation of DNA–polycation nanoparticles using a single PEG-b-polyphosphoramidate (PEG-b-PPA) block copolymer and plasmid DNA by tuning solvent polarity (Figure 1c).125 When polyelectrolyte complexes were formed in water–dimethylformamide (DMF) or dimethyl sulfoxide (DMSO) mixture solvents with different volume ratios, PEG-b-PPA/DNA micelles assumed different shapes ranging from worm-like shapes in water to rod-like and spherical shapes in lower polarity solvents. A similar reversible shape transformation process as shown for amphiphilic block copolymers 88 was also observed for these DNA-containing micelles by titrating solvent with water or DMF after the complexation and micelle formation, allowing shape tuning from spheres to rod-like and worm-like shapes, and vice versa. More interestingly, this shape tuning technique yielded micellar particles with a high degree of shape uniformity. It is important to point out that despite of the similarity in shape control with amphiphilic block copolymer, the mechanism for micelle assembly and shape control is distinctly different for these complex core micelles. Molecular dynamics simulations revealed that the DNA plasmid serves as both a functional payload and as a shape template as the conformation of DNA in each solvent condition dictated the final shape of the micelle, and that the DNA chain rigidity and solvent-DNA interaction are key factors influencing micelle shape control in this micelle system. In addition, maximization of PEG entropy in the solvent is essential to ensure shape conversion in these micelles, as nanoparticles prepared without the PEG block only exhibited more condensed spherical shapes in all solvents.
It is important to note that this micelle assembly strategy is not limited to block copolymer of PEG and polycations; polycation-g-PEG graft copolymers are also able to condense plasmid DNA into micellar nanoparticles with distinct shapes.126 Compared with block copolymer carriers, graft copolymers offer much wider parameter space for method optimization to control micelle shapes and stability, including for example, PEG graft density and the molecular weight of the PEG grafts.
One common limitation of polyelectrolyte micelles is their stability in physiological medium. One strategy to stabilize the micelles in ionic buffers is to introduce reversible crosslinks.127, 128 This method was employed to effectively preserve the size and shape of PEG-b-PPA/DNA micelles following solvent exchange to remove organic solvent.125
Shape Transforming Nanoparticles
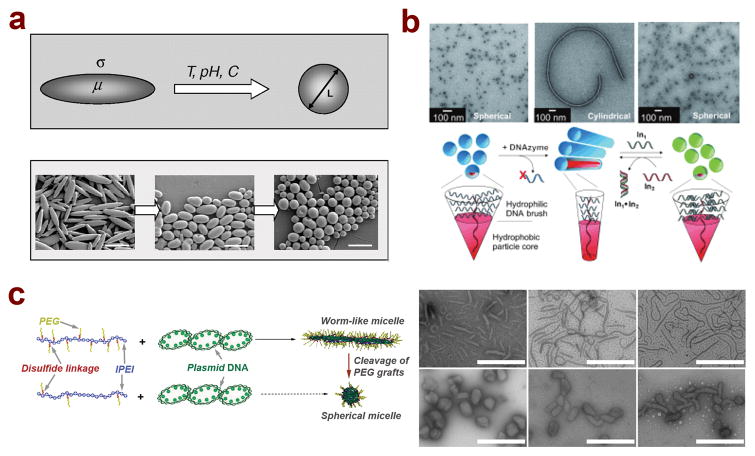
Nanoparticles with discrete shapes and the capability to transform their shape in response to an external stimulus are particularly attractive, as it is possible to combine different sets of properties into one nanoparticle system. A recent study by Yoo and Mitragotri showed that rod-like PLGA particles generated by the particle stretch method were able to transform their shape to spheres in response to a change of external stimulus including pH, temperature, or chemical, during time scales ranging from minutes to hours (Figure 2a).33 Mechanistic studies revealed that a balance between the viscoelastic property of the polymer and the interfacial tension between the polymer and surrounding media were the two main factors influencing shape transformation kinetics. Another study by Chien et al. utilized DNA-amphiphile micelles to demonstrate controllable shape transformation (Figure 2b).91 The DNA brush component of the micelle contained a sequence specific cleavage site, and upon treatment with an enzyme specific for this sequence, the micelles transformed from 25-nm spheres to micron-length cylinders. On the other hand, this shape transition could be reversed by addition of a single strand DNA sequence complementary to the DNA remaining in the micelle, leading to the reformation of 25-nm spheres. Surprising, such a shape transformation process can also be achieved for micellar nanoparticles with a complex core prepared from plasmid DNA and PEG-polycation copolymer. A recent study by Williford et al. reported the worm-like micelles that were prepared by complexation between plasmid DNA and lPEI-g-PEG with a 2 mol% PEG (MW10 KDa) grafted to lPEI (MW17 KDa) through disulfide linkages. When these micellar nanoparticles were subjected to a reducing environment such as that in cytosol and cell nuclei, the PEG chains could be cleaved, which triggered the shape transformation from worm-like particles to more condensed spherical and short rod shapes (Figure 2c).126 Zeta potential measurements confirmed the cleavage of PEG grafts, highlighting the importance of PEG for controlling and transforming the shape of PEG-polycation/DNA nanoparticles.
Figure 2.
Several recent studies have developed polymeric nanoparticles capable of shape transformation in response to external stimuli. (A) PLGA polymer particles can transform their shape from rod-like to spherical on time scales ranging from minutes to hours in response to external triggers such as various chemicals, pH changes, and temperature changes;33 (B) Self-assembled DNA brush polymer micelles undergo shape transformation from spherical to cylindrical particles upon enzymatic cleavage of a fraction of the brush segment, which can be reversed by re-introducing a similar DNA segment through complementary base pairing;91 (reprinted with permission, ©2010 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim) (C) Shape transformation of polymer/DNA micelles can be achieved through cleavage of a fraction of the PEG chains on the micelle surface, leading to a transition from worm-like shapes to more condensed spherical and short rod shapes.126 (Reproduced by permission of The Royal Society of Chemistry.)
Comparative Analyses of Different Methods
While several different methods have been developed to generate nanoparticles with controlled shapes, certain considerations must be given to best utilize these materials for therapeutic delivery applications. Lithography-based methods have significant advantages in the generation of highly monodisperse particles, but limitations can exist in the range of shapes possible with the need to design molds for particle preparation. Drug loading capacity must continue to be studied, particularly for biologics such as siRNA. Membrane stretching provides a robust method for generating a range of aspect ratios over a large size range, from hundreds of nanometers to tens of microns. This method, however, limits the materials choice, as the polymer must be heated above the glass transition temperature during the stretching process. The heating process may also hinder the potential cargo for therapeutic delivery. Finally, self-assembly methods require no complex particle manufacturing processes, relying solely on intermolecular forces to generate complex shapes. The self-assembly process, however, has not been well-studied, and few mechanistic studies have been performed to identify the key driving forces for shape control. Molecular dynamics simulation may be an attractive tool for researchers to better understand the mechanisms that drive shape control of self-assembled polymeric nanoparticles. Stability of nanoparticles generated through each method must also continue to be optimized in physiological media for delivery applications. For example, self-assembled micelles may require additional core crosslinking strategies to preserve the size and shape under in vivo conditions. Membrane-stretched particles, comprised of PLA or PLGA, may require some surface conjugation strategies to minimize serum protein adsorption. DNA nanostructures must be designed to minimize susceptibility to nucleases in serum. Finally, the choice of shape control method may be determined by both the materials choice and the payload being delivered. Each method, although versatile for a range of materials and cargos, has specific set of design constraints and optimal conditions that may be tailored for a specific therapeutic molecule and application (see Table 1).
Table 1.
Design Constraints and Preparation Parameters in Shape Control for Different Nanoparticle Preparation Methods
| Nanoparticle Preparation Method | Shapes Prepared | Range of Aspect Ratio | Polymers† | Cargos | |
|---|---|---|---|---|---|
| Lithography | Rods, Cylinders, Cubes, Filamentous 14, 15, 18, 19 | 1 – 60 18, 19 | Polyethylene glycol, poly(lactic acid), poly(lactic acid-co- glycolic acid), proteins 15, 20, 21, 24 | Small molecule drugs, contrast agents, siRNA 17, 21, 25 | |
| S-FIL | Disks, Rods, Squares, Triangles 28–30 | 1 – 8 28, 29 | Polyethylene glycol 28, 29 | Antibodies, nucleic acids, small molecule drugs 28, 30 | |
| Membrane stretching | Rods, Disks, Ellipsoids, Pills, Barrels, Bullets, Lenses 32, 33, 35 | 1.5 – 15 32, 37, 38 | Polystyrene, poly(lactic acid-co- glycolic acid) 33, 35, 38 | Antibodies, small molecule drugs 32, 35, 38 | |
| Self assembly | Amphiphilic copolymers | Spheres, Rods, Filamentous, Vesicles, Toroids, Disks 43–45, 65, 88 | 1 – >100 44, 65, 88 | Polyethylene-block-polyethylene glycol, polycaprolactone-block- polyethylene glycol, poly(lactic acid-co-glycolic acid)- polyethylene glycol, poly(lactic acid)-block-polyethylene glycol, polystyrene-block-poly(acrylic acid) 39, 42, 44, 47, 65, 88 | Hydrophobic drugs loaded in the core 65, 66, 129 |
| Nucleic acid base-pairing | Cages, Polyhedrons, Nanotubes 92, 95, 96, 106 | 1 – 7 92, 95, 106 | Single-stranded DNA 92, 97 | Imaging agents, small molecule drugs, siRNA, proteins 102–106 | |
| Complex core micelles | Spheres, Rods, Worms 120–122, 125, 126 | 1 – 30 122, 125, 126 | Polyphosphoramidate-block- polyethylene glycol, linear polyethylenimine-graft- polyethylene glycol, poly(glutamic acid)-block-polyethylene glycol, polylysine-block-polyethylene glycol 5, 115, 116, 125, 126 | Nucleic acids 5, 116, 125 | |
Effect of Nanoparticle Shape on Their Biological Activities
With the development of several methods for precise control of nanoparticle shape, it has been possible to interrogate the role of shape in nanoparticle-mediated drug and gene delivery. Studies have highlighted the role of shape at each step of the delivery process, including extending nanoparticle circulation time, enhancing tissue-specific delivery, transport and retention, and influencing cellular uptake and intracellular trafficking. The following sections will provide a brief overview on the effect of nanoparticle shape at different delivery steps.
Shape Effect on Systemic Circulation of Nanoparticles
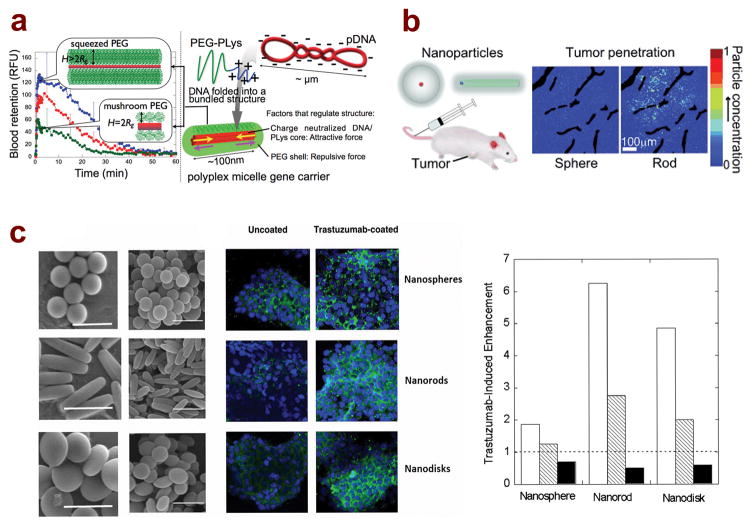
Based on the observation that flexible and long worm-like micelles (> 1 μm) became elongated under shear in flow condition, which may prevent significant interaction with macrophages in circulation, Geng et al. tested the effect of shape on self-assembled nanoparticle circulation time following systemic injection by comparing 3.5-μm long filomicelles with 200-nm spherical micellar nanoparticles.65 They showed that a significant fraction of both the non-degradable polyethylene-b-PEG filomicelles and the degradable PCL-b-PEG micelles circulated for up to one week in rodents after i.v. injection, whereas spherical nanoparticles were cleared within 2 days. The circulation half-life decreased as the initial length of the micelle decreased. The extended circulation time of filomicelles was primarily attributed to their ability to reduce binding and clearance by circulating macrophages, in contrast to spherical particles. More efficient delivery of paclitaxel using these filomicelles was demonstrated in a mouse lung cancer model compared to free paclitaxel. At the same dose of paclitaxel, filomicelles showed significant tumor size decrease with about half the amount of free paclitaxel. Furthermore, the animals were able to tolerate a higher dose of the drug, which could potentially translate to higher degree of tumor killing. Tockary et al. reported a study comparing circulation time of a series of rod-like PEG-b-PLL/DNA micellar nanoparticles with different lengths (Figure 3a).122 From blood circulation profile analysis, nanoparticles with longer rod lengths of 162 nm had a greater percentage of particles remaining in circulation compared to shorter, 70-nm rods. For instance, at 3 minutes post-injection, approximately 60% of the longer rods remained in circulation compared with only 30% of the shorter rods. While these numbers do not show such a striking differences in nanoparticle circulation time as the previous study, it is possible that the differences arise from the fact that these particles are much shorter than the micron-sized filomicelles. These results still highlight the potential benefit of elongated particles in providing extended circulation time.
Figure 3.
Nanoparticle shape influences the biological response both in vitro and in vivo for drug and gene delivery applications. (A) Nanoparticle shape is influenced by the crowdedness of the PEG layer on the surface of polymer/DNA micelles. Dense PEG layer leads to the formation of longer rod shapes that, upon systemic administration via tail vein injection, leads to extended circulation compared to shorter, rod shaped micelles;122 (reprinted with permission, ©2013 American Chemical Society) (B) Greater tumor penetration is observed following i.v. injection of spherical and rod-shaped silica-quantum dot nanoparticles;137 (reprinted with permission, ©2011 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim) (C) Scanning electron microscopy images of polystyrene nanospheres, nanorods, and nanodisks, as well as fluorescent microscopy images of in vitro cellular uptake of shaped nanoparticles (green) in BT-474 breast cancer cells (blue) comparing uncoated and antibody (trastuzumab)-coated particles. Graph shows that trastuzumab coating enhancement is greatest for nanorods, followed by nanodisks and nanospheres in both BT-474 cells (white bars) and SK-BR-3 cells (dashed bars). No enhancement was observed for any shape in MDA-MB-231 cells (black bars).35
Nanoparticle shape may also play a role in margination i.e. drifting of a particle from the center of the vessel close to the vessel wall.130 Nanoparticle margination dynamics may be important in binding to the endothelium and escaping leaky vasculature in tumor tissue. Theoretical analysis indicates that spherical particles under flow conditions tend to follow a streamline parallel to a vessel wall, whereas non-spherical ellipsoidal particles exhibit more complex rotational and translation trajectories, drifting from one side of the vessel to the other during flow.131, 132 Using a detailed model of particle dynamics in microcirculation where low shear rates are present, elongated, discoidal particles display the greatest ability to marginate; however, the lateral velocity of the particle depends significantly on its Stokes number, which is influenced by the size and the density of the material.133 Additionally, when comparing a nanosphere and nanorod under identical shear conditions, the binding probability was three-fold higher for the nanorod.132 A study looking specifically at the effect of shear rate on the margination of non-spherical, micron-sized silica particles found that for the ranges tested, discoidal shapes marginated the most, followed by quasi-hemispherical and spherical particles.134 In vitro studies on effect of nanoparticle shape under flow condition using microfluidic flow chamber experiments, also using the silica-based particles,134 confirmed that spherical particles exhibit significantly lower sedimentation and binding to the walls of the chamber compared to hemispherical and discoidal particles. It will be interesting to see if the results hold for shaped, polymeric nanoparticles as well.
Shape Effect on Tissue Binding and Distribution of Nanoparticles
Nanoparticle shape has been shown to significantly influence tissue binding and distribution in vivo. Using the membrane-stretched PS nanoparticles surface-conjugated with antibodies against ICAM-1 and transferrin for lung and brain targeting, respectively, Kolhar et al. demonstrated that anti-ICAM-1-conjugated rods (500 nm length × 125 nm diameter) yielded twice the accumulation in lungs compared to 200-nm spherical particles.38 Lung to liver accumulation was also 1.7-fold higher for the rods, compared with 0.7 for the spheres, showing that rod-shaped particles improved tissue-specific binding. Similarly for anti-transferrin-conjugated nanoparticles, rod-shaped particles showed a nearly 7-fold higher accumulation in brain compared to spherical particles. In vitro studies using a synthetic microvascular network model complemented the in vivo results, showing higher binding to the walls of the chamber for the rod-shaped, antibody-conjugated particles, compared with antibody-conjugated spherical particles. Additional studies incorporating bifurcation junctions into the microvascular networks found that particles accumulated more at the branching sites compared to the straight segments, which may have application in certain disease states.135 Another study by Shuvaev et al. using PEG-b-polyethylene filomicelles decorated with endothelium-targeting antibodies also showed combined advantages of extended circulation time and improved tissue binding for these worm-like particles, highlighting the importance of nanoparticle shape for improving transport and targeting capabilities.136
In addition to tissue targeting, nanoparticle shape has the potential to influence their penetration and distribution once particles transport from the vessels to the tissue. Chu et al. reported that PLGA particles formulated by the PRINT method exhibited significantly different tissue distribution of the encapsulated drug, docetaxel, in a particle shape-dependent manner.17 Rod-like particles (80 nm diameter × 320 nm length) delivered a higher concentration of docetaxel in the tumor tissue and lower spleen and liver deposition, compared with cube particles (200 nm × 200 nm). Such an improved delivery with rod-like particles was correlated with better permeation or diffusion of these particles in the tumor tissue and a reduction in clearance by macrophages. These results are consistent with a recent study by Chauhan et al. using silica nanoparticles and nanorods containing a quantum dot core (Figure 3b). Tumor tissue distribution for the rod-like nanoparticles (15 nm × 54 nm) was greater than the 35-nm spherical particles, as a result of enhanced penetration and diffusion property for the rods in tissue or gel-like medium.137
While the literature is not as extensive for gene therapy applications, several studies have begun to focus on the effect of DNA nanoparticle shape on in vivo gene delivery efficiency. Dirisala et al., using self-assembled PEG-b-PLL/DNA nanoparticles carrying plasmid DNA encoding anti-angiogenic proteins for pancreatic cancer delivery, identified a critical rod length of 200 nm that is capable of mediating efficient transfection efficiency both in vitro and in vivo.121 Above a 200-nm length, rod-shaped nanoparticles showed lower cell uptake in BxPC3 cells and minimal antitumor effect in mice following i.v. injection, even though these nanoparticles displayed extended circulation time. Rods shorter than 200 nm, on the other hand, exhibited significant antitumor response through reduction in tumor volume as well as decreased tumor vascular density upon treatment. Jiang et al. used retrograde intrabiliary infusion to deliver shaped, PEG-b-PPA/DNA micelles to the rat liver.125 In this study, worm-like particles (20 nm diameter × 581 nm length) mediated 126-fold higher transfection efficiency than rod-like particles (30 nm diameter × 130 nm length), and 1,680-fold higher expression than the spherical particles with an average diameter of 40 nm.
Shape Effect on Cellular Uptake of Nanoparticles
The shape effect of nanoparticles has been most extensively characterized at the cellular level. A study by Gratton et al. using cationic PEG particles prepared by the PRINT method found that longer rod particles (150 nm diameter × 450 nm length) had faster internalization rates in HeLa cells compared to short rods (200 nm diameter × 200 nm length).18 As the particles were positively charged, it is likely that the higher aspect ratio particles were able to experience multivalent interactions with the cells, leading to faster uptake. Results for hyaluronic acid (HA)-coated PLGA PRINT particles also showed similar trends: 320 nm × 80 nm rod particles showed nearly 10-fold higher uptake than 200 nm × 200 nm short rods in BT-20 triple negative breast cancer cells.27 Agarwal et al. similarly studied the uptake of hydrogel nanorod and nanodisc particles in several epithelial and endothelial cell lines.29 In all cell types, the nanodiscs are internalized more than the nanorods. From the results, the authors proposed three key factors that dictate cell uptake in vitro: contact force between the particle and the cell surface, sedimentation, and strain energy needed for membrane deformation to uptake the particles. The strain energy, in particular, is hypothesized to be the reason for the increased uptake of nanodiscs compared to nanorods, as more energy is needed to engulf the elongated rod-shaped particles. Polystyrene rods with a size of 367 nm × 126 nm, prepared by the membrane stretching method, displayed higher specific uptake and lower nonspecific uptake compared to 200 nm spherical and disk-shaped (236 nm diameter × 88 nm thickness) nanoparticles in three breast cancer cell lines following surface conjugation of trastuzumab, a HER2-targeted antibody (Figure 3c).35 On the other hand, a study comparing cellular uptake of spherical, short cylinder, and long cylinder, PAA-b-PS block copolymer nanoparticles conjugated with TAT cell adhesion peptide found that spherical particles exhibited the highest amount of uptake in Chinese hamster ovary cells.78 The results suggest that it may be difficult for the micron-length cylindrical particles to be internalized by the cell; in addition, particle uptake was compared at the same molar concentration of polymer, meaning that there was a higher number of spherical particles compared to worm-like particles, which may affect the measurements. It is also possible that uptake of different shapes will vary significantly depending on material choice or cell type being investigated.
Several studies have also begun to investigate shape-dependent uptake and intracellular trafficking of DNA-containing nanoparticles. Shi et al., using HPMA-b-PLL copolymers to condense DNA, compared cellular uptake, intracellular trafficking, and transfection efficiency for 25 nm × 74 nm oblong complex core micelles and 18 nm × 102 nm rod-like micelles.124 The transfection efficiency for the oblong micelles was 42-fold higher than the rod-like micelles with a higher aspect ratio. Mechanistic studies showed that cellular uptake for the more condensed particles was approximately 4-fold higher, whereas the rod-like particles, after internalization, tended to accumulate more in endosomal/lysosomal compartments. Other factors, such as charge density, DNA unpacking, and intracellular trafficking kinetics, did not differ significantly for the two nanoparticles, suggesting that shape-dependent cellular uptake may be a determining factor in the observed transfection efficiency differences. Our recent studies also showed that nanoparticle shape may similarly influence cell uptake and transfection efficiency.125, 126 In addition, as polymer/DNA micelles undergoing shape transformation from worm-like to condensed spherical and short rod shapes, a significant increase in in vitro transfection efficiency was observed, whereas control particles without shape transformation did not increase transfection efficiency.126 However, it is important to note that in this study shape transformation occurred concomitantly with PEG cleavage, therefore both shape transformation and increase in nanoparticle surface charge may contribute to the improved transfection results.
Significant discoveries have been made on shape-dependent uptake of microparticles and nanoparticles in phagocytic cells. Using high aspect ratio disc and worm-like particles, results highlight that shape, but not necessarily size, dictates the degree of particle internalization by macrophages.36, 37 For example, for 14 μm × 3 μm ellipsoidal particles, when the macrophage attaches to the end of the particle, it can be effectively internalized within 3 min.36 When it attaches to the flat side of the particle, however, no internalization was observed for the duration of the 110 min study. Extending from these observations, a follow-up study compared macrophage internalization of 1–3 μm spherical particles with fiber-like particles stretched from the same set of spherical particles to an aspect ratio of 22.5.37 The fiber-like particles displayed almost no internalization, whereas approximately half of the spherical particles that attached to the macrophages were internalized. This study highlights the ability to use shape as sole factor to generate a particle that can effectively evade phagocytosis. As another example highlighting the utility of shape to influence macrophage uptake, PLGA particles capable of shape transformation from elliptical disks with an aspect ratio of 5 to spherical particles were compared to control particles unable to transform their shape.33 When incubated in vitro with macrophages, particles were internalized quickly upon shape transformation to near-spherical shapes. In contrast, cells could not phagocytose elliptical disks without shape transformation.
Conclusions
Using the methods presented here, it has been shown that precise shape control can be achieved for polymeric nanoparticles, obtaining a range of nanoparticle shapes including spherical and cylindrical particles, all the way to micron sized, high aspect ratio rod and filamentous particles. Methods are amenable to a wide range of polymeric systems, both non-degradable and degradable, and can deliver cargo ranging from chemotherapeutics, nucleic acids, and contrast agents. Applications of these shape-controlled nanoparticles in vitro and in vivo has revealed shape-dependent nanoparticle transport properties, allowing particle shape optimization to extend circulation time following systemic injection, improve tissue penetration and distribution, enhance binding to target cells and tissues, and direct cellular uptake and intracellular trafficking behavior. While these findings highlight the promise of shape control for improving the delivery efficiency of polymeric nanoparticles, significant challenges remain to be addressed. As shape is one of the fundamental attributes of nanoparticles, along with size, surface chemistry, charge, surface energetics, etc., it may be difficult to isolate the specific shape effect from other parameters on biological transport properties and bioactivities of the nanoparticles. New strategies are needed to independently control nanoparticle shape and other parameters in order to identify shape as the sole governing factor.
Further evaluation of the shape in a biological context is also necessary; it is possible that, upon exposure to the complex biological environment, the shape factor could be masked by serum proteins or biological media. Finally, reproducibility, uniformity, and scalability must continue to be addressed in order to identify key formulations for potential clinical development. These considerations aside, it is clear that nanoparticle shape holds great potential as an additional design parameter for improving therapeutic delivery. The continued development of new methods and materials for such shape control provides for an exciting future for nanomedicine.
Supplementary Material
Acknowledgments
We would like to acknowledge funding support provided by NIH grants R21EB015152, R21EB013274, and U54CA151838, as well as the Defense Threat Reduction Agency Grant W81XWH-13-2-0037.
Biographies
John-Michael Williford is a Ph.D. candidate in the Biomedical Engineering Program at Johns Hopkins University School of Medicine. His thesis research in Prof. Hai-Quan Mao’s Lab involves developing methods and mechanism of shape control of polymer/DNA nanoparticles and engineering shape tunability in DNA nanoparticles for gene delivery applications. John-Michael received his B.S. in biomedical engineering from The University of Akron in 2010.
Dr. Jose Luis Santos completed his Ph.D. in Materials Chemistry at the University of Madeira in Portugal in 2009. His graduate research focused on the development of dendrimers to deliver nucleic acids to stem cells for regenerative medicine applications. He conducted his postdoctoral research on dynamic self-assembly of block copolymers for nanoparticle engineering with Prof. Margarita Herrera-Alonso in the Department of Materials Science and Engineering at Johns Hopkins University from 2010 to 2014. Currently, Dr. Santos is a research associate in Prof. Hai-Quan Mao’s Lab at the Institute for NanoBioTechnology developing polyelectrolyte nanoparticles for therapeutic delivery.
Rishab Shyam is a Ph.D. candidate in the Department of Biomedical Engineering at Johns Hopkins University School of Medicine under the mentorship of Prof. Philip Wong and Prof. Hai-Quan Mao. His thesis research focuses on the development of polymer-based siRNA nanoparticles for gene knockdown in the central nervous system for therapeutic targets implicated in the Alzheimer’s Disease. He received his B.S. in chemical engineering from Purdue University in 2006.
Dr. Hai-Quan Mao is a professor at the Department of Materials Science and Engineering and the Whitaker Biomedical Engineering Institute at Johns Hopkins University and is a member of the Translational Tissue Engineering Center and the Institute for NanoBioTechnology at Johns Hopkins University. Dr. Mao’s research focuses on engineering nanomaterials for therapeutic delivery and regenerative medicine applications. Dr. Mao received his B.S. in chemistry in 1988 and Ph.D. in polymer chemistry in 1993 from Wuhan University in China and completed his postdoctoral training in the Department of Biomedical Engineering at Johns Hopkins University School of Medicine in 1998.
References
- 1.Davis ME, Chen Z, Shin DM. Nat Rev Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 2.Parveen S, Misra R, Sahoo SK. Nanomedicine. 2012;8:147–166. doi: 10.1016/j.nano.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 3.Shi JJ, Votruba AR, Farokhzad OC, Langer R. Nano Lett. 2010;10:3223–3230. doi: 10.1021/nl102184c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perfezou M, Turner A, Merkoci A. Chem Soc Rev. 2012;41:2606–2622. doi: 10.1039/c1cs15134g. [DOI] [PubMed] [Google Scholar]
- 5.Pack DW, Hoffman AS, Pun S, Stayton PS. Nat Rev Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 6.Peer D, Karp JM, Hong S, FaroKHzad OC, Margalit R, Langer R. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 7.Williford JM, Wu J, Ren Y, Archang MM, Leong KW, Mao HQ. Annu Rev Biomed Eng. 2014;16:347–370. doi: 10.1146/annurev-bioeng-071813-105119. [DOI] [PubMed] [Google Scholar]
- 8.Pisal DS, Kosloski MP, Balu-Iyer SV. J Pharm Sci. 2010;99:2557–2575. doi: 10.1002/jps.22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janib SM, Moses AS, MacKay JA. Adv Drug Deliver Rev. 2010;62:1052–1063. doi: 10.1016/j.addr.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bae YH, Park K. J Control Release. 2011;153:198–205. doi: 10.1016/j.jconrel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park K. ACS Nano. 2013;7:7442–7447. doi: 10.1021/nn404501g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baron S. Medical Microbiology. 4. University of Texas Medical Branch at Galveston; Galveston, Tex: 1996. [PubMed] [Google Scholar]
- 13.Murray Baron PR, Pfaller EJ, Tenover MA, YRH FC American Society for Microbiology. Manual of Clinical Microbiology. 7. ASM Press; Washington, D.C: 1999. [Google Scholar]
- 14.Xu J, Wong DH, Byrne JD, Chen K, Bowerman C, DeSimone JM. Angew Chem Int Ed Engl. 2013;52:6580–6589. doi: 10.1002/anie.201209145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rolland JP, Maynor BW, Euliss LE, Exner AE, Denison GM, DeSimone JM. J Am Chem Soc. 2005;127:10096–10100. doi: 10.1021/ja051977c. [DOI] [PubMed] [Google Scholar]
- 16.Gratton SE, Pohlhaus PD, Lee J, Guo J, Cho MJ, Desimone JM. J Control Release. 2007;121:10–18. doi: 10.1016/j.jconrel.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu KS, Hasan W, Rawal S, Walsh MD, Enlow EM, Luft JC, Bridges AS, Kuijer JL, Napier ME, Zamboni WC, DeSimone JM. Nanomedicine. 2013;9:686–693. doi: 10.1016/j.nano.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gratton SEA, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. Proc Natl Acad Sci USA. 2008;105:11613–11618. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kersey FR, Merkel TJ, Perry JL, Napier ME, DeSimone JM. Langmuir. 2012;28:8773–8781. doi: 10.1021/la301279v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunn SS, Tian S, Blake S, Wang J, Galloway AL, Murphy A, Pohlhaus PD, Rolland JP, Napier ME, DeSimone JM. J Am Chem Soc. 2012;134:7423–7430. doi: 10.1021/ja300174v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasan W, Chu K, Gullapalli A, Dunn SS, Enlow EM, Luft JC, Tian S, Napier ME, Pohlhaus PD, Rolland JP, DeSimone JM. Nano Lett. 2012;12:287–292. doi: 10.1021/nl2035354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enlow EM, Luft JC, Napier ME, DeSimone JM. Nano Lett. 2011;11:808–813. doi: 10.1021/nl104117p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly JY, DeSimone JM. J Am Chem Soc. 2008;130:5438–5439. doi: 10.1021/ja8014428. [DOI] [PubMed] [Google Scholar]
- 24.Xu J, Wang J, Luft JC, Tian S, Owens G, Jr, Pandya AA, Berglund P, Pohlhaus P, Maynor BW, Smith J, Hubby B, Napier ME, DeSimone JM. J Am Chem Soc. 2012;134:8774–8777. doi: 10.1021/ja302363r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nunes J, Herlihy KP, Mair L, Superfine R, DeSimone JM. Nano Lett. 2010;10:1113–1119. doi: 10.1021/nl904152e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Nunes JK, Gratton SEA, Herlihy KP, Pohlhaus PD, DeSimone JM. New J Phys. 2009:11. [Google Scholar]
- 27.Morton SW, Herlihy KP, Shopsowitz KE, Deng ZJ, Chu KS, Bowerman CJ, Desimone JM, Hammond PT. Adv Mater. 2013;25:4707–4713. doi: 10.1002/adma.201302025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glangchai LC, Caldorera-Moore M, Shi L, Roy K. J Control Release. 2008;125:263–272. doi: 10.1016/j.jconrel.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 29.Agarwal R, Singh V, Jurney P, Shi L, Sreenivasan SV, Roy K. Proc Natl Acad Sci USA. 2013;110:17247–17252. doi: 10.1073/pnas.1305000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwal R, Singh V, Jurney P, Shi L, Sreenivasan SV, Roy K. ACS Nano. 2012;6:2524–2531. doi: 10.1021/nn2049152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caldorera-Moore M, Kang MK, Moore Z, Singh V, Sreenivasan SV, Shi L, Huang R, Roy K. Soft Matter. 2011;7:2879–2887. [Google Scholar]
- 32.Champion JA, Katare YK, Mitragotri S. Proc Natl Acad Sci USA. 2007;104:11901–11904. doi: 10.1073/pnas.0705326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo JW, Mitragotri S. Proc Natl Acad Sci USA. 2010;107:11205–11210. doi: 10.1073/pnas.1000346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoo JW, Doshi N, Mitragotri S. Macromol Rapid Comm. 2010;31:142–148. doi: 10.1002/marc.200900592. [DOI] [PubMed] [Google Scholar]
- 35.Barua S, Yoo JW, Kolhar P, Wakankar A, Gokarn YR, Mitragotri S. Proc Natl Acad Sci USA. 2013;110:3270–3275. doi: 10.1073/pnas.1216893110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Champion JA, Mitragotri S. Proc Natl Acad Sci USA. 2006;103:4930–4934. doi: 10.1073/pnas.0600997103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Champion JA, Mitragotri S. Pharm Res. 2009;26:244–249. doi: 10.1007/s11095-008-9626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolhar P, Anselmo AC, Gupta V, Pant K, Prabhakarpandian B, Ruoslahti E, Mitragotri S. Proc Natl Acad Sci USA. 2013;110:10753–10758. doi: 10.1073/pnas.1308345110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim JK, Yang SY, Lee Y, Kim Y. Prog Polym Sci. 2010;35:1325–1349. [Google Scholar]
- 40.Kuila BK, Stamm M. J Mater Chem. 2010;20:6086–6094. [Google Scholar]
- 41.Orilall MC, Wiesner U. Chem Soc Rev. 2011;40:520–535. doi: 10.1039/c0cs00034e. [DOI] [PubMed] [Google Scholar]
- 42.Park C, Yoon J, Thomas EL. Polymer. 2003;44:7779–7779. [Google Scholar]
- 43.Cui HG, Chen ZY, Zhong S, Wooley KL, Pochan DJ. Science. 2007;317:647–650. doi: 10.1126/science.1141768. [DOI] [PubMed] [Google Scholar]
- 44.Hayward RC, Pochan DJ. Macromolecules. 2010;43:3577–3584. [Google Scholar]
- 45.Holder SJ, Sommerdijk NAJM. Polym Chem. 2011;2:1018–1028. [Google Scholar]
- 46.Broz P, Driamov S, Ziegler J, Ben-Haim N, Marsch S, Meier W, Hunziker P. Nano Lett. 2006;6:2349–2353. doi: 10.1021/nl0619305. [DOI] [PubMed] [Google Scholar]
- 47.Tyrrell ZL, Shen YQ, Radosz M. Prog Polym Sci. 2010;35:1128–1143. [Google Scholar]
- 48.D’Addio SM, Prud’homme RK. Adv Drug Deliver Rev. 2011;63:417–426. doi: 10.1016/j.addr.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 49.Valencia PM, Farokhzad OC, Karnik R, Langer R. Nat Nanotechnol. 2012;7:623–629. doi: 10.1038/nnano.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang CW, Sinton D, Moffitt MG. ACS Nano. 2013;7:1424–1436. doi: 10.1021/nn305197m. [DOI] [PubMed] [Google Scholar]
- 51.Wang CW, Sinton D, Moffitt MG. J Am Chem Soc. 2011;133:18853–18864. doi: 10.1021/ja2067252. [DOI] [PubMed] [Google Scholar]
- 52.Wang CW, Bains A, Sinton D, Moffitt MG. Langmuir. 2012;28:15756–15761. doi: 10.1021/la303655s. [DOI] [PubMed] [Google Scholar]
- 53.Luo HY, Santos JL, Herrera-Alonso M. Chem Commun. 2014;50:536–538. doi: 10.1039/c3cc46834h. [DOI] [PubMed] [Google Scholar]
- 54.Thiele J, Steinhauser D, Pfohl T, Forster S. Langmuir. 2010;26:6860–6863. doi: 10.1021/la904163v. [DOI] [PubMed] [Google Scholar]
- 55.Bae Y, Fukushima S, Harada A, Kataoka K. Angew Chem Int Ed Engl. 2003;42:4640–4643. doi: 10.1002/anie.200250653. [DOI] [PubMed] [Google Scholar]
- 56.Tong R, Cheng JJ. Angew Chem Int Ed Engl. 2008;47:4830–4834. doi: 10.1002/anie.200800491. [DOI] [PubMed] [Google Scholar]
- 57.Tong R, Cheng JJ. J Am Chem Soc. 2009;131:4744–4754. doi: 10.1021/ja8084675. [DOI] [PubMed] [Google Scholar]
- 58.Tong R, Cheng JJ. Bioconjugate Chem. 2010;21:111–121. doi: 10.1021/bc900356g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yen HC, Cabral H, Mi P, Toh K, Matsumoto Y, Liu X, Koori H, Kim A, Miyazaki K, Miura Y, Nishiyama N, Kataoka K. ACS Nano. 2014;8:11591–11602. doi: 10.1021/nn504836s. [DOI] [PubMed] [Google Scholar]
- 60.Yu Y, Chen CK, Law WC, Mok J, Zou J, Prasad PN, Cheng C. Mol Pharm. 2013;10:867–874. doi: 10.1021/mp3004868. [DOI] [PubMed] [Google Scholar]
- 61.Aguirre-Chagala YE, Santos JL, Huang Y, Herrera-Alonso M. ACS Macro Lett. 2014;3:1249–1253. doi: 10.1021/mz500594m. [DOI] [PubMed] [Google Scholar]
- 62.Zhang SY, Zou J, Elsabahy M, Karwa A, Li A, Moore DA, Dorshow RB, Wooley KL. Chem Sci. 2013;4:2122–2126. doi: 10.1039/C3SC50252J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zou J, Zhang FW, Zhang SY, Pollack SF, Elsabahy M, Fan JW, Wooley KL. Adv Healthc Mater. 2014;3:441–448. doi: 10.1002/adhm.201300235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu XL, Hu JM, Tian J, Ge ZS, Zhang GY, Luo KF, Liu SY. J Am Chem Soc. 2013;135:17617–17629. doi: 10.1021/ja409686x. [DOI] [PubMed] [Google Scholar]
- 65.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Nat Nanotechnol. 2007;2:249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim S, Shi Y, Kim JY, Park K, Cheng JX. Expert Opin Drug Deliv. 2010;7:49–62. doi: 10.1517/17425240903380446. [DOI] [PubMed] [Google Scholar]
- 67.Rijcken CJ, Snel CJ, Schiffelers RM, van Nostrum CF, Hennink WE. Biomaterials. 2007;28:5581–5593. doi: 10.1016/j.biomaterials.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 68.Talelli M, Iman M, Varkouhi AK, Rijcken CJF, Schiffelers RM, Etrych T, Ulbrich K, van Nostrum CF, Lammers T, Storm G, Hennink WE. Biomaterials. 2010;31:7797–7804. doi: 10.1016/j.biomaterials.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 69.Zhang XL, Liu K, Huang YX, Xu JN, Li J, Ma XC, Li S. Bioconjugate Chem. 2014;25:1689–1696. doi: 10.1021/bc500292j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O’Reilly RK, Hawker CJ, Wooley KL. Chem Soc Rev. 2006;35:1068–1083. doi: 10.1039/b514858h. [DOI] [PubMed] [Google Scholar]
- 71.Nystrom AM, Wooley KL. Acc Chem Res. 2011;44:969–978. doi: 10.1021/ar200097k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jin J, Zhang MM, Xiong QQ, Sun PC, Zhao HY. Soft Matter. 2012;8:11809–11816. [Google Scholar]
- 73.Li Y, Xiao W, Xiao K, Berti L, Luo J, Tseng HP, Fung G, Lam KS. Angew Chem Int Ed Engl. 2012;51:2864–2869. doi: 10.1002/anie.201107144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang H, Tang L, Tu CL, Song ZY, Yin Q, Yin LC, Zhang ZH, Cheng JJ. Biomacromolecules. 2013;14:3706–3712. doi: 10.1021/bm401086d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shuai XT, Merdan T, Schaper AK, Xi F, Kissel T. Bioconjugate Chem. 2004;15:441–448. doi: 10.1021/bc034113u. [DOI] [PubMed] [Google Scholar]
- 76.Nystrom AM, Wooley KL. Tetrahedron. 2008;64:8543–8552. doi: 10.1016/j.tet.2008.04.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fang H, Zhang K, Shen G, Woolley KL, Taylor JSA. Mol Pharm. 2009;6:615–626. doi: 10.1021/mp800199w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang K, Fang HF, Chen ZY, Taylor JSA, Wooley KL. Bioconjugate Chem. 2008;19:1880–1887. doi: 10.1021/bc800160b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao P, Liu LX, Feng XQ, Wang C, Shuai XT, Chen YM. Macromol Rapid Comm. 2012;33:1351–1355. doi: 10.1002/marc.201200172. [DOI] [PubMed] [Google Scholar]
- 80.Guo J, Hong H, Chen G, Shi S, Nayak TR, Theuer CP, Barnhart TE, Cai W, Gong S. ACS Appl Mater Interfaces. 2014;6:21769–21779. doi: 10.1021/am5002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sowers MA, McCombs JR, Wang Y, Paletta JT, Morton SW, Dreaden EC, Boska MD, Ottaviani MF, Hammond PT, Rajca A, Johnson JA. Nat Commun. 2014;5:5460. doi: 10.1038/ncomms6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee HI, Pietrasik J, Sheiko SS, Matyjaszewski K. Prog Polym Sci. 2010;35:24–44. [Google Scholar]
- 83.Sheiko SS, Sumerlin BS, Matyjaszewski K. Prog Polym Sci. 2008;33:759–785. [Google Scholar]
- 84.Johnson JA, Lu YY, Burts AO, Lim YH, Finn MG, Koberstein JT, Turro NJ, Tirrell DA, Grubbs RH. J Am Chem Soc. 2011;133:559–566. doi: 10.1021/ja108441d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Johnson JA, Lu YY, Burts AO, Xia Y, Durrell AC, Tirrell DA, Grubbs RH. Macromolecules. 2010;43:10326–10335. doi: 10.1021/ma1021506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zou J, Jafr G, Themistou E, Yap Y, Wintrob ZA, Alexandridis P, Ceacareanu AC, Cheng C. Chem Commun. 2011;47:4493–4495. doi: 10.1039/c0cc05531j. [DOI] [PubMed] [Google Scholar]
- 87.Shen HW, Eisenberg A. J Phys Chem B. 1999;103:9473–9487. [Google Scholar]
- 88.Choucair A, Eisenberg A. Eur Phys J E Soft Matter. 2003;10:37–44. doi: 10.1140/epje/e2003-00002-5. [DOI] [PubMed] [Google Scholar]
- 89.Kwak M, Herrmann A. Chem Soc Rev. 2011;40:5745–5755. doi: 10.1039/c1cs15138j. [DOI] [PubMed] [Google Scholar]
- 90.Alemdaroglu FE, Alemdaroglu NC, Langguth P, Herrmann A. Macromol Rapid Comm. 2008;29:326–329. [Google Scholar]
- 91.Chien MP, Rush AM, Thompson MP, Gianneschi NC. Angew Chem Int Ed Engl. 2010;49:5076–5080. doi: 10.1002/anie.201000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li J, Fan C, Pei H, Shi J, Huang Q. Adv Mater. 2013;25:4386–4396. doi: 10.1002/adma.201300875. [DOI] [PubMed] [Google Scholar]
- 93.Goodman RP, Heilemann M, Doose S, Erben CM, Kapanidis AN, Turberfield AJ. Nat Nanotechnol. 2008;3:93–96. doi: 10.1038/nnano.2008.3. [DOI] [PubMed] [Google Scholar]
- 94.Aldaye FA, Lo PK, Karam P, McLaughlin CK, Cosa G, Sleiman HF. Nat Nanotechnol. 2009;4:349–352. doi: 10.1038/nnano.2009.72. [DOI] [PubMed] [Google Scholar]
- 95.Qian H, Tian C, Yu J, Guo F, Zheng MS, Jiang W, Dong QF, Mao C. Small. 2014;10:855–858. doi: 10.1002/smll.201301891. [DOI] [PubMed] [Google Scholar]
- 96.Zhang C, Li X, Tian C, Yu G, Li Y, Jiang W, Mao C. ACS Nano. 2014;8:1130–1135. doi: 10.1021/nn406039p. [DOI] [PubMed] [Google Scholar]
- 97.Torring T, Voigt NV, Nangreave J, Yan H, Gothelf KV. Chem Soc Rev. 2011;40:5636–5646. doi: 10.1039/c1cs15057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hahn J, Wickham SFJ, Shih WM, Perrault SD. ACS Nano. 2014;8:8765–8775. doi: 10.1021/nn503513p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Keum J-W, Bermudez H. Chem Commun. 2009:7036–7038. doi: 10.1039/b917661f. [DOI] [PubMed] [Google Scholar]
- 100.Mei Q, Wei X, Su F, Liu Y, Youngbull C, Johnson R, Lindsay S, Yan H, Meldrum D. Nano Lett. 2011;11:1477–1482. doi: 10.1021/nl1040836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhao YX, Shaw A, Zeng X, Benson E, Nystrom AM, Hogberg B. ACS Nano. 2012;6:8684–8691. doi: 10.1021/nn3022662. [DOI] [PubMed] [Google Scholar]
- 102.Ko S, Liu H, Chen Y, Mao C. Biomacromolecules. 2008;9:3039–3043. doi: 10.1021/bm800479e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Douglas SM, Bachelet I, Church GM. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 104.Liu X, Xu Y, Yu T, Clifford C, Liu Y, Yan H, Chang Y. Nano Lett. 2012;12:4254–4259. doi: 10.1021/nl301877k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Crawford R, Erben CM, Periz J, Hall LM, Brown T, Turberfield AJ, Kapanidis AN. Angew Chem Int Ed Engl. 2013;52:2284–2288. doi: 10.1002/anie.201207914. [DOI] [PubMed] [Google Scholar]
- 106.Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, Peng CG, Charisse K, Borodovsky A, Manoharan M, Donahoe JS, Truelove J, Nahrendorf M, Langer R, Anderson DG. Nat Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grabow WW, Jaeger L. Acc Chem Res. 2014;47:1871–1880. doi: 10.1021/ar500076k. [DOI] [PubMed] [Google Scholar]
- 108.Shu D, Moll WD, Deng Z, Mao C, Guo P. Nano Lett. 2004;4:1717–1723. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ohno H, Kobayashi T, Kabata R, Endo K, Iwasa T, Yoshimura SH, Takeyasu K, Inoue T, Saito H. Nat Nanotechnol. 2011;6:116–120. doi: 10.1038/nnano.2010.268. [DOI] [PubMed] [Google Scholar]
- 110.Afonin KA, Kasprzak WK, Bindewald E, Kireeva M, Viard M, Kashlev M, Shapiro BA. Acc Chem Res. 2014;47:1731–1741. doi: 10.1021/ar400329z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim T, Afonin KA, Viard M, Koyfman AY, Sparks S, Heldman E, Grinberg S, Linder C, Blumenthal RP, Shapiro BA. Mol Ther Nucleic Acids. 2013;2:e80. doi: 10.1038/mtna.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shopsowitz KE, Roh YH, Deng ZJ, Morton SW, Hammond PT. Small. 2014;10:1623–1633. doi: 10.1002/smll.201302676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. Nat Mater. 2012;11:316–322. doi: 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Voets IK, de Keizer A, Cohen Stuart MA. Adv Colloid Interface Sci. 2009;147–148:300–318. doi: 10.1016/j.cis.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 115.Kataoka K, Harada A, Nagasaki Y. Adv Drug Deliver Rev. 2001;47:113–131. doi: 10.1016/s0169-409x(00)00124-1. [DOI] [PubMed] [Google Scholar]
- 116.Osada K, Christie RJ, Kataoka K. J R Soc Interface. 2009;6(Suppl 3):S325–339. doi: 10.1098/rsif.2008.0547.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen JH, Seeman NC. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 118.Goodman RP, Schaap IA, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 119.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 120.Osada K, Shiotani T, Tockary TA, Kobayashi D, Oshima H, Ikeda S, Christie RJ, Itaka K, Kataoka K. Biomaterials. 2012;33:325–332. doi: 10.1016/j.biomaterials.2011.09.046. [DOI] [PubMed] [Google Scholar]
- 121.Dirisala A, Osada K, Chen Q, Tockary TA, Machitani K, Osawa S, Liu X, Ishii T, Miyata K, Oba M, Uchida S, Itaka K, Kataoka K. Biomaterials. 2014;35:5359–5368. doi: 10.1016/j.biomaterials.2014.03.037. [DOI] [PubMed] [Google Scholar]
- 122.Tockary TA, Osada K, Chen Q, Machitani K, Dirisala A, Uchida S, Nomoto T, Toh K, Matsumoto Y, Itaka K, Nitta K, Nagayama K, Kataoka K. Macromolecules. 2013;46:6585–6592. [Google Scholar]
- 123.Osada K. Polym J. 2014;46:469–475. [Google Scholar]
- 124.Shi J, Choi JL, Chou B, Johnson RN, Schellinger JG, Pun SH. ACS Nano. 2013;7:10612–10620. doi: 10.1021/nn403069n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jiang X, Qu W, Pan D, Ren Y, Williford JM, Cui H, Luijten E, Mao HQ. Adv Mater. 2013;25:227–232. doi: 10.1002/adma.201202932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Williford JM, Ren Y, Huang K, Pan D, Mao HQ. J Mater Chem B Mater Biol Med. 2014;2:8106–8109. doi: 10.1039/C4TB00967C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jiang X, Zheng Y, Chen HH, Leong KW, Wang TH, Mao HQ. Adv Mater. 2010;22:2556–2560. doi: 10.1002/adma.200903933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li J, Yu X, Wang Y, Yuan Y, Xiao H, Cheng D, Shuai X. Adv Mater. 2014;26:8217–8224. doi: 10.1002/adma.201403877. [DOI] [PubMed] [Google Scholar]
- 129.D’Addio SM, Prud’homme RK. Adv Drug Deliver Rev. 2011;63:417–426. doi: 10.1016/j.addr.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 130.Decuzzi P, Pasqualini R, Arap W, Ferrari M. Pharm Res. 2009;26:235–243. doi: 10.1007/s11095-008-9697-x. [DOI] [PubMed] [Google Scholar]
- 131.Decuzzi P, Lee S, Bhushan B, Ferrari M. Ann Biomed Eng. 2005;33:179–190. doi: 10.1007/s10439-005-8976-5. [DOI] [PubMed] [Google Scholar]
- 132.Shah S, Liu YL, Hu W, Gao JM. J Nanosci Nanotechnol. 2011;11:919–928. doi: 10.1166/jnn.2011.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lee SY, Ferrari M, Decuzzi P. Nanotechnology. 2009;20:495101. doi: 10.1088/0957-4484/20/49/495101. [DOI] [PubMed] [Google Scholar]
- 134.Gentile F, Chiappini C, Fine D, Bhavane RC, Peluccio MS, Cheng MM, Liu X, Ferrari M, Decuzzi P. J Biomech. 2008;41:2312–2318. doi: 10.1016/j.jbiomech.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 135.Doshi N, Prabhakarpandian B, Rea-Ramsey A, Pant K, Sundaram S, Mitragotri S. J Control Release. 2010;146:196–200. doi: 10.1016/j.jconrel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shuvaev VV, Ilies MA, Simone E, Zaitsev S, Kim Y, Cai S, Mahmud A, Dziubla T, Muro S, Discher DE, Muzykantov VR. ACS Nano. 2011;5:6991–6999. doi: 10.1021/nn2015453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chauhan VP, Popovic Z, Chen O, Cui J, Fukumura D, Bawendi MG, Jain RK. Angew Chem Int Ed Engl. 2011;50:11417–11420. doi: 10.1002/anie.201104449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.