Abstract
The aim of this study was to investigate the prevalence of ESBL and MBL encoding genes among A. baumannii isolates. In this cross sectional study, 100 A. baumannii strains were isolated from ICU wards of 3 educational hospitals of Hamadan City, Iran in 2011. Phenotypic identification of the production of ESBLs and MBLs has been carried out by using E-test and DDST methods, respectively. PCR technique was used for amplification of the ESBL and MBL encoding genes, namely: CTX-M, SHV, TEM, OXA-51, VIM-Family, IMP-Family, SPM-1, SIM-1, and GIM-1. Eighty seven (87%), 95 (95%), 98 (98%) and 95 (95%) out of 100 A. baumannii isolates were resistant to imipenem, meropenem, ceftazidime and cefotaxime, respectively. Also, 99% and 7% of the isolates were MBLs and ESBLs produced phenotypically. Thirty (30%), 20 (20%) and 58 (58%) out of 100 A. baumannii isolates have been confirmed to harbor the blaVIM-family, TEM and SHV genes, respectively. Our results show no significant relationship between the detected gens with production of MBLs and ESBLs in spite of high prevalence of MBL encoding and drug resistant A. baumannii. Probably some other genes rather than what we studied are involved in phenotypic production of MBLs and ESBLs and subsequent drug resistance in Hamadan area, Iran.
Keywords: Acinetobacter baumannii, Drug resistance, MBL, ESBL
1. Introduction
Acinetobacter baumannii is an opportunistic pathogen, with the following characteristics of being Gram-negative, oxidase-negative, non-fermentative, nonmotile coccobacilli and is an unknown natural reservoir and has broad range of antibiotic resistance. The bacterium is prevalent in most places especially in hospitals and other health care institutes (Perez et al., 2007; Yeom et al., 2013; Poirel et al., 2011). Over the last 30 years, Acinetobacter genus has been faced with considerable changes in its taxonomic place (Peleg et al., 2008). High morbidity and mortality are the characteristics of nosocomial infections caused by Acinetobacter spp., which included urinary tract, skin and soft tissue infections, pneumonia and bacteremia especially in patients with severe health conditions (Karageorgopoulos et al., 2008; Safari et al., 2013; Kuo et al., 2012). Most studies conducted by researchers on this bacterium have been carried out concentrating on its drug resistant aspects, which is a major factor limiting the treatment of nosocomial infections (Poirel et al., 2011; Cerqueira and Peleg, 2011). During the last two decades, the advent and widespread dissemination of bacterial infections resistant to beta-lactams, especially to 3rd generation of cephalosporins and carbapenems, has become a globally significant problem (Pfeifer et al., 2010). Metallo beta lactamases (MBLs) are sorts of powerful enzymes called carbapenemases responsible for antibiotic resistance (Bush, 2001). Four groups of these enzymes have been described in A. baumannii, including IMP-like, SIM-1, NDM-type and VIM-like carbapenemases (Poirel et al., 2011, 2010). Genetic characteristic of MBLs-encoding A. baumannii isolates revealed the presence of blaSIM, blaIMP and blaVIM genes (Poirel et al., 2011). Extended-spectrum ß-lactamases (ESBLs) are encoded by TEM-type, SHV-type and CTX-M-type genes, which are subjected to phonotypical resistance to penicillins and 3rd generation cephalosporins (Pfeifer et al., 2010). ESBLs are mostly plasmid-mediated and most are parts of the TEM and SHV families of enzymes (Mehrgan and Rahbar, 2008).
One of the notable carbapenem-resistance mechanisms is caused by carbapenem-hydrolyzing β-lactamases, carbapenemases. In addition, the metallo-β-lactamases (MβLs) paly a crucial role in drug resistance against carbapenems (Poirel and Nordmann, 2006). Also, the extended-spectrum β-lactamases, ESBLs, play an important role in resistance against later generation cephalosporins such as cefepime, cefotaxime and ceftazidime (Zhanel et al., 2013).
The aim of this study was to investigate the prevalence of ESBLs and MBLs encoding genes and drug resistance against meropenem, imipenem, ceftazidime and cefotaxime among A. baumannii isolates.
2. Materials and methods
2.1. Sampling and isolation of bacteria
The study was a cross sectional study initiated in June 2011 by collecting 100 non-duplicate A. baumannii isolates from clinical specimens from ICU ward patients hospitalized in three educational hospitals of Hamadan City in Iran. The sampling was conducted for 17 months and the isolates were almost from 74 tracheal aspirate, 16 blood, 5 urine, 4 sputum and 1 wound samples. The identification of the isolates has been accomplished by biochemical tests and confirmed by tracking the blaOXA-51 -like carbapenemase gene, which is intrinsic to this species, using single PCR (Turton et al., 2006). The confirmed A. baumannii isolates were kept frozen at −70 °C for further tests.
2.2. Antibiogram
Susceptibility to meropenem, imipenem, ceftazidime and cefotaxime (Mast CO, UK) was tested by Kirby-Bauer disk diffusion method. The antibiogram procedure was performed as the manufacturer constructed. In brief, 1.5 × 108 CFU of bacterial suspension, equivalent to McFarland Turbidity Standard No. 0.5, was transferred on Muller-Hinton agar medium (Merck, Germany) and antibiogram disks containing meropenem (10 μg), imipenem (10 μg), ceftazidime (30 μg) and cefotaxime (30 μg) were placed on the medium. Then, the media were incubated for 18 h at 35 °C. The results were interpreted according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2007). A control strain of Pseudomonas aeruginosa ATCC 27853 was used for quality control of susceptibility testing.
2.3. Phenotypic MBL detection
For determination of phenotypic MβL production among the bacterial isolates, MIC Test Strips (Lioflichem® Italy) containing imipenem and imipenem plus EDTA were used. The E-test procedure was performed according to the manufacturer’s manual. Reduction in the MIC of imipenem of ⩾3 dilutions in the presence of EDTA is regarded as a positive result. Also, A. baumannii strain was considered MβL producer if a phantom zone or deformation of the ellipse was obviously observed.
2.4. Phenotypic identification of ESBL producing isolates
Phenotypic identification of ESBL producing isolates have been carried out using DDST screening method. Antibiogram disks containing ceftazidime (30 μg), cefotaxime (30 μg), ceftazidime (30 μg) + clavulanic acid (10 μg) and cfotaxime (30 μg) + clavulanic (10 μg) were used. Pairs of disks (ceftazidime with ceftazidime/clavulanic acid and cefotaxime with cfotaxime /clavulanic) were placed on Muller-Hinton agar medium (Merck, Germany) with 20 mm space between them. According to the CLSI criteria and manufacturer instruction, the ⩾5 mm inhibition zone of growth in ceftazidime/clavulanic acid and cfotaxime/clavulanic than ceftazidime and cfotaxime was regarded as an isolate that is producing ESBLs.
2.5. Amplification of blaOXA-51-like gene
The bacterial DNA was extracted by the alkaline lysis method (Kheyrodin and Ghazvinian, 2012). The pair of blaOXA-51 primers (Bioneer® Korea); OXA-F 5′-TAATGCTTTGATCGGCCTTG-3′, and OXA-R 5′-TGGATTGCACTTCATCTTGG-3′ were used for PCR detection of the genes (Turton et al., 2006). PCR amplification procedure was performed using 25 μl of master mix containing 0.2 μl of Taq polymerase 5 U/μl, 2.5 μl of 10XPCR buffer along with MgCl2, 1 μl of 10 pM from each reverse and forward primers, 2.5 μl of dNTPs MIX (2 Mm), 3 μl of DNA template, 14.8 μl of DNase-Free and RNase-Free Distilled Water. PCR amplification was done in the thermal cycler device. Agarose gel electrophoresis of the amplified DNA product with 100 bp size marker (Fermentas®, Korea) was carried out in a 2% agarose gel for 2 h at 80 V and stained with ethidium bromide to manifest and detect the 353 bp band. Also, PCR reaction contained positive and negative control. The A. baumannii carrying blaOXA-51-like gene (obtained from Dr. Bahador, Tehran University of Medical Sciences, Tehran, Iran) was used as the positive control.
2.6. Amplification of ESBL genes
The specific primers (Bioneer® Korea) including CTX-M, SHV and TEM (Table 1) were used for PCR amplification of the genes (Ellington et al., 2007; Kalai Blagui et al., 2007; Kolar et al., 2010; Turton et al., 2006). PCR amplification procedure was performed with 25 μl of master mix containing 0.2 μl of Taq polymerase 5 U/μl, 2.5 μl of 10× PCR buffer along with MgCl2, 1 μl of 10 pM from each reverse and forward primers, 2.5 μl of dNTPs MIX (2 Mm), 3 μl of DNA template,14.8 μl of DNase-Free and RNase-Free Distilled Water. PCR amplification was done in the thermal cycler device. Agarose gel electrophoresis of the amplified DNA product with 100 bp size marker (Fermentas®, Korea) was carried out in a 2% agarose gel for 2 h at 80 V and stained with ethidium bromide.
Table 1.
Primers used for PCR amplification of the studied genes.
| Primer name | Primer sequence (5′ to 3′) | Annealing temp (°C) | Product size (bp) | Reference |
|---|---|---|---|---|
| CTX-M | F:TCTTCCAGAATAAGGAATCCC | 51 | 909 | Kalai Blagui et al. (2007) |
| R:CCGTTTCCGCTATTACAAAC | ||||
| SHV | F: CTTTACTCGCTTTATCG | 53 | 868 | Kolar et al. (2010) |
| R: TCCCGCAGATAAATCAC | ||||
| TEM | F:ATGAGTATTCAACATTTCCG | 53 | 931 | Kalai Blagui et al. (2007) |
| R:CCAATGCTTAATCAGTGAGC | ||||
| OXA-51 | F:TAATGCTTTGATCGGCCTTG | 52 | 353 | Turton et al. (2006) |
| R:TGG ATTGCACTTCATCTTGG | ||||
| VIM-Family | F: GATGGTGTTTGGTCGCATA | 52 | 390 | Ellington et al. (2007) |
| R: CGA ATGCGCAGCACCAG | ||||
| IMP-Family | F: GGAATAGAGTGGCTTAAYTCTC | 52 | 188 | Ellington et al. (2007) |
| R: CCA AACYACTASGTTATCT | ||||
| SPM-1 | F:AAAATCTGGGTACGCAAACG | 54 | 271 | Ellington et al., 2007 |
| R:ACATTATCCGCTGGAACAGG | ||||
| SIM-1 | F:TAC AAGGGATTCGGCATCG | 54 | 570 | Ellington et al. (2007) |
| R: TAATGGCCTGTTCCCATGTG | ||||
| GIM-1 | F: TCG ACACACCTTGGTCTGAA | 54 | 477 | Ellington et al. (2007) |
2.7. Amplification of MBL genes
The specific primers (Bioneer® Korea) including VIM-Family, IMP-Family, SPM-1, SIM-1 and GIM-1 (Table 1) were used for PCR amplification of the genes (Ellington et al., 2007). PCR amplification procedure was performed with 25 μl of master mix containing 0.2 μl of Taq polymerase 5 U/μl, 2.5 μl of 10X PCR buffer along with MgCl2, 1 μl of 10 pM from each reverse and forward primers, 2.5 μl of dNTPs MIX (2 Mm), 3 μl of DNA template,14.8 μl of DNase-Free and RNase-Free Distilled Water. PCR amplification was done in the thermal cycler device. Agarose gel electrophoresis of the amplified DNA product with 100 bp size marker (Fermentas®, Korea) was carried out in a 2% agarose gel for 2 h at 80 V and stained with ethidium bromide.
2.8. Statistical analysis
Statistical results were calculated by the Statistical Package for the Social Sciences (SPSS Inc., USA) version 16.0 for windows and also, using McNemar and Chi square tests regarding P ⩽ 0.05 as significance level.
3. Results
A. baumannii isolates were confirmed with microbiological and PCR (bla-OXA-51) methods and entered to the study (Fig. 1).
Figure 1.
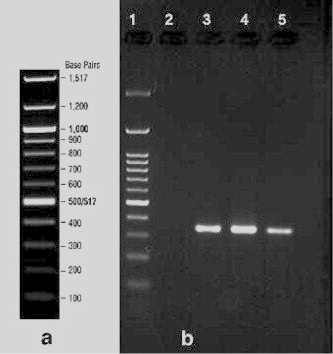
PCR results for amplification of blaOXA-51 gene in A. baumannii isolates; Lane 1:100 bp DNA ladder; Lane 2: negative control; Lane 3: Positive control; Lanes 4, 5 different isolates harboring blaOXA-51-like gene.
3.1. Antibiogram
Susceptibility to meropenem, imipenem, ceftazidime and cefotaxime was evaluated by the Kirby-Bauer disk diffusion method. Eighty seven percent, 95%, 98% and 95% out of 100 A. baumannii isolates were resistant to imipenem, meropenem, ceftazidime and cefotaxime, respectively.
3.2. Phenotypic MBL detection
For determination of phenotypic MβL production among isolates, MIC Test strips containing imipenem and imipenem along with EDTA were used. Ninety nine percent out of 100 A. baumannii isolates were MBL producing.
3.3. Phenotypic identification of ESBL producing isolates
Phenotypic identification of ESBL producing isolates have been carried out using DDST screening method. From total of 100 samples, 7% A. baumannii isolates identified to be produce ESBL enzymes.
3.4. Detection of ESBL–SHV, -CTX-M and -TEM genes
Of all 100 A. baumannii isolates, 58%, and 20% isolates were harboring HSV, and CTX-M genes, respectively. The TEM gene was not found in the studied strains. There was no statistically significant relationship between the cefotaxime and ceftazidime resistance and presence of SHV, CTX-M genes in the isolates.
3.5. Detection of VIM-Family, IMP-Family, SPM-1, SIM-1 and GIM-1genes
Thirty percent out of 100 A. baumannii isolates has been confirmed to harbor the blaVIM-family genes (Fig. 2), but the other genes including IMP-Family, SPM-1, SIM-1 and GIM-1 have not been detected (Table 2). No significant relationship was observed between the presence and absence of blaVIM-family genes in the isolates’ resistance to imipenem and meropenem.
Figure 2.
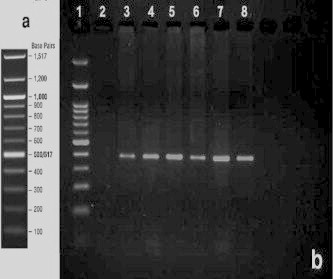
PCR product gel electrophoresis; a: 100 bp DNA ladder template of the company; b: PCR results for amplification of blaVIM-family genes in A. baumannii isolates; Lane 1:100 bp DNA ladder; Lane 2: negative control; Lane 3, Positive control; Lanes 4, 8 different isolates harboring blaVIM-family gene.
Table 2.
Frequency of the studied genes among 100 A. baumannii isolates.
| Gene | PCR results |
|
|---|---|---|
| Positive N (%) | Negative N (%) | |
| SHV | 58 (58) | 42 (42) |
| TEM | 20 (20) | 80 (80) |
| CTX-M | 0 (0) | 100 (100) |
| VIM-Family | 30 (30) | 70 (70) |
| IM-Family | 0 (0) | 100 (100) |
| SIM | 0 (0) | 100 (100) |
| SPM | 0 (0) | 100 (100) |
| GIM | 0 (0) | 100 (100) |
4. Discussion
Production of carbapenem-hydrolyzing β-lactamases, also called carbapenemases, is one of the significant mechanisms of carbapenem resistance, in which Methalo β-lactamases (MβLs) possess the principal role in drug resistance against carbapenems (Poirel and Nordmann, 2006). Also, the extended-spectrum β-lactamases, ESBLs, play an important role in resistance against later generation cephalosporins such as cefotaxime, ceftazidime, and cefepime (Zhanel et al., 2013). In this study, the prevalence of ESBLs and MBLs encoding genes and drug resistance against meropenem, imipenem, ceftazidime and cefotaxime among A. baumannii isolates has been investigated showing a high resistance rate among the antibiotics.
Totally 100 A. baumannii isolates have been examined for 3 ESBLs and 5 MBLs encoding genes. Three out of eight genes have been detected including SHV (58%), TEM (20%) and VIM (30%). None of the other studied genes has been detected among 100 isolates of A. baumannii, which were isolated from ICU wards of 3 educational Hospitals, Hamadan City, Iran, 2011. Also, no significant relationship has been observed about the presence of the detected ESBL and MBL encoding genes with phonotypical resistance against imipenem, meropenem, cefotaxime and ceftazidime. Eighty seven percent, 95%, 98% and 95% out of 100 A. baumannii isolates were resistant to imipenem, meropenem, ceftazidime and cefotaxime, respectively.
The results showed that most of the A. baumannii isolates were producing MBLs (99%), but not ESBLs (7%). There was not any significant relationship about phonotypic ESBL and MBL producing and the detected genes. Reports from Iran illustrate the high prevalence of drug resistance and multi drug resistance in A. baumannii especially against most effective antibiotics such as imipenem and meropenem (Feizabadi et al., 2008; Peymani et al., 2011). Ting et al. (2013) investigated the drug resistance genes in 7 strains of imipenem-resistant A. baumannii including TEM, SHV, CTX-M, DHA, CIT, IMP, VIM, KPC, OXA-23. They detected TEM (100%) and OXA-23 (100%) genes among the isolates, but the other genes such as SHV, CTX-M, DHA, CIT, IMP, VIM, KPC could not be detected from 7 strains of imipenem-resistant A. baumannii. In the present study, consistent with Ting et al. (2013), just some of the genes have been detected including SHV (58%), TEM (20%) and VIM (30%). In another study by Shahcheraghi et al. (2011) in Tehran, Iran, they showed that the MBL encoding genes included bla VIM-2, bla SPM-1, bla IMP-2, bla GES-1, bla OXA-51, bla OXA-23 genes among 203 A. baumannii isolates. They reported that 6 isolates produce MBLs and 94 isolates produce OXA-type carbapenemase. Their finding suggests that in Tehran the prevalence of MBLs producing A. baumannii strains is lower than that of the present study from Hamadan City. They detect blaSPM-1, blaGES-1, blaOXA-51, blaOXA-23 genes among 6, 2, 94 and 84 isolates of the bacterium, respectively (Shahcheraghi et al., 2011). The previous research by Rezaee et al. (2013) revealed genes coding for IMP, SPM-1, VIM, PER-1, VEB-1, TEM, SHV, GES-1, and CTX-M among 76 Acinetobacter spp. Also, they reported that 37% of isolates carried at least one of the blaPER-1 or blaTEM-1 genes and 13.15% of their studied isolates reported to harbor blaTEM-1 gene, which is similar to that of the present study (20%). Also, none of their studied A. baumannii isolates were harboring for blaVEB-1, blaSHV-, blaCTX-M-2 and blaGES-1 (Rezaee et al., 2013). In our study, none of the following genes, CTX-M, IMP, SIM, SPM, and GIM, has been detected among 100 isolates of A. baumannii.
Our results show that there was not any significant relationship between the detected genes with production of MBLs and ESBLs. Probably, some other genes rather than what we studied are involved in phenotypic production of MBLs and ESBLs and the subsequent drug resistance in Hamadan, Iran.
5. Conclusion
Despite the high prevalence of phenotypic MBL production and high resistance rate against imipenem and meropenem A. baumannii isolates, lower rates of MBL encoding genes have been detected. Also, the high resistance rate against ceftazidime and cefotaxime was not in relation with phenotypic and genotypic ESBL production. Probably some other genes rather than what we studied are involved in phenotypic production of MBLs and ESBLs and subsequent high drug resistance in Hamadan, Iran.
Acknowledgments
The authors would like to thank the Vice-chancellor of Research of Hamadan University of Medical Sciences for supporting this project.
Footnotes
Peer review under responsibility of King Saud University.
References
- Bush K. New beta-lactamases in gram-negative bacteria: diversity and impact on the selection of antimicrobial therapy. Clin. Infect. Dis. 2001;32:1085–1089. doi: 10.1086/319610. [DOI] [PubMed] [Google Scholar]
- Cerqueira G.M., Peleg A.Y. Insights into Acinetobacter baumannii pathogenicity. IUBMB Life. 2011;63:1055–1060. doi: 10.1002/iub.533. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Eighteenth Informational Supplement. M100-S18. Wayne, PA, 2007.
- Ellington M.J., Kistler J., Livermore D.M., Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J. Antimicrob. Chemother. 2007;59:321–322. doi: 10.1093/jac/dkl481. [DOI] [PubMed] [Google Scholar]
- Feizabadi M.M., Fathollahzadeh B., Taherikalani M., Rasoolinejad M., Sadeghifard N., Aligholi M., Soroush S., Mohammadi-Yegane S. Antimicrobial susceptibility patterns and distribution of blaOXA genes among Acinetobacter spp. isolated from patients at Tehran hospitals. Jpn. J. Infect. Dis. 2008;61:274–278. [PubMed] [Google Scholar]
- Kalai Blagui S., Achour W., Abbassi M.S., Bejaoui M., Abdeladhim A., Ben Hassen A. Nosocomial outbreak of OXA-18-producing Pseudomonas aeruginosa in Tunisia. Clin. Microbiol. Infect. 2007;13:794–800. doi: 10.1111/j.1469-0691.2007.01757.x. [DOI] [PubMed] [Google Scholar]
- Karageorgopoulos D.E., Kelesidis T., Kelesidis I., Falagas M.E. Tigecycline for the treatment of multidrug-resistant (including carbapenem-resistant) Acinetobacter infections: a review of the scientific evidence. J. Antimicrob. Chemother. 2008;62:45–55. doi: 10.1093/jac/dkn165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheyrodin H., Ghazvinian K. DNA purification and isolation of genomic DNA from bacterial species by plasmid purification system. Afr. J. Agric. Res. 2012;7:433–442. [Google Scholar]
- Kolar M., Bardon J., Chroma M., Hricova K., Stosova T., Sauer P., Koukalova D. ESBL and AmpC beta-lactamase-producing Enterobacteriaceae in poultry in the Czech Republic. Vet. Med. Czech. 2010;55:119–124. [Google Scholar]
- Kuo S.C., Chang S.C., Wang H.Y., Lai J.F., Chen P.C., Shiau Y.R., Huang1 I.-W., Yang Lauderdale T.-L., TSAR Hospitals Emergence of extensively drug-resistant Acinetobacter baumannii complex over 10 years: nationwide data from the Taiwan Surveillance of Antimicrobial Resistance (TSAR) program. BMC Infect. Dis.2012;12:200. doi: 10.1186/1471-2334-12-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrgan H., Rahbar M. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli in a tertiary care hospital in Tehran, Iran. Int. J. Antimicrob. Agents. 2008;31:147–151. doi: 10.1016/j.ijantimicag.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Peleg A.Y., Seifert H., Paterson D.L. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F., Hujer A.M., Hujer K.M., Decker B.K., Rather P.N., Bonomo R.A. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007;51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peymani A., Nahaei M.R., Farajnia S., Hasani A., Mirsalehian A., Sohrabi N., Abbasi L. High prevalence of metallo-beta-lactamase-producing Acinetobacter baumannii in a teaching hospital in Tabriz, Iran. Jpn. J. Infect. Dis. 2011;64:69–71. [PubMed] [Google Scholar]
- Pfeifer Y., Cullik A., Witte W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int. J. Med. Microbiol. 2010;300:371–379. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Poirel L., Bonnin R.A., Nordmann P. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life. 2011;63:1061–1067. doi: 10.1002/iub.532. [DOI] [PubMed] [Google Scholar]
- Poirel L., Naas T., Nordmann P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob. Agents Chemother. 2010;54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 2006;12:826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- Yeom J., Shin J.H., Yang J.Y., Kim J., Hwang G.S. H NMR-based metabolite profiling of planktonic and biofilm cells in Acinetobacter baumannii 1656-2. PLoS One. 2013;8:e57730. doi: 10.1371/journal.pone.0057730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaee M.A., Pajand O., Nahaei M.R., Mahdian R., Aghazadeh M., Ghojazadeh M., Hojabri Z. Prevalence of Ambler class A beta-lactamases and ampC expression in cephalosporin-resistant isolates of Acinetobacter baumannii. Diagn. Microbiol. Infect. Dis. 2013;76:330–334. doi: 10.1016/j.diagmicrobio.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Safari M., Saidijam M., Bahador A., Jafari R., Alikhani M.Y. High prevalence of multidrug resistance and metallo-beta-lactamase (MbetaL) producing Acinetobacter baumannii isolated from patients in ICU Wards, Hamadan, Iran. J. Res. Health Sci. 2013;13:162–167. [PubMed] [Google Scholar]
- Shahcheraghi F., Abbasalipour M., Feizabadi M., Ebrahimipour G., Akbari N. Isolation and genetic characterization of metallo-beta-lactamase and carbapenamase producing strains of Acinetobacter baumannii from patients at Tehran hospitals. Iran. J. Microbiol. 2011;3:68–74. [PMC free article] [PubMed] [Google Scholar]
- Ting C., Jun A., Shun Z. Detection of the common resistance genes in Gram-negative bacteria using gene chip technology. Indian J. Med. Microbiol. 2013;31:142–147. doi: 10.4103/0255-0857.115230. [DOI] [PubMed] [Google Scholar]
- Turton J.F., Woodford N., Glover J., Yarde S., Kaufmann M.E., Pitt T.L. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 2006;44:2974–2976. doi: 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhanel G.G., Lawson C.D., Adam H., Schweizer F., Zelenitsky S., Lagace-Wiens P.R., Denisuik A., Rubinstein E., Gin A.S. Ceftazidime–avibactam: a novel cephalosporin/beta-lactamase inhibitor combination. Drugs. 2013;73:159–177. doi: 10.1007/s40265-013-0013-7. [DOI] [PubMed] [Google Scholar]


