Abstract
In the present study, a series of copolymers (PAMD-Ch) was synthesized by grafting polymeric Plerixafor/AMD3100 (PAMD) with different amounts of cholesterol and the effect of cholesterol modification on siRNA delivery was investigated. PAMD-Ch/siRNA polyplexes exhibited improved colloidal and enzymatic stability when compared with PAMD/siRNA polyplexes containing no cholesterol. PAMD-Ch with low (17 wt%) and medium (25 wt%) cholesterol content exhibited CXCR4 antagonism comparable to unmodified PAMD. Cholesterol modification increased cell uptake of siRNA polyplexes and significantly decreased sensitivity of siRNA transfection to the presence of serum. When used to deliver anticancer siRNA against polo-like kinase 1 (PLK1), polyplexes based on PAMD-Ch with 17 wt% cholesterol exhibited the highest cancer cell killing activity both in serum-free and serum-containing conditions. Overall, the results of this study validate cholesterol modified PAMD as dual-function delivery vectors suitable for efficient delivery of anticancer siRNA and simultaneous CXCR4 inhibition for combined anticancer therapies.
Keywords: polyplexes, siRNA, CXCR4, PLK-1, cancer
1. Introduction
Nucleic acids represent a class of promising agents with therapeutic potential in a broad range of diseases – from simple monogenic disorders to complex conditions like cancer. Despite recent progress towards the therapeutic delivery of nucleic acids, there remains a compelling need for development of novel delivery systems for various types of nucleic acids and diseases. In particular, small interfering RNA (siRNA) capable of achieving sequence-specific gene silencing remain one of the most promising nucleic acid therapeutics with potential to treat various diseases.1, 2
Polyelectrolyte complexes of nucleic acids with polycations (polyplexes) have received significant attention in the development of siRNA delivery systems.3 Polyplexes offer multiple advantages in siRNA delivery, including low toxicity, minimal immunogenicity, and versatility in introducing various functional modifications.4, 5 Yet, despite ongoing efforts, the use of siRNA polyplexes remains hindered by a relatively low efficacy when compared with lipid based delivery methods.6, 7 It has been realized that polyplex formulations optimized for delivery of large DNA often perform poorly when delivering siRNA.8 Taking advantage of the chemical versatility of polyplexes, multiple approaches have been developed to improve siRNA delivery by polyplexes and to overcome critical biological hurdles.9, 10 Among the most successful approaches has been modification of polycations with hydrophobic moieties (e.g., cholesterol).11, 12 Cholesterol is a naturally occurring lipid that is readily metabolized in the body and can play an important role in controlling stability and interaction of polyplexes with cell membranes during uptake and intracellular trafficking.13–16 Modification of poly(ethylene imine) (PEI) with cholesterol allowed formulation of polyplexes with high serum compatibility and enhanced cellular uptake due to favorable interactions between cholesterol moiety and cell membrane.17 In another example, bioreducible cholesterol-modified poly(amido amines) were successfully employed for delivery of anti-angiogenic siRNA with promising antitumor activity in vivo.18
In tumors, a complex network of chemokines and chemokine receptors controls cell trafficking into and out of the tumor microenvironment.19 The tumor chemokine network also participates in angiogenesis and generation of the fibroblast stroma. Importantly, chemokines and chemokine receptors are directly involved in the molecular control of cancer metastasis and govern organ-specific homing of metastatic cancer cells.20 Although malignant cells from different types of cancer have different expression profiles of chemokine receptors, CXCR4 is the most widely expressed chemokine receptor in human cancers, making it, and its ligand SDF-1, among the most-promising targets within the chemokine network for novel therapies. Binding of SDF-1 to CXCR4 activates multiple intracellular signaling transduction pathways that regulate survival, proliferation, adhesion, and invasion of cancer cells.21, 22 Evidence supporting the exploration of CXCR4 as a therapeutic target in various cancers stems from experimental in vitro and in vivo studies as well as retrospective clinical studies. The studies have documented increased invasive and metastatic potential in CXCR4-expressing tumor cells.21, 22 The effect of CXCR4 expression on poor clinical outcomes of cancer is also well documented by multiple retrospective clinical studies.23, 24 Available evidence points to the involvement of the CXCR4/SDF-1 axis in both cancer metastasis and primary tumor growth. CXCR4 expression is increased in general in tumor tissues of multiple cancers (e.g., breast, pancreatic, prostate, and lung) and in tumors of patients with metastatic disease.
Current evidence strongly supports antimetastatic potential of CXCR4 inhibition, in particular in combination with other treatment modalities.25 We have recently developed a new approach to the design of nucleic acid delivery systems that takes advantage of the crucial role of CXCR4 in cancer progression. The delivery systems rely on polymeric CXCR4 antagonists for simultaneous nucleic acid delivery and CXCR4 inhibition. We have used a commercial bicyclam CXCR4 antagonist AMD3100 (Plerixafor) as the main building block of the polymers (named PAMD in this study). Due to their cationic nature, PAMD formed polyplexes with DNA and facilitated efficient transfection in various types of cancer cells. Importantly, the PAMD/DNA polyplexes exhibited CXCR4 antagonism demonstrated by their ability to inhibit cancer cell invasion and metastasis.26–28 Although efficient in DNA delivery, the original PAMD exhibited poor siRNA delivery activity.
The goal of the present study was to further develop PAMD as siRNA delivery vectors to achieve combined antimetastatic and antitumor effect. Based on available evidence, we proposed that modification of PAMD with cholesterol will improve overall stability and improve cell uptake and intracellular trafficking of siRNA polyplexes. We investigated the effect of cholesterol modification on siRNA complexation, colloidal and enzymatic stability of polyplexes, and the ability to inhibit CXCR4 and deliver anticancer siRNA against PLK1.
2. Materials and methods
2.1. Materials
Cholesteryl chloroformate and branched poly(ethylene imine) (PEI, 25 kDa) were from Sigma-Aldrich (St. Louis, MO). N,N′-Hexamethylenebisacrylamide (HMBA) was purchased from Polysciences, Inc. (Warrington, PA), N,N-diisopropylethylamine (DIPEA) was from Acros Organics (New Jersey, US), AMD3100 (base form) was from Biochempartner (Shanghai, China). Dulbecco’s Modified Eagle Medium (DMEM), Dulbecco’s Phosphate Buffered Saline (PBS), Fetal Bovine Serum (FBS) and RNase I were from Thermo Scientific (Waltham, MA). Human SDF-1 was from Shenandoah Biotechnology, Inc. (Warwick, PA). Scrambled siRNA (siScr, 5’-ACGUGACACGUUCGGAGAAUU-3’) and siGENOME human polo-like kinase 1 (PLK1) siRNA Smartpool (siPLK1) were purchased from GE Healthcare Dharmacon, Inc. (Lafayette, CO). Succinimidyl ester of Alexa Fluor® 647 carboxylic acid was from Life Technologies (Eugene, OR). All other reagents were from Fisher Scientific and used as received unless otherwise noted.
2.2. Polymer synthesis
PAMD was synthesized by Michael-type polyaddition of equal molar ratio of AMD3100 and HMBA. Typically, 0.66 mmol of each reactant was dissolved in a glass scintillation vial containing 6 mL MeOH/water (7/3 v/v) mixture. Polymerization was carried out under nitrogen protection in dark at 37 °C for 3 days. Then, additional 0.066 mmol of AMD3100 was added to the reaction mixture and stirred for another day to consume any unreacted acrylamides. Product was isolated by double precipitation in diethyl ether, collected by centrifugation, and dried in vacuum (yield 84%).
To synthesize PAMD-Ch with various degrees of cholesterol substitution, PAMD was first dissolved in a mixture of anhydrous methylene chloride and DIPEA. Different calculated amounts of cholesteryl chloroformate in anhydrous methylene chloride were added drop wise to the ice-cold PAMD solution over 1 h. The reaction was continued under stirring for another 24 h. The product was obtained by evaporating the solvent, and washing with diethyl ether three times to remove unreacted cholesteryl chloroformate. The product was further dissolved in ethanol/water (v/v 1/1) mixture, followed by adjusting the pH to 4.0 using HCl. The polymers then underwent extensive dialysis against ethanol/water mixture (v/v 1/1) for 2 days and distilled water for another day (membrane molecular weight cut-off 3.5 kDa) before lyophilization. Typical yield of PAMD-Ch ranged from 61% to 88%.
2.3. Polymer characterization
The molar mass of PAMD was analyzed by gel permeation chromatography (GPC) operated in 0.3 M sodium acetate buffer (pH 5) using Agilent 1260 Infinity LC system equipped with a miniDAWN TREOS multi-angle light scattering (MALS) detector and a Optilab T-rEX refractive index detector from Wyatt Technology (Santa Barbara, CA). Polycation-compatible TSKgel G3000PWXL-CP column (Part No. 0021873, Tosoh Bioscience LLC, King of Prussia, PA) was used at a flow rate of 0.5 mL/min. Results were analyzed using Astra 6.1 software from Wyatt Technology. The content of cholesterol in PAMD-Ch was determined using 1H-NMR on Varian INOVA (500 MHz). The molecular weights of PAMD-Ch were calculated using the determined molar mass of PAMD and the known cholesterol substitution degree determined by 1H-NMR.
2.4. Critical Micelle Concentration (CMC)
Fluorescence spectroscopy was used to determine CMC of PAMD-Ch polymers using pyrene as a hydrophobic fluorescent probe. Different concentrations of PAMD-Ch in water were allowed to equilibrate with 600 nM pyrene overnight at room temperature after 1 h sonication. Each fluorescence intensity index ratio at 335ex/384em nm (I3) vs. 335ex/373em nm (I1) (I3/I1) was measured, and plotted against the logarithmic concentration of the polymer. The concentration at the inflection point was determined as CMC.29
2.5. Preparation and characterization of PAMD-Ch/siRNA
PAMD-Ch/siRNA polyplexes were formed by mixing equal solution volumes of siRNA (20 µg/mL) and polymer, followed by incubation at room temperature for 20 min before use. Complexation of siRNA by PAMD was examined by agarose gel electrophoresis. PAMD-Ch/siRNA polyplexes were prepared at various polymer/siRNA w/w ratios (w/w 1 corresponds to N/P ratio of ~3), loaded onto a 2% agarose gel containing 0.5 µg/mL ethidium bromide and run at 75 V in 0.5× Tris/Borate/EDTA (TBE) buffer for 30 min. The gel was then imaged under UV. Hydrodynamic diameter and zeta potential of the polyplexes in 10 mM HEPES buffer (pH 7.4) were determined by dynamic light scattering using a ZEN3600 Zetasizer Nano-ZS (Malvern Instruments Ltd., Worcestershire, UK).
2.6. Colloidal and enzymatic stability of polyplexes
Colloidal stability of PAMD-Ch/siRNA polyplexes was evaluated by measuring hydrodynamic diameter of the polyplexes in PBS at 25 °C for up to 12 h. Results were expressed as mean ± SD of three measurements. To study the resistance to RNase I, PAMD-Ch/siRNA polyplexes containing a total amount of 0.2 µg siRNA were incubated with 2.5 units of RNase I at 37 °C for 30 min, followed by incubation at 87°C for 30 min to inactivate the enzyme. Heparin (200 µg/mL) was added to the samples and the mixture was incubated for additional 30 min to release the siRNA. Gel electrophoresis was then used to determine siRNA integrity.
2.7. Cell Culture
Human epithelial osteosarcoma U2OS cells stably expressing functional EGFP-CXCR4 fusion protein were purchased from Fisher Scientific and cultured in DMEM supplemented with 2 mM L-glutamine, 1% Pen-Strep, 0.5 mg/mL G418 and 10% FBS. The cells were maintained at 37°C with 5% CO2 in a humidified incubator.
2.8. Cytotoxicity
Cytotoxicity of the synthesized polycations was tested by MTS assay in U2OS cells. The cells were seeded in 96-well plates at a density of 8000 cells/well and incubated overnight. Culture medium was then replaced by 150 µL of serial dilutions of a polymer in serum-supplemented medium and the cells were incubated for 24 h. The medium was then aspirated and replaced by a mixture of 100 µL serum-free media and 20 µL of MTS reagent (CellTiter 96®AQueous Non-Radioactive Cell Proliferation Assay, Promega, Madison, WI). After 1 h incubation, the absorbance [A] was measured using SpectraMax®M5e Multi-Mode Microplate Reader at a wavelength of 490 nm. The relative cell viability (%) was calculated as [A]sample/[A]untreated × 100%. The IC50 were calculated in GraphPad Prism using a built-in dose-response analysis as the polymer concentration that achieves 50% growth inhibition relative to untreated cells (n = 3).
2.9. CXCR4 antagonism
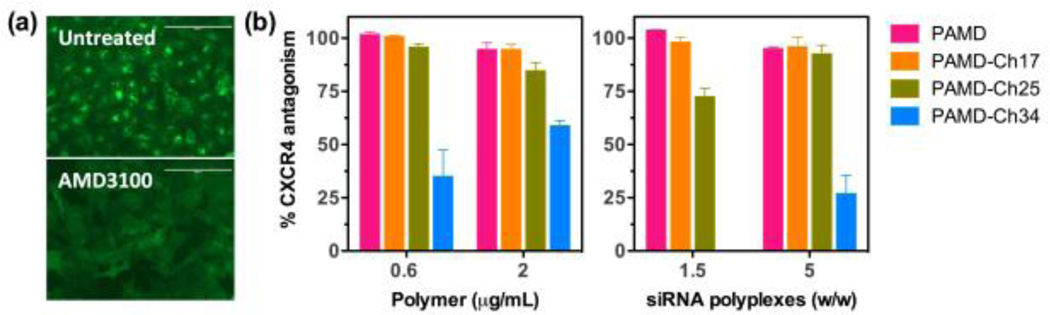
CXCR4 antagonism of the polycations and their siRNA polyplexes was determined by CXCR4 redistribution assay. U2OS cells were seeded at a density of 8,000 cells/well in 96-well black plates with optical bottom 24 h before the experiment. On the day of the assay, cells were washed twice with 100 µL assay buffer (DMEM supplemented with 2 mM L-glutamine, 1% FBS, 1% Pen-Strep, and 10 mM HEPES) and incubated with different concentrations of the polycations, polyplexes, or AMD3100 in the assay buffer containing 0.25% DMSO at 37 °C for 30 min. Then, SDF-1 was added to each well to make final concentration of 10 nM and the cells were incubated at 37 °C for 1 h. Cells were then fixed with 4% paraformaldehyde at room temperature for 20 min, washed 4 times with PBS and stained in 1 µM Hoechst 33258 solution for 30 min before imaging by EVOS fl microscope. To quantify the internalization/redistribution levels of the CXCR4 receptors, high-content analysis was conducted using Cellomics ArrayScan VT1 Reader and SpotDetectorV3 BioApplication software. CXCR4 antagonism was determined based on % CXCR4 internalization inhibition calculated relative to the positive (AMD3100, 100%) and negative (SDF-1 only, 0%) controls, and the results were expressed as mean % inhibition ± SD (n = 3).
2.10. siRNA transfection
siRNA transfection efficiency of the polyplexes was evaluated in U2OS cells using human siPLK1 as a therapeutic siRNA. Cells were seeded at a density of 2,500 cells/well in 96-well plates one day before. On the day of experiment, culture medium was carefully removed and replaced with 50 µL medium (+/− 10% FBS) and 12.5 µL polyplexes (siRNA dose: 5 pmol per well). After 4 h of incubation, polyplexes were removed and cells were maintained in 200 µL fresh culture medium for another 44 h. CellTiter-Blue Cell Viability Assay (Promega) was used to measure % cell viability. Activity was expressed as % cell death induced by PLK1 gene silencing compared with scrambled siRNA.
2.11. Cellular uptake and intracellular distribution of polyplexes
PAMD-Ch polymers were fluorescently labelled with AlexaFluor 647 following manufacturer’s instructions and purified by dialysis to remove unreacted dye. Fluorescently labelled siRNA (Block-iT™ Alexa Fluor® Red) was purchased from Invitrogen (Carlsbad, CA). 100,000 of U2OS cells were seeded in a 23 mm glass-bottom dish (Nioptechs Inc. Cat# 0420041500C) 24 h before the experiment. Cells were then incubated with PAMD-Ch/siRNA polyplexes (siRNA concentration 25 nM) for 1 h, washed twice with PBS, fixed with 4% paraformaldehyde, washed with PBS for additional 4 times and stained in 1 µM Hoechst 33258 solution All the images were taken using Zeiss 710 confocal laser scanning microscope equipped with a 63× oil objective and 4 lasers (Blue Diode 405 nm, Argon 458/488/514 nm, DPSS 561 nm and He-Ne 633 nm).
3. Results and discussion
We have previously reported PAMD as a new class of dual-function poly(amidoamine)s based on the CXCR4 antagonist AMD3100. We showed that PAMD are capable of inhibiting CXCR4/SDF-1 signaling to limit cancer invasion, while simultaneously delivering DNA for gene therapy.26–28 While suitable for delivery of DNA, the first generations of PAMD showed limited ability to deliver other types of therapeutic nucleic acids such as siRNA and microRNA.
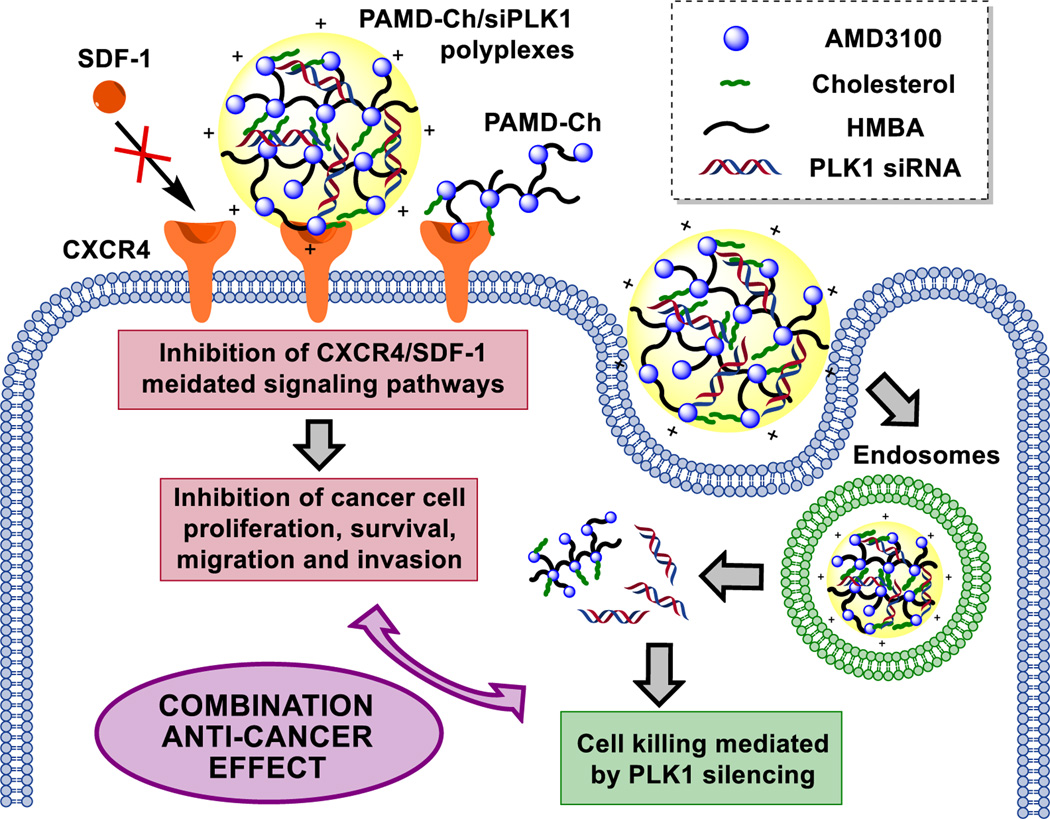
To further explore and develop the potential of PAMD as a versatile platform for delivery of nucleic acids in cancer treatment, the aim of this study was to adapt PAMD for efficient delivery of siRNA. Based on available evidence with polycations used in siRNA delivery, the hypothesis was that simple modification of PAMD with cholesterol would improve siRNA delivery.12 Modification with cholesterol is a well-established method to improve polycation-medicated siRNA delivery and in vivo stability.30 This modification is expected to improve siRNA delivery due to stabilization through hydrophobic interactions within the interior of the polyplexes and enhanced interaction of the polyplexes with endosomal membranes during intracellular trafficking (Scheme 1).17, 31
Scheme 1.
Proposed mechanism of action of the dual-function PAMD-Ch as polymeric CXCR4 antagonists and siRNA (PLK1) delivery vectors.
3.1. Synthesis and characterization of PAMD-Ch
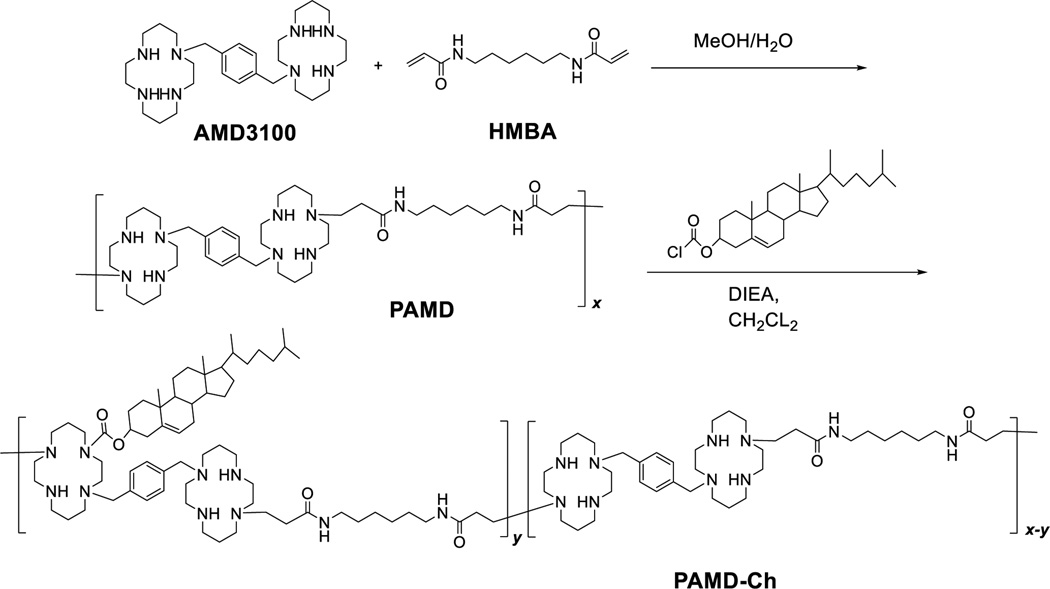
PAMD was synthesized by Michael-type polyaddition of equal molar ratio of AMD3100 and HMBA (Scheme 2). AMD3100 contains six secondary amines and functions as a hexafunctional monomer in this reaction. The acrylamide groups in HMBA react randomly with the AMD3100 amines, which results in the formation of branched water-soluble polymers when conducting the reaction at low temperature.32, 33 The weight-average molar mass (Mw) of the PAMD synthesized and used in this study was 13.9 kg/mol and polydispersity index (Mw/Mn) was 1.9.
Scheme 2.
Synthesis of PAMD-Ch. (*please note that any of the cyclam secondary amines could participate in the Michael-type addition)
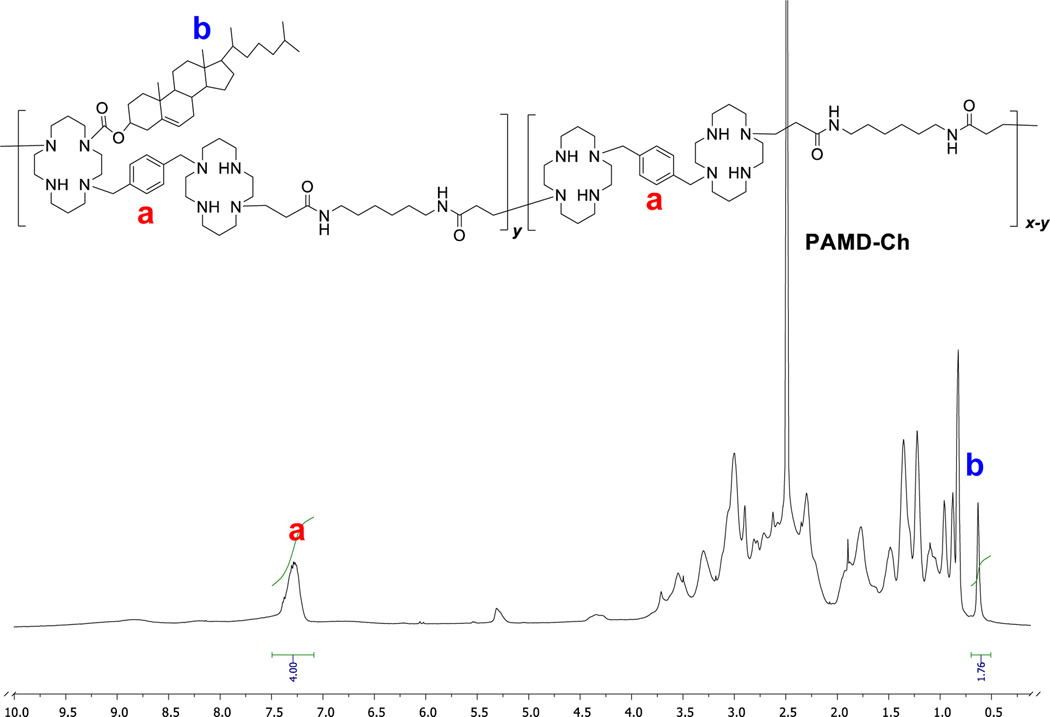
PAMD-Ch copolymers were synthesized by reaction of cholesteryl chloroformate with the available secondary amines in PAMD (Scheme 2). The content of cholesterol in the copolymers could be easily tuned by changing the ratio of cholesteryl chloroformate to PAMD in the reaction. The content of cholesterol in the copolymers was determined from 1H-NMR integral intensity of the cholesterol methyl group b at 0.65 ppm and the aromatic phenylene protons in AMD3100 at 7.1–7.5 ppm (Figure 1). The molar mass of the PAMD-Ch copolymers was calculated based on the known molar mass of PAMD and the degree of cholesterol modification from the 1H-NMR analysis. We have synthesized three copolymers with increasing content of cholesterol (Table 1). The copolymers are named according to the weight percentage of cholesterol.
Figure 1.
Typical 1H-NMR spectrum of PAMD-Ch used in the determination of the cholesterol content (spectrum of PAMD-Ch25 in DMSO shown).
Table 1.
Characterization of PAMD-Ch copolymers.
| Polymer | Cholesterol content (wt %) |
Mw (kg/mol) |
|
|---|---|---|---|
| In Feed | In copolymera | ||
| PAMD | 0 | 0 | 13.9b |
| PAMD-Ch17 | 15 | 17 | 16.7c |
| PAMD-Ch25 | 25 | 25 | 18.5c |
| PAMD-Ch34 | 36 | 34 | 21.1c |
From 1H-NMR.
From GPC.
Calculated from the Mw of PAMD and cholesterol content.
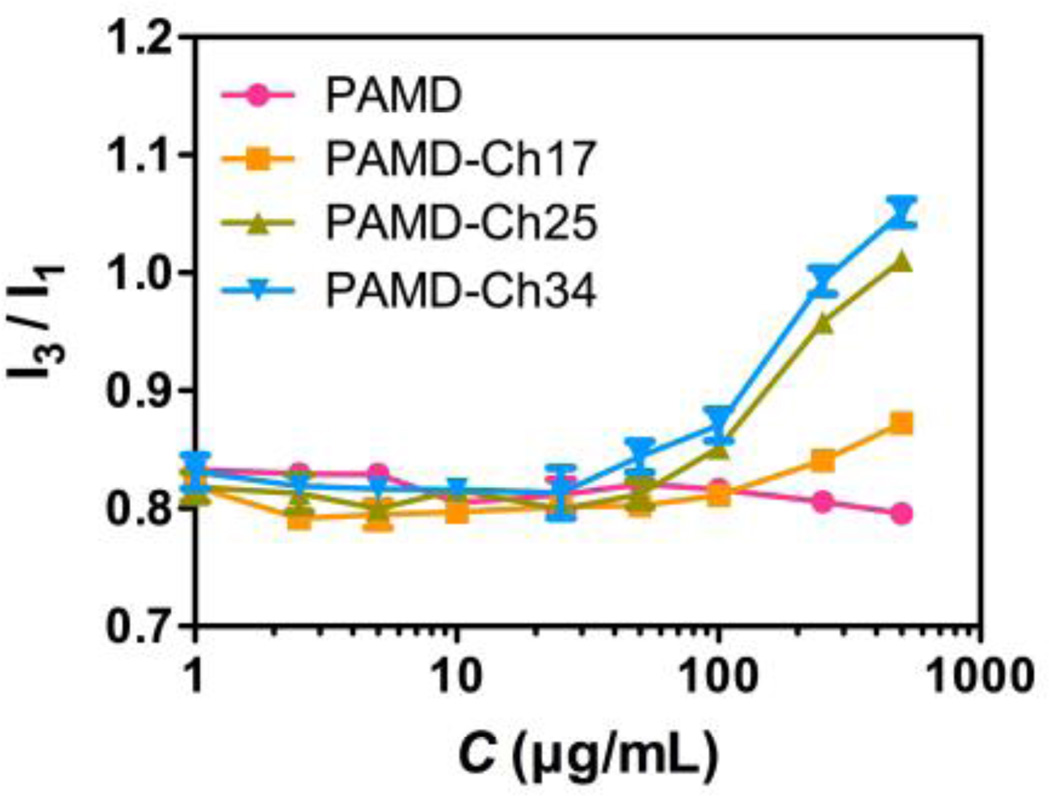
The introduction of hydrophobic cholesterol moieties to the hydrophilic PAMD polycation backbone results in the formation of amphiphilic PAMD-Ch copolymers. The potential of these copolymers to self-assemble into micelles in aqueous media could change the nature and dynamic of the complexation with siRNA depending on whether the copolymers bind the nucleic acid as a unimer or as assembled micelles. Here we determined the critical micelle concentration (CMC) values for the PAMD-Ch in distilled water by fluorescence spectroscopy using pyrene as a hydrophobic fluorescent probe. In the presence of micelles (above CMC), pyrene could be incorporated into the hydrophobic core in the micelles, resulting in the increase in the ratio of two fluorescence intensity peaks (I3/I1). By plotting the ratio of I3/I1 against the polymer concentration, CMC of each PAMD-Ch could be determined (Figure 2). The results showed that all PAMD-Ch could form micelles with CMC ranging from 63.1 to 89.1 µg/mL in water. As expected, increasing the cholesterol content in the copolymers resulted in decreasing CMC.
Figure 2.
Critical micelle concentration (CMC) of PAMD-Ch determined by fluorescence spectroscopy. CMC was determined as the concentration at the inflection point of the curve where I3/I1 was plotted against PAMD-Ch concentration (n=3).
3.2. Preparation and characterization of PAMD-Ch/siRNA polyplexes
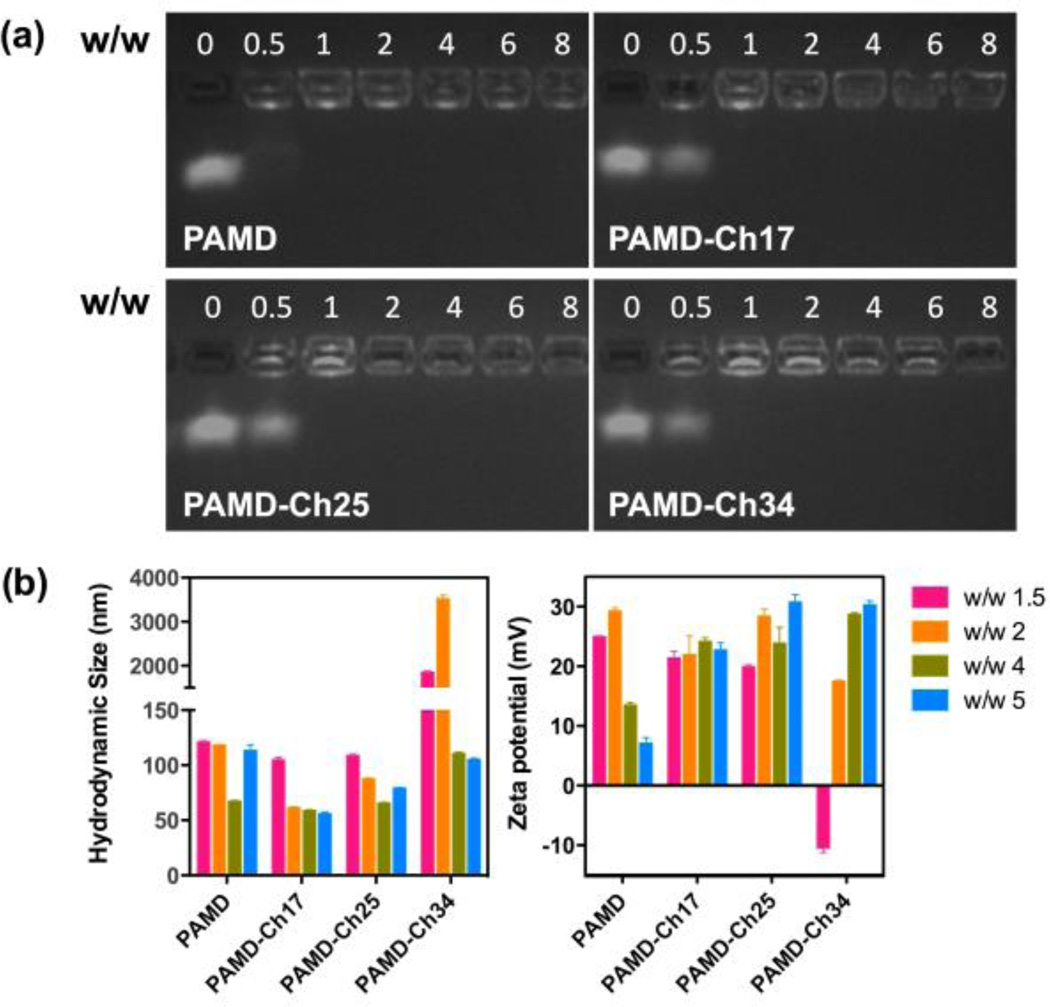
The influence of cholesterol grafting on the ability of PAMD to complex siRNA into polyplexes was investigated by agarose gel electrophoresis (Figure 3a). All the PAMD-Ch copolymers were able to fully complex siRNA at or above w/w ratios equivalent to PAMD/siRNA w/w = 1. The siRNA binding ability of the parent PAMD was slightly better than PAMD-Ch, as suggested by nearly complete siRNA complexation at w/w ratio 0.5. The better complexation ability of PAMD is likely due to a decreased number of protonated amines in PAMD-Ch due to cholesterol conjugation. Therefore all siRNA polyplexes used in the following studies were prepared above w/w = 1 (N/P ~3) to assure complete siRNA complexation. All w/w ratios in this study are expressed as equivalent PAMD/siRNA ratios (i.e., not taking into account cholesterol content).
Figure 3.
siRNA complexation and physicochemical characterization of siRNA polyplexes. (a) siRNA binding ability of the PAMD-Ch copolymers. (b) Hydrodynamic size and zeta potential of PAMD-Ch/siRNA polyplexes at various w/w ratios (equivalent PAMD/siRNA). Results are shown as mean ± SD of three measurements.
Hydrodynamic size and zeta potential of PAMD-Ch/siRNA polyplexes were evaluated by dynamic light scattering (Figure 3b). Polyplexes were prepared in 10 mM HEPES buffer (pH 7.4) at various equivalent w/w ratios and were allowed to stabilize for 20 min before measurement. Except for PAMD-Ch34/siRNA prepared at lower w/w ratios, all the other polyplexes displayed small particle size ranging from 56 to 121 nm. All the PAMD-Ch/siRNA polyplexes prepared at higher w/w ratios showed significantly smaller sizes than polyplexes prepared at lower w/w ratios, perhaps suggesting tighter binding. At w/w ratios above 2, PAMD-Ch17 with the lowest cholesterol content exhibited the smallest sizes compared with other PAMD-Ch/siRNA polyplexes. All PAMD-Ch/siRNA polyplexes exhibited positive surface charge indicated by zeta potentials ranging from 18 to 31 mV.
3.3. Stability of PAMD-Ch/siRNA polyplexes
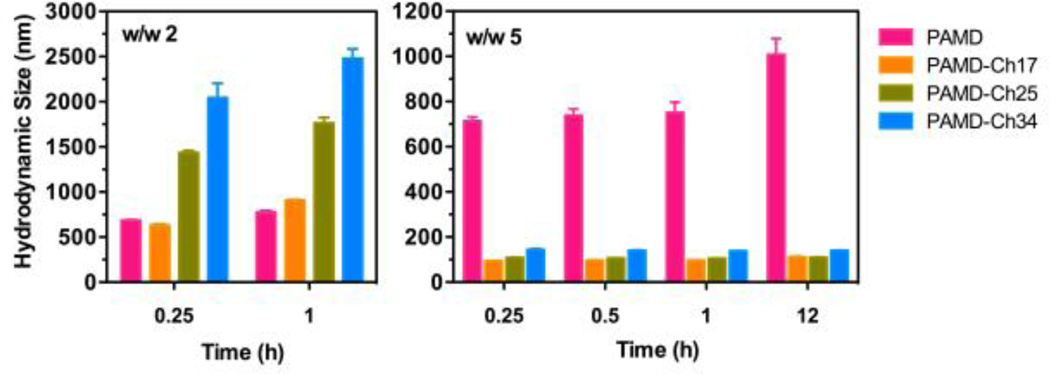
Colloidal and enzymatic stability is an important prerequisite for successful application of PAMD/Ch/siRNA polyplexes. Similar to other polycation/siRNA polyplexes, PAMD/siRNA polyplexes maintained stable size only when prepared in low concentration buffer (e.g., 10 mM HEPES, pH 7.4) and addition of salts to reach physiologically relevant levels resulted in aggregation (Figure 3b vs. 4). Aggregation of polyplexes depends on various parameters including the chemical structure, molar mass and hydrophobicity of the used polycations.34
Figure 4.
Colloidal stability of PAMD-Ch/siRNA polyplexes in PBS up to 12 h. Results are shown as mean ± SD of three measurements.
To evaluate the colloidal stability of PAMD-Ch/siRNA polyplexes, we prepared the polyplexes at two different equivalent w/w ratios (2 and 5) and incubated them in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) while monitoring changes in hydrodynamic diameter over time (Figure 4). At w/w 2, all the PAMD-Ch/siRNA polyplexes aggregated and reached sizes ranging from ~690 nm to ~2 µm within 15 min of incubation in PBS. Polyplexes based on PAMD-Ch with higher cholesterol content (PAMD-Ch25 and 34) showed significantly faster rate of aggregation than PAMD-Ch with low (PAMD-Ch17) or no (PAMD) cholesterol. Interestingly, when we prepared PAMD-Ch/siRNA polyplexes at w/w ratio 5, all the cholesterol-containing polyplexes exhibited markedly improved colloidal stability with sizes <150 nm maintained for at least 12 h incubation in PBS. This observation was fully consistent with similar findings on the behavior observed in DNA polyplexes where increasing the amount of cholesterol resulted in polyplexes with enhanced colloidal stability.35 In contrast, siRNA polyplexes prepared with the parent PAMD showed similar aggregation behavior as polyplexes prepared at w/w ratio of 2 and rapidly formed large aggregates. We propose that as the surface positive charge of polyplexes is reduced at physiological salt concentrations, more PAMD-Ch can bind to the particle surface via hydrophobic interactions and increase colloidal stability by forming an additional shell of a polycation. In all of the above experiments, the PAMD-Ch concentrations were below their CMC. Nevertheless, local copolymer concentration within each polyplex particle was likely significantly higher than CMC, thus providing another contributing factor to the improved stability.
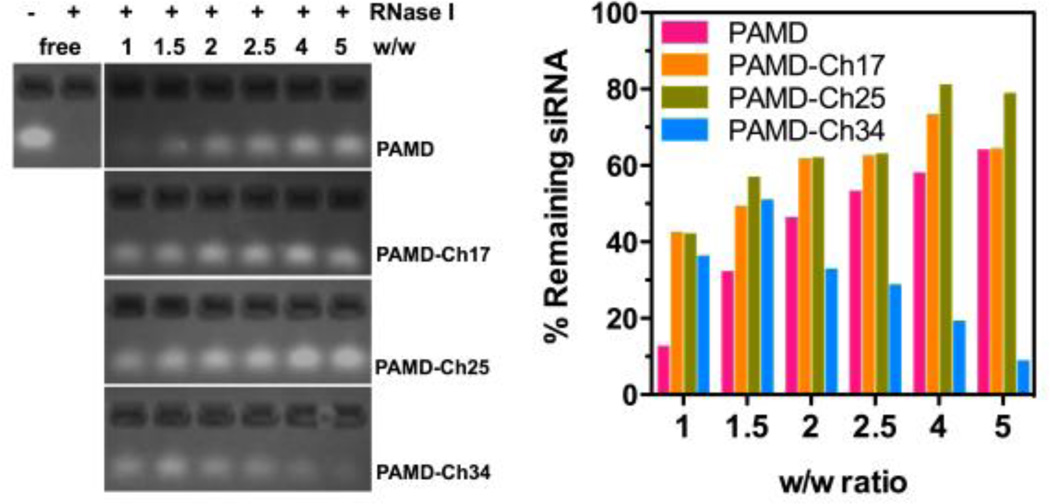
Enzymatic degradation is one of the main factors hindering effective siRNA delivery in vivo. Here we evaluated the effect of cholesterol modification on the stability of PAMD-Ch/siRNA polyplexes against RNase I degradation. siRNA polyplexes were formed at various equivalent w/w ratios, and incubated with 0.5 U RNase I for 30 min. In order to evaluate siRNA integrity, heparin was then added to dissociate the polyplexes and release the siRNA. Gel electrophoresis was used to examine the siRNA integrity and the intensity of each band was quantified and normalized to free siRNA. As shown in Figure 5, free siRNA completely degraded once exposed to RNase I. All the polymers, including parent PAMD, were able to provide protection of the siRNA against RNase I. PAMD with lower cholesterol modification (PAMD-Ch17 and 25) exhibited improved ability to protect siRNA when compared with the parent PAMD at the same equivalent w/w ratio. In addition, PAMD-Ch17 and PAMD-Ch25 also displayed increasing protecting ability with increasing w/w ratio, similar to PAMD, indicating important role of excess polycations in properties of polyplexes. PAMD-Ch25/siRNA polyplexes prepared above w/w 4 demonstrated the best protection against RNase I degradation with ~80% siRNA remaining intact after exposure. In contrast, PAMD-Ch34 with the highest cholesterol content, showed decreasing ability to protect siRNA above w/w 2, while ~50% siRNA remained intact when the polyplexes were prepared at w/w 1.5.
Figure 5.
Stability of PAMD-Ch/siRNA polyplexes against RNase I. Polyplexes prepared at various w/w were exposed to RNase I, followed by incubation with heparin to release the siRNA for agarose gel electrophoresis. siRNA band intensity was quantified to calculate % siRNA remaining compared with untreated free siRNA.
3.4. Cytotoxicity of PAMD-Ch
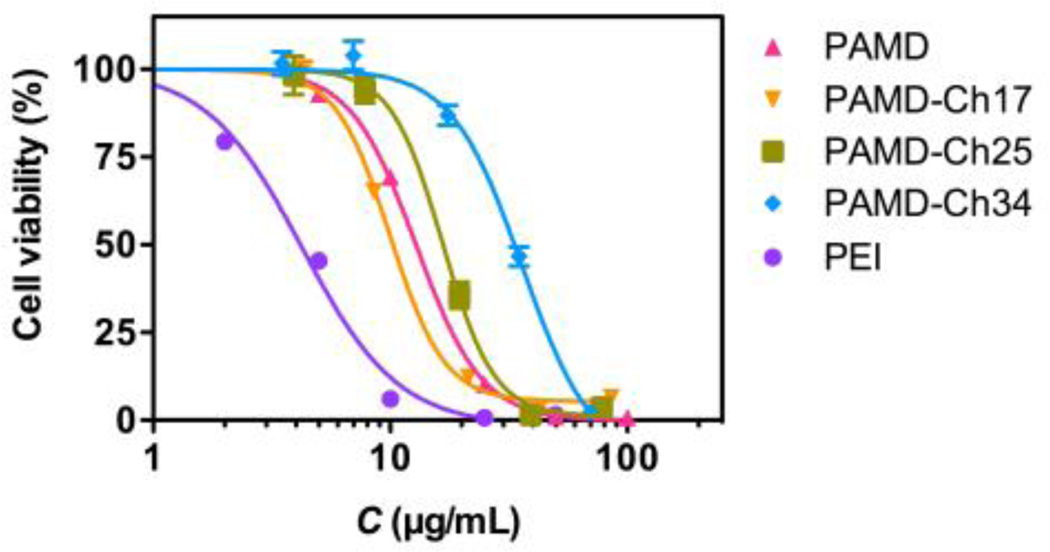
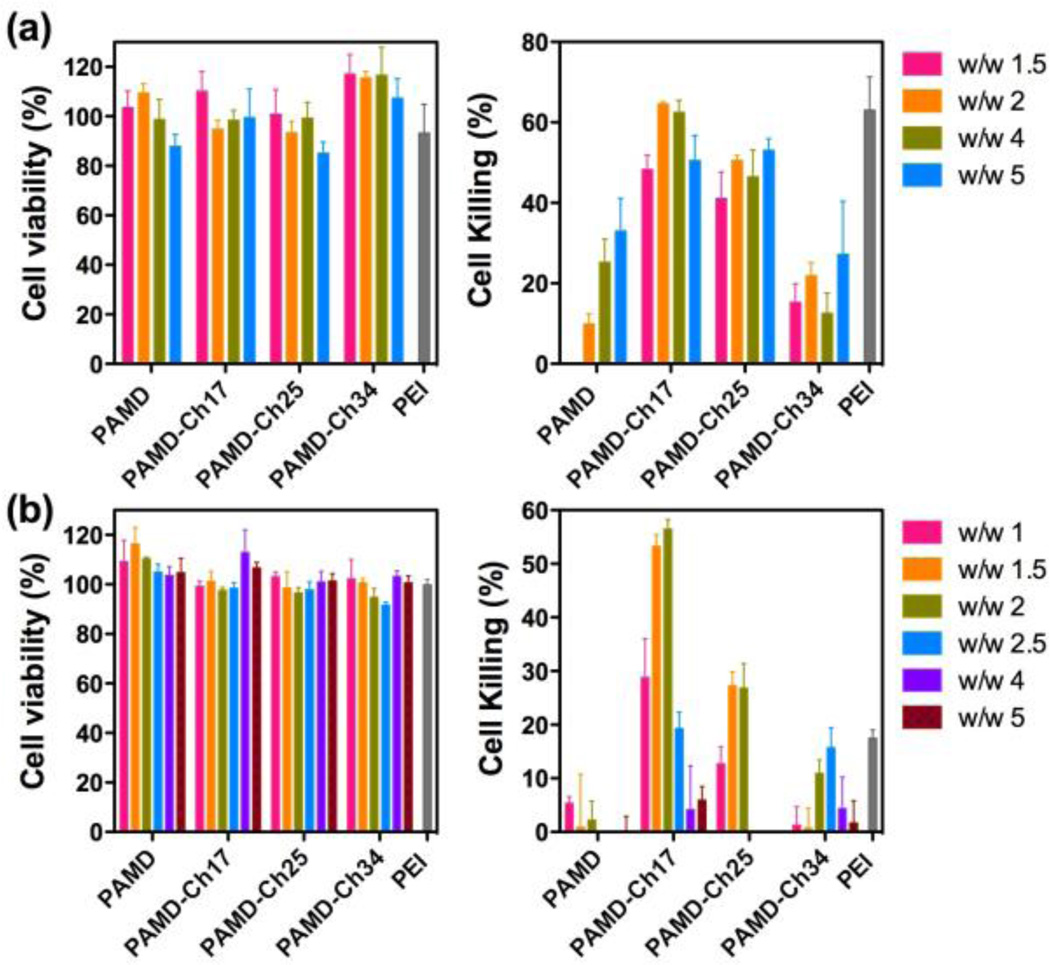
Cytotoxicity of polycation-based gene delivery systems is known to correlate with several key factors including the polymer structure, molar mass of the polycations, cationic charge density and biodegradability.36–38 Both positive and negative effects of hydrophobic modification on polycations have been reported previously.12 In some cases, high content of hydrophobic chains could cause cell membrane disruption, which resulted in cell death.39 Thus it was important to evaluate how cholesterol affects toxicity of PAMD to avoid or minimize any undesired toxic side effects. Here we investigated the cytotoxicity of PAMD-Ch by MTS assay in U2OS osteosarcoma cells, as these cells were then used throughout this study to assess the biological activity of the PAMD-Ch/siRNA polyplexes (Figure 6). In all toxicity experiments, PAMD-Ch concentrations are expressed as PAMD concentration only (i.e., excluding cholesterol), and the IC50 values were also calculated considering only the PAMD content to allow direct evaluation of the effect of cholesterol modification on the toxicity of the polycation. Unmodified PAMD showed IC50 12.8 µg/mL, which was significantly higher than the benchmark 25 kDa branched PEI used here as a control (4.2 µg/mL). IC50 of PAMD-Ch17 was 10.3 µg/mL, which was only slightly lower than PAMD. However the IC50 values increased to 16.7 µg/mL and 33.4 µg/mL in the case of PAMD-Ch25 and PAMD-Ch34. The results demonstrated that higher content of cholesterol leads to safer polycations, most likely due to decreased number of exposed positive charges in the polymer available for interaction with cell membranes.
Figure 6.
Cytotoxicity of PAMD-Ch. % Cell viability was measured by MTS assay after 24 h incubation with increasing concentrations of polymers. PAMD-Ch concentrations are expressed as PAMD concentration only (i.e., excluding cholesterol). Results are expressed as mean cell viability ± SD (n = 3).
3.5. CXCR4 antagonism of PAMD-Ch and PAMD-Ch/siRNA
CXCR4 antagonism of PAMD requires accessibility of the receptor-binding cyclam moieties in the polycation structure. Although not all of the eight amino groups in the AMD3100 are required for CXCR4 binding and inhibition,40 the activity of the cyclam residues in PAMD is decreased when compared with the parent small molecule drug.41 The conjugation of cholesterol to the remaining secondary amines of PAMD could further negatively impact the CXCR4 antagonism of PAMD-Ch. Furthermore, the presence of the hydrophobic cholesterol residues in the polyplex structure could alter the particle morphology and surface properties and thus the ability to bind the CXCR4 receptor. It was therefore important to investigate the effect of cholesterol on the ability of free PAMD-Ch and PAMD-Ch/siRNA polyplexes to bind CXCR4 receptor and inhibit the CXCR4/SDF-1 pathway in order to assure that the proposed dual functionality of the vector is preserved.
We evaluated the CXCR4 inhibition of all the PAMD-Ch copolymers and their siRNA polyplexes to investigate the effect of cholesterol grafting on the CXCR4 antagonism. We used a CXCR4 redistribution assay based on the inhibition of SDF1-triggered endocytosis of EGFP-CXCR4 receptors in U2OS osteosarcoma cells. This is a phenotypic assay that uses automatic image analysis to quantify the extent of EGFP-tagged CXCR4 internalization into the cells. Untreated cells display punctate fluorescence indicative of EGFP-CXCR4 internalization into endosomes in response to SDF-1 stimulation. In contrast, CXCR4 antagonists like AMD3100 inhibit receptor internalization, as documented by the diffuse pattern of fluorescence. The distinct fluorescence patterns of EGFP-CXCR4 in untreated and AMD3100-treated cells are shown in Figure 7a. To allow direct evaluation of the effect of cholesterol moieties on CXCR4 antagonism of PAMD, we tested CXCR4 inhibitory activity of PAMD-Ch at equal concentrations of the PAMD part of the copolymers using two different concentrations (0.6 and 2 µg/mL). We also evaluated PAMD-Ch/siRNA polyplexes prepared at w/w ratios (1.5 and 5). The selected w/w ratios allowed us to achieve the same polymer concentrations as in the experiment with free polymers. All the results are expressed as % CXCR4 antagonism relative to the positive control (300 nM AMD3100) (Figure 7b). We found that CXCR4 antagonism of PAMD-Ch copolymers and their siRNA polyplexes display similar concentration-dependent behavior. In general, polyplexes showed slightly decreased CXCR4 inhibition when compared with the free polymer at the same concentration, a likely result of sequestration of a portion of the copolymers in the core of the siRNA polyplexes. Increasing the content of cholesterol in PAMD-Ch resulted in a decrease in CXCR4 inhibition. However, PAMD-Ch17/siRNA polyplexes were able to attain nearly complete CXCR4 inhibition at a low w/w ratio of 1.5. PAMD-Ch25/siRNA polyplexes required w/w 5 to achieve similar levels of CXCR4 antagonism. Copolymer with the highest cholesterol content (PAMD-Ch34) showed the lowest CXCR4 antagonism among all the tested polymers in all the tested conditions. This finding shows that there is a fine balance when introducing the cholesterol moieties into dual-function polymeric CXCR4 antagonists that require specific receptor binding for their activity.
Figure 7.
CXCR4 antagonism of PAMD-Ch and PAMD-Ch/siRNA polyplexes. (a) Illustration of EGFP-CXCR4 receptor redistribution assay: untreated cells (0% CXCR4 antagonism) and cells treated with 300 nM AMD3100 (100% CXCR4 antagonism). (b) CXCR4 antagonism of PAMD-Ch and their siRNA polyplexes. The results are shown as mean % CXCR4 inhibition relative to positive control 300 nM AMD3100 ± SD (n = 3).
3.6. Delivery of anti-PLK1 siRNA (siPLK1) by PAMD-Ch
PLK1 – a key mitotic regulator in mammalian cells – is an attractive target in cancer treatment.42, 43 PLK1 expression is elevated in multiple types of human cancers and it has a prognostic value for predicting aggressiveness of cancer.44 Inhibition of PLK1 by small molecule inhibitors or using PLK1 gene silencing with siRNA results in cell apoptosis and inhibition of tumor growth in vivo.45–47 Thus, combining the antitumor activity of PLK1 silencing with antimetastatic effect of CXCR4 antagonism by PAMD-Ch represents a promising therapeutic approach to treat cancer.
We have evaluated the effect of cholesterol modification on the ability of PAMD-Ch to deliver siPLK1 in U2OS osteosarcoma cells. Scrambled siRNA (siScr) was used in control experiments to assess toxicity of the studied polyplexes. PEI/siPLK1 polyplexes prepared at w/w 1.5 were used as a positive control. We first established safety of the selected polyplex formulations in serum-free conditions using siScr (Figure 8a). All the tested polyplex formulations exhibited acceptable levels of toxicity with cell viability above 85%. Then, anticancer activity of PAMD-Ch/siPLK1 polyplexes was evaluated from their ability to trigger cell death as a result of PLK1 gene silencing. As shown in Figure 8a (right), multiple of the tested polyplex formulations were able to kill substantial fraction of the cancer cells. PAMD-Ch17 and PAMD-Ch24 polyplexes exhibited significantly higher anticancer activity than control PAMD and PAMD-Ch34 polyplexes. The best performing PAMD-Ch17/siPLK1 polyplexes showed cell killing activity (48–62%) fully comparable to the PEI/siPLK1 control.
Figure 8.
siRNA delivery by PAMD-Ch in U2OS cells. Transfections were conducted either in the absence (a) or the presence of 10% serum (b). Polyplexes were prepared with control siScr (left) or siPLK1 (right) at various equivalent PAMD/siRNA w/w ratios and cell killing mediated by PLK1 knockdown was measured (n = 4).
While useful for mechanistic studies, experiments conducted in the absence of serum have limited value for practical application of PAMD-Ch/siRNA polyplexes. Thus, we evaluated the anticancer activity also in the presence of 10% serum (Figure 8b). As expected, safety of the polyplex formulations assessed with siScr improved markedly in the presence of serum as indicated by negligible effect of the PAMD-Ch/siScr treatment on cell viability (Figure 8b left). However, the presence of serum had detrimental effect on the ability of most of the tested polyplexes to deliver siPLK1 as indicated by nearly-background levels of cell killing. Notably, both PAMD/siPLK1 and PEI/siPLK1 lost nearly all their anticancer activity when compared with the serum-free conditions. In contrast, PAMD-Ch17/siPLK1 polyplexes retained significant cell killing activity. When prepared at w/w 2, the cell killing activity of PAMD-Ch17/siPLK1 polyplexes was comparable to the activity in serum-free conditions. Unlike the results in serum-free conditions, the cell killing activity in the presence of serum was highly dependent on the w/w ratios. For PAMD-Ch17 and PAMD-Ch25, optimal activity was achieved when the polyplexes were prepared at w/w 1.5 and 2, while in the case of PAMD-Ch34 the highest activity was reached at w/w 2.5. These findings suggest that content of cholesterol in PAMD-Ch has to be carefully controlled and optimized to achieve maximum anticancer activity.
3.7. Intracellular distribution of PAMD-Ch/siRNA polyplexes
Appropriate intracellular trafficking is critical for successful delivery of functional siRNA by polyplexes. In general, siRNA has to be protected in the polyplexes, internalized by the cells and released in the cytoplasm to gain the desired silencing effect. Multiple previous reports have shown positive effects of introducing hydrophobic moieties into polycations on enhancing nucleic acid delivery due to favorable effects on cell membrane adsorption, alleviation of serum inhibition, and facilitation of nucleic acid dissociation from polycations.12, 18
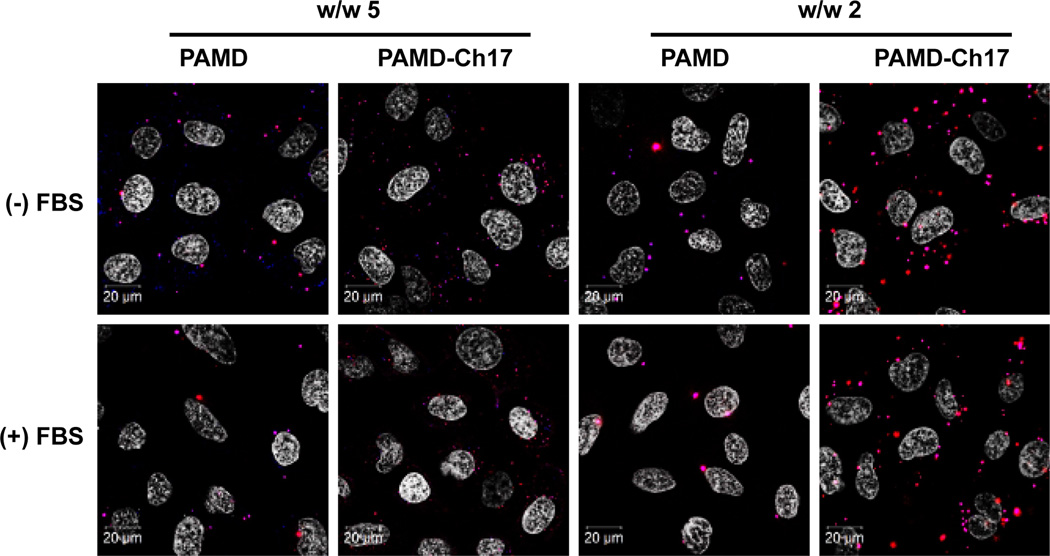
In order to better illuminate the mechanism of action of the dual-function PAMD-Ch copolymers, we employed confocal microscopy to investigate the influence of cholesterol modification on the cellular uptake and intracellular distribution of the PAMD-Ch/siRNA polyplexes. In order to visualize both components of the polyplexes, we labelled the copolymers with AlexaFluor 647 and used commercially available siRNA labelled with AlexaFluor 555 to conduct side-by-side comparison between the best performing PAMD-Ch17/siRNA polyplexes and parent PAMD/siRNA polyplexes (Figure 9). Not surprisingly, our results revealed that polyplexes that exhibited high transfection activity (Figure 8) also showed high levels of cellular internalization, confirming that cellular uptake is one of the main factors determining the success of siRNA delivery by the dual-function PAMD polyplexes. We then evaluated the effect of serum on the polyplex uptake and intracellular distribution. Serum is rich in anionic proteins that can bind to cationic polyplexes and affect the extent and mechanism of cell uptake and intracellular trafficking.48 Several studies have shown that hydrophobic modification of polycations can enhance serum compatibility.49, 50 While the presence of serum greatly decreased the uptake of PAMD/siRNA polyplexes, the adverse effect on uptake of PAMD-Ch17/siRNA was markedly reduced. In particular, PAMD-Ch17/siRNA polyplexes prepared at w/w 2 exhibited considerably higher cellular uptake and distribution to the cytoplasm than PAMD/siRNA polyplexes. We also observed less co-localization of the PAMD-Ch17 and siRNA signal (bright pink) when compared with PAMD/siRNA where nearly all siRNA was associated with the polycation. The decreased co-localization points to increased intracellular dissociation of the cholesterol-containing polyplexes and release of free siRNA. Such facilitation of intracellular polyplex dissociation by incorporating hydrophobic moieties has also been reported in other studies.51, 52 It is also worth noting that PAMD-Ch17/siRNA polyplexes prepared at w/w 2 appeared larger inside the cells than the ones prepared at w/w 5 (Figure 9). This could be due to lower colloidal stability (Figure 4) or due to the enhanced intracellular dissociation.
Figure 9.
Intracellular distribution of PAMD/siRNA and PAMD-Ch17/siRNA polyplexes in U2OS cells using siRNA labelled with AlexaFluor 555 (red) and polymers labelled with AlexaFluor 647 (blue) (cell nuclei stained with Hoechst 33258 (shown as white)).
CXCR4 antagonists compete with chemokine ligand SDF-1 and bind to CXCR4 receptors at the cell membrane, leading to inhibition of receptor internalization. Such receptor inhibition, however, would not be productive for siRNA delivery to the cytoplasm. Our previous studies with PAMD/DNA polyplexes have shown the internalization to be independent of the CXCR4 trafficking pathway. In a competitive binding experiment, we have shown that saturating CXCR4 receptors by preincubation with a small molecule inhibitor had no effect on cell uptake or transfection activity of the DNA polyplexes.27, 28 However, because cholesterol modification may alter the mechanism of interaction of polyplexes with cell membranes and membrane-bound receptors, it was essential to determine if the CXCR4 trafficking was involved and if any significant amount of siRNA polyplexes was trapped at the cell membrane.
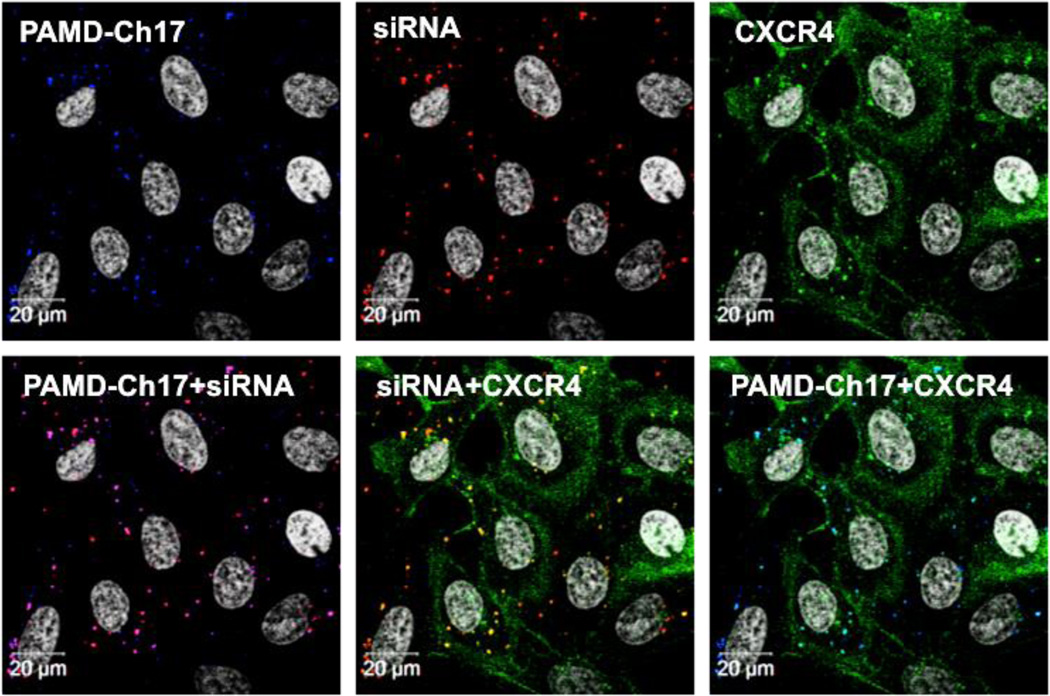
We tracked fluorescently labelled polyplexes and EGFP-CXCR4 receptors in U2OS cells by confocal microscopy to study the intracellular distribution of the polyplexes, as well as their interactions with the CXCR4 receptors in a single experimental setting. Cells were treated with PAMD-Ch17/siRNA polyplexes (w/w 5) and 10 nM SDF-1 for 1 h, which allowed PAMD-Ch17 polyplexes to directly compete with the chemokine ligand for binding with CXCR4. As shown in Figure 10, the polyplexes effectively antagonized CXCR4, as indicated by the mostly diffuse pattern of the EGFP-CXCR4 fluorescence (green) and only a limited amount of internalized receptors (discrete green puncta). In contrast to our previous finding with PAMD/DNA polyplexes,27 intracellular trafficking of at least a fraction of the PAMD-Ch/siRNA polyplexes overlapped with the CXCR4 trafficking as demonstrated by the colocalization of siRNA and CXCR4 (yellow), as well as the PAMD-Ch17 and CXCR4 (bright blue). The significance of this observation for the mechanism of siRNA delivery by PAMD-Ch is not clear and remains to be fully elucidated.
Figure 10.
Intracellular distribution of fluorescently labelled PAMD-Ch17/siRNA polyplexes in U2OS cells expressing EGFP-CXCR4 receptors. Cells were incubated with polyplexes and 10 nM SDF-1 for 1 h and imaged using a confocal microscope: siRNA (red), PAMD-Ch17 (blue), EGFP-CXCR4 (green), cell nuclei (white).
4. Conclusion
This study examined the effect of cholesterol modification on the ability PAMD to simultaneously deliver siRNA and inhibit CXCR4 chemokine receptor. Our results show that cholesterol modification provided the PAMD-Ch/siRNA polyplexes with increased colloidal stability and greatly enhanced siRNA transfection in the presence of serum, all the while preserving strong CXCR4 inhibitory activity. Studies with therapeutic siPLK1 demonstrated promising anticancer activity suggesting potential of PAMD-Ch/siRNA polyplexes as a novel combination treatment of cancer.
Acknowledgments
This work was supported in part by grants R01EB015216 and R21EB014570 from the National Institutes of Health. We thank Dr. Kelly from the UNMC high-throughput facility for help with the CXCR4 inhibition analysis and Janice Taylor and James Talaska of the UNMC Advanced Microscopy Core Facility for help with the confocal microscopy experiments.
References
- 1.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Nat. Biotech. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 2.Kim DH, Rossi JJ. Nat. Rev. Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 3.Wagner E. Biomaterials Sci. 2013;1:804. doi: 10.1039/c3bm60071h. [DOI] [PubMed] [Google Scholar]
- 4.Thomas CE, Ehrhardt A, Kay MA. Nat. Rev. Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 5.Pack DW, Hoffman AS, Pun S, Stayton PS. Nat. Rev. Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 6.Whitehead KA, Langer R, Anderson DG. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, Molema G, Lu PY, Scaria PV, Woodle MC. Nucleic Acids Res. 2004;32:e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng M, Pavan GM, Neeb M, Schaper AK, Danani A, Klebe G, Merkel OM, Kissel T. ACS Nano. 2012;6:9447–9454. doi: 10.1021/nn301966r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merkel OM, Kissel T. J. Controlled Rel. 2014;190:415–423. doi: 10.1016/j.jconrel.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Kozielski KL, Tzeng SY, Hurtado De Mendoza BA, Green JJ. ACS Nano. 2014;8:3232–3241. doi: 10.1021/nn500704t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson CE, Kintzing JR, Hanna A, Shannon JM, Gupta MK, Duvall CL. ACS Nano. 2013;7:8870–8880. doi: 10.1021/nn403325f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, Zhang Z, Zhou C, Jiao Y. Prog. Polym. Sci. 2010;35:1144–1162. [Google Scholar]
- 13.Oe Y, Christie RJ, Naito M, Low SA, Fukushima S, Toh K, Miura Y, Matsumoto Y, Nishiyama N, Miyata K, Kataoka K. Biomaterials. 2014;35:7887–7895. doi: 10.1016/j.biomaterials.2014.05.041. [DOI] [PubMed] [Google Scholar]
- 14.Ranucci E, Suardi MA, Annunziata R, Ferruti P, Chiellini F, Bartoli C. Biomacromolecules. 2008;9:2693–2704. doi: 10.1021/bm800655s. [DOI] [PubMed] [Google Scholar]
- 15.Bajaj A, Kondaiah P, Bhattacharya S. Bioconjugate Chem. 2008;19:1640–1651. doi: 10.1021/bc700381v. [DOI] [PubMed] [Google Scholar]
- 16.Kim WJ, Chang CW, Lee M, Kim SW. J. Controlled Rel. 2007;118:357–363. doi: 10.1016/j.jconrel.2006.12.026. [DOI] [PubMed] [Google Scholar]
- 17.Wang DA, Narang AS, Kotb M, Gaber AO, Miller DD, Kim SW, Mahato RI. Biomacromolecules. 2002;3:1197–1207. doi: 10.1021/bm025563c. [DOI] [PubMed] [Google Scholar]
- 18.Chen CJ, Wang JC, Zhao EY, Gao LY, Feng Q, Liu XY, Zhao ZX, Ma XF, Hou WJ, Zhang LR, Lu WL, Zhang Q. Biomaterials. 2013;34:5303–5316. doi: 10.1016/j.biomaterials.2013.03.056. [DOI] [PubMed] [Google Scholar]
- 19.Balkwill FR. J. Pathol. 2012;226:148–157. doi: 10.1002/path.3029. [DOI] [PubMed] [Google Scholar]
- 20.Singh S, Sadanandam A, Singh R. Cancer Metastasis Rev. 2007;26:453–467. doi: 10.1007/s10555-007-9068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zlotnik A, Burkhardt AM, Homey B. Nat. Rev. Immunol. 2011;11:597–606. doi: 10.1038/nri3049. [DOI] [PubMed] [Google Scholar]
- 22.Burger M, Glodek A, Hartmann T, Schmitt-Graff A, Silberstein LE, Fujii N, Kipps TJ, Burger JA. Oncogene. 2003;22:8093–8101. doi: 10.1038/sj.onc.1207097. [DOI] [PubMed] [Google Scholar]
- 23.Chu QD, Holm NT, Madumere P, Johnson LW, Abreo F, Li BDL. Surgery. 2011;149:193–199. doi: 10.1016/j.surg.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 24.Hiller DJ, Meschonat C, Kim R, Li BD, Chu QD. Surgery. 2011;150:459–465. doi: 10.1016/j.surg.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Duda DG, Kozin SV, Kirkpatrick ND, Xu L, Fukumura D, Jain RK. Clin. Cancer Res. 2011;17:2074–2080. doi: 10.1158/1078-0432.CCR-10-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Li J, Oupicky D. Pharm. Res. 2014;31:3538–3548. doi: 10.1007/s11095-014-1440-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Oupicky D. Biomaterials. 2014;35:5572–5579. doi: 10.1016/j.biomaterials.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Zhu Y, Hazeldine ST, Li C, Oupicky D. Angew. Chem. Int. Ed. Engl. 2012;51:8740–8743. doi: 10.1002/anie.201203463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Gao S, Ye WH, Yoon HS, Yang YY. Nat. Mat. 2006;5:791–796. doi: 10.1038/nmat1737. [DOI] [PubMed] [Google Scholar]
- 30.Chen C-J, Wang J-C, Zhao E-Y, Gao L-Y, Feng Q, Liu X-Y, Zhao Z-X, Ma X-F, Hou W-J, Zhang L-R, Lu W-L, Zhang Q. Biomaterials. 2013;34:5303–5316. doi: 10.1016/j.biomaterials.2013.03.056. [DOI] [PubMed] [Google Scholar]
- 31.Furgeson DY, Chan WS, Yockman JW, Kim SW. Bioconjugate Chem. 2003;14:840–847. doi: 10.1021/bc0340565. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Zhu Y, Hazeldine ST, Firestine SM, Oupicky D. Biomacromolecules. 2012;13:3220–3227. doi: 10.1021/bm3009999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong CY, You YZ, Wu DC, Liu Y, Pan CY. J. Am. Chem. Soc. 2007;129 doi: 10.1021/ja070871+. 5354-+. [DOI] [PubMed] [Google Scholar]
- 34.Reschel T, Konak C, Oupicky D, Seymour LW, Ulbrich K. J. Controlled Rel. 2002;81:201–217. doi: 10.1016/s0168-3659(02)00045-7. [DOI] [PubMed] [Google Scholar]
- 35.Filippov SK, Konak C, Kopeckova P, Starovoytova L, Spirkova M, Stepanek P. Langmuir. 2010;26:4999–5006. doi: 10.1021/la9036716. [DOI] [PubMed] [Google Scholar]
- 36.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. Biomaterials. 2003;24:1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 37.Parhamifar L, Larsen AK, Hunter AC, Andresen TL, Moghimi SM. Soft Matter. 2010;6:4001–4009. [Google Scholar]
- 38.Wu C, Li J, Zhu Y, Chen J, Oupicky D. Biomaterials. 2013;34:8843–8850. doi: 10.1016/j.biomaterials.2013.07.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neamnark A, Suwantong O, Bahadur RK, Hsu CY, Supaphol P, Uludag H. Mol. Pharm. 2009;6:1798–1815. doi: 10.1021/mp900074d. [DOI] [PubMed] [Google Scholar]
- 40.Bridger GJ, Skerlj RT, Hernandez-Abad PE, Bogucki DE, Wang Z, Zhou Y, Nan S, Boehringer EM, Wilson T, Crawford J, Metz M, Hatse S, Princen K, De Clercq E, Schols D. J. Med. Chem. 2010;53:1250–1260. doi: 10.1021/jm901530b. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Hazeldine ST, Li J, Oupicky D. Adv. Healthc. Mater. doi: 10.1002/adhm.201400608. (in press). [DOI] [PubMed] [Google Scholar]
- 42.Degenhardt Y, Lampkin T. Clin. Cancer Res. 2010;16:384–389. doi: 10.1158/1078-0432.CCR-09-1380. [DOI] [PubMed] [Google Scholar]
- 43.McInnes C, Wyatt MD. Drug Discov. Today. 2011;16:619–625. doi: 10.1016/j.drudis.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 44.Strebhardt K. Nat. Rev. Drug Discov. 2010;9:643–660. doi: 10.1038/nrd3184. [DOI] [PubMed] [Google Scholar]
- 45.Yao YD, Sun TM, Huang SY, Dou S, Lin L, Chen JN, Ruan JB, Mao CQ, Yu FY, Zeng MS, Zang JY, Liu Q, Su FX, Zhang P, Lieberman J, Wang J, Song E. Sci. Transl. Med. 2012;4:130ra148. doi: 10.1126/scitranslmed.3003601. [DOI] [PubMed] [Google Scholar]
- 46.Liu X, Lei M, Erikson RL. Mol. Cell. Biol. 2006;26:2093–2108. doi: 10.1128/MCB.26.6.2093-2108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seth S, Matsui Y, Fosnaugh K, Liu Y, Vaish N, Adami R, Harvie P, Johns R, Severson G, Brown T, Takagi A, Bell S, Chen Y, Chen F, Zhu T, Fam R, Maciagiewicz I, Kwang E, McCutcheon M, Farber K, Charmley P, Houston ME, Jr, So A, Templin MV, Polisky B. Mol. Ther. 2011 [Google Scholar]
- 48.Thomas M, Klibanov AM. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14640–14645. doi: 10.1073/pnas.192581499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bajaj A, Kondaiah P, Bhattacharya S. Bioconjugate Chem. 2008;19:1640–1651. doi: 10.1021/bc700381v. [DOI] [PubMed] [Google Scholar]
- 50.Eliyahu H, Makovitzki A, Azzam T, Zlotkin A, Joseph A, Gazit D, Barenholz Y, Domb A. Gene Ther. 2004;12:494–503. doi: 10.1038/sj.gt.3302395. [DOI] [PubMed] [Google Scholar]
- 51.Kurisawa M, Yokoyama M, Okano T. J. Controlled Rel. 2000;68:1–8. doi: 10.1016/s0168-3659(00)00246-7. [DOI] [PubMed] [Google Scholar]
- 52.Gabrielson NP, Pack DW. Biomacromolecules. 2006;7:2427–2435. doi: 10.1021/bm060300u. [DOI] [PubMed] [Google Scholar]