Abstract
Imaging genetics has identified many contributions of DNA sequence variation to individual differences in brain function, behavior, and risk for psychopathology. Recent studies have extended this work beyond the genome by mapping epigenetic differences, specifically gene methylation in peripherally assessed DNA, onto variability in behaviorally and clinically relevant brain function. These data have generated understandable enthusiasm for the potential of such research to illuminate biological mechanisms of risk. Here, we use our research on effects of genetic and epigenetic variation in the human serotonin transporter on brain function to generate a guardedly optimistic opinion that available data encourages continued research in this direction, and suggest strategies to promote faster progress moving forward.
Imaging Genetics
Nearly two decades since its introduction [1–3], the field of imaging genetics has provided novel insights into the fundamental nature through which variation in our DNA code shapes brain function and, consequently, our behavior [4]. By leveraging the common language of DNA, imaging genetics has further facilitated rapid translational discoveries across pre-clinical animal models and clinical human research [5]. Simultaneously, by identifying readily accessible genetic markers that reliably predict variability in behaviorally and clinically relevant brain function [6], imaging genetics has encouraged the development of fast and affordable methods for identifying specific mechanisms of risk for psychopathology positioned to inform novel strategies for targeted intervention and, possibly, prevention [4,7,8].
One example of how imaging genetics can drive the interdisciplinary research described above comes from studies of an association between common alleles of the serotonin transporter linked polymorphic region (5-HTTLPR) and variability in threat-related amygdala activity [3,9], which represents a risk phenotype for stress-related psychopathology including depression, anxiety, and posttraumatic stress disorder [10,11]. Specifically, we and others have demonstrated that carriers of the 5-HTTLPR short (S) allele typically express relatively increased threat-related amygdala activity in comparison to individuals homozygous for the long (L) allele [3,12]. This observed association in human imaging genetics studies has been remarkably convergent with findings not only from human pharmacological and multimodal neuroimaging research but also with studies of orthologous genetic variation non-human primate or genetic manipulation in rodent models [13]. Collectively, this research suggests that a relatively decreased capacity for serotonin (5-hydroxytryptamine, 5-HT) reuptake is associated with heightened neural and behavioral responding to threat. These basic mechanistic findings, in turn, align with epidemiologic data, which, despite some initial inconsistencies [14], suggests the S allele is associated with increased risk for psychopathology subsequent to stress exposure [15,16]. Thus, the 5-HTTLPR may usefully contribute to the search for biomarkers in precision medicine [17].
The association between 5-HTTLPR genotype and amygdala activity, although reliable, has remained frustratingly small ultimately explaining a meager 1–5% of inter-individual variability in this neural phenotype [18]. Although small effects of individual common polymorphisms are to be expected when examining complex biological and behavioral phenotypes, inconsistent methods for assessing amygdala activity [18] as well as unmeasured epistatic interactions [19], and environmental effects [20,21], may all obscure possibly greater contributions of the 5-HTTLPR to variability in amygdala activity. Accordingly, consideration of these factors, in addition to utilizing larger samples to minimize false positives and better characterize relatively small anticipated or observed effects [12], has been advanced as a future direction for studies of the 5-HTTLPR specifically and imaging genetics broadly [22,23]. In addition to these factors, however, there is increasing recognition that variability in the functional properties of the human genome beyond the DNA sequence likely contributes to individual differences in behaviorally and clinically relevant brain function.
The Epigenome
Amongst non-sequence based sources of variability in the genome, epigenetic modifications have been of specific interest. Broadly, epigenetic modifications encompass conformational changes in DNA and/or chromatin that do not alter the underlying nucleotide sequence, but regulate the intricate molecular machinery through which the spatiotemporal dynamics of gene expression are regulated (Box 1). Thus, unlike the basic DNA sequence, epigenetic “marks” not only vary among cell and tissue types, but also are often part of the very mechanism that drives cell and tissue differentiation. Within the mammalian genome, DNA methylation, or the chemical addition of a methyl group at the 5-carbon position of cytosine, most frequently within the context of a cytosine-guanine dinucleotide or CpG site (Box 2), is a stable and well-characterized form of epigenetic modification [24,25].
Box 1. Epigenetic Mechanisms.
The term epigenetics (from Greek: epi- over, outside of, around) refers to the study of chemical processes that regulate gene expression by modifying DNA or its associated proteins, but without altering the underlying DNA sequence. The epigenetic landscape can be dynamic, capturing genetic, environmental, and cell lineage effects, and has been shown to be at least partially heritable [41]. Epigenetic processes can be broadly divided into three groups, comprising DNA methylation, histone modifications, and the actions of non-coding RNA molecules.
Histone modifications
In the nucleus of a eukaryotic cell, DNA is typically organized into chromatin, a multimolecular complex whose basic unit is a nucleosome. Within a nucleosome, a stretch of DNA (146 base pairs) is wrapped around an octamer comprised of two molecules each of four core histone proteins. Nucleosomes are further organized into higher-order chromatin structures [82]. Chemical and physical interactions between DNA and histones, as well as among histones, determine how accessible a stretch of DNA is to transcription-regulating molecules and thus how likely a particular gene is to be transcribed at any moment. The more tightly a stretch of DNA is bound to histones, the less likely it is that genes in this stretch are transcriptionally active. Histones have amino acid tails that “stick out” beyond the compact nucleosome structure and can be modified post-translationally by the addition of one or more chemical groups. There are more than 100 different histone modifications which can alter interactions among histones, between histones and DNA, or facilitate the recruitment of additional chromatin-modifying proteins [83].
Non-coding RNAs
Functional RNA molecules whose structure is encoded in DNA, but which are not translated into protein are referred to as non-coding RNAs. Depending on each molecule’s unique properties, non-coding RNAs can participate in epigenetic regulation by recruiting DNA-or chromatin-modifying enzymes, or directly modifying other RNA molecules or RNA-protein complexes [84–86]. Accumulating research in animal models suggests that both histone modifications and non-coding RNAs in the brain play an important role in memory formation and other forms of normal or pathological neuroplasticity [58,87–89]. However, due to the highly dynamic and cell-specific [84,90,91] nature of these epigenetic processes, relatively little is known about their specific role in human brain function and behavior.
DNA methylation
As detailed in Box 2, methylation of DNA is the best understood epigenetic modification.
Box 2. DNA Methylation.
DNA methylation refers to the covalent addition of a methyl (-CH3) group to the fifth position carbon in the cytosine carbon ring, usually in the context of a CG dinucleotide (i.e., CpG site), to form 5-methylcytosine (5-mC). 5-mC can be further hydroxylated at select locations to form 5-hydroxymethylcytosine (5-hmC) [92]. Non-CpG methylation also occurs in the mammalian genome [93] and may be particularly common in the brain [93]; however, its functional implications, as well as those of 5-hmC, are less well understood.
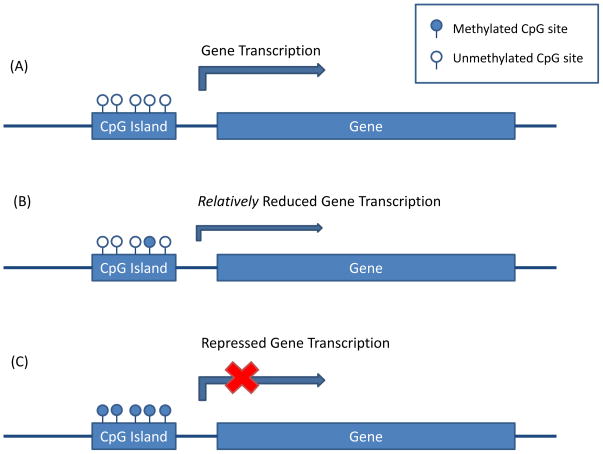
Most of the human genome is relatively depleted of CG content, with the majority of CpG sites being methylated [94]. However, CG content is enriched in distinct stretches of DNA called CpG islands, which tend to occur near transcriptional start sites of genes and which are generally hypomethylated in differentiated cells. Notably, the majority of known gene promoters have an associated CpG island [95,96]. Methylation of promoter-associated CpG islands tends to have an inhibitory effect on transcription initiation and thus correlates with reduced gene expression ([24], Figure I). However, studies suggest that methylation within gene bodies may have the opposite or no effect on gene expression [24].
It has been demonstrated that DNA methylation can directly impede the binding of transcription factors to recognition sequences containing CpG sites [97,98]. However, DNA methylation can also impact gene expression by recruiting methyl-CpG binding domain proteins (MBDs), which can in turn facilitate chromatin remodeling [99].
DNA methylation patterns across the genome are erased and re-established during gametogenesis and shortly after fertilization, but are otherwise maintained throughout development [100]. The enzyme DNA (cytosine-5)-methyltransferase 1 (DNMT1) is responsible for methylation maintenance during cell division, ensuring that methylation patterns are replicated between the two daughter cells [101]. The enzymes DNA (cytosine-5)-methyltransferase 3a and 3b (DNMT3A and DNMT3B) are responsible for de novo methylation, particularly early in development [102]. The removal of methylation marks can happen passively, when DNMT1 is absent or inhibited and methylation marks are not copied during cell division. However, given the dynamic and flexible nature of demethylation, it is more likely that the removal of methylation marks is enzymatically catalyzed. Since large amounts of energy are required to break the C-C bond within 5mC, there are likely several lower-energy steps likely involving 5-hmC as an intermediate product [92] in the demethylation process (See [103] for an extensive review).
Although the effects of DNA methylation on gene expression are complex and may vary depending on genomic location [24], numerous studies suggest that methylation occurring within CpG-rich regions near the transcription start site of a gene (i.e., CpG islands) tends to have a repressive effect on gene expression across tissues [24,26]. Furthermore, the existence of variability in DNA methylation independent of the underlying nucleotide sequence [27,28], suggests that epigenetic modifications can moderate the impact of genetic variation (i.e., genotypes) on biological processes including brain function. Thus, it is not just reasonable but imperative to consider the possibility that unaccounted for differences in DNA methylation or other epigenetic marks may contribute to unexplained variability within genotype groups in imaging genetics studies. Conversely, the consideration of epigenetic mechanisms offers a valuable opportunity to account for previously unexplained variability in neural phenotypes, and shed much needed additional light on the precise molecular mechanisms that may drive the emergence of inter-individual variability in brain function.
Epigenetics of Human Brain Function
We recently demonstrated that individual differences in the methylation of the proximal promoter of the serotonin transporter gene (SLC6A4) accounts for 6.7%–10.4% of variability in threat-related amygdala activity across two independent cohorts [29]. This association was independent of the 5-HTTLPR, which is near but not overlapping with the proximal promoter, and present across windows of development (i.e., adolescence and young adulthood). Moreover, the association was robust to assessment of DNA methylation from two different peripheral tissues (i.e., blood and saliva). We further demonstrated that methylation of the CpG site with the strongest association with amygdala activity was negatively correlated with serotonin transporter mRNA concentrations in postmortem amygdala tissue, providing a potential molecular mechanism that may drive the observed in vivo associations.
Other studies from the emergent field of imaging epigenetics [30], both prior to and following our report, have similarly focused on methylation within or near candidate genes that have been the target of previous imaging and psychiatric genetic studies (Table 1). Results from these studies have largely confirmed the notion that increased methylation, particularly within or near the promoter of a gene, is associated with reduced gene expression, and downstream neural phenotypes consistent therewith. As has been generally true in cutting edge research, results from these early epigenetics studies raise more questions than they answer. For example, where and how do these methylation patterns originate, and how do methylation patterns in peripheral tissues map onto patterns in brain? Although prior studies have provided partial answers to these questions (e.g., [31,32]), below we outline specific strategies that may be particularly useful in advancing this burgeoning field further, while accounting for specific limitations.
Table 1.
Summary of imaging epigenetics findings to date.
| Method | Authors | Year | Gene | Finding |
|---|---|---|---|---|
| Functional MRI | Ursini et al [31] | 2011 | COMT | Reduced methylation of the Val158 COMT allele was associated with less cortical efficiency in a working memory task, particularly in the context of stress. |
| Jack et al [65] | 2012 | OXTR | Increased methylation of the OXTR gene promoter was associated with greater activity in the temporal-parietal junction and dorsal anterior cingulate cortex during a social perception task. | |
| Walton et al [66] | 2014 | COMT | Increased COMT promoter methylation was associated with greater left dorsolateral prefrontal cortex activity during a working-memory task across patients with schizophrenia and controls. | |
| Vukojevic et al [32] | 2014 | NR3C1 | Increased NR3C1 promoter methylation was associated with greater activity in the right ventrolateral prefrontal cortex and cuneus, as well as reduced performance, in a memory task in healthy men. | |
| Nikolova et al [29] | 2014 | SLC6A4 | Increased SLC6A4 promoter methylation was associated with greater amygdala reactivity to threatening faces. | |
| Puglia et al [67] | 2015 | OXTR | Increased methylation of the OXTR gene promoter was associated with greater activity in the amygdala, insula and fusiform gyrus, as well as decreased amygdala connectivity with regulatory regions during threatening face processing. | |
| Frodl et al [68] | 2015 | SLC6A4 | SLC6A4 methylation was associated with differential modulation of activity in the insula, operculum, hippocampus and amygdala in an emotional attention-shifting task. | |
| Ziegler et al [69] | 2015 | OXTR | Decreased OXTR methylation was associated with increased amygdala activity during social-phobia related word processing in individuals with social anxiety disorder. | |
| Structural MRI | Klengel et al. [70] | 2013 | FKBP5 | Increased FKBP5 methylation was associated with decreased volume of the right hippocampal head. |
| Dannlowski et al [71] | 2014 | SLC6A4 | Increased methylation in a functional element of the SLC6A4 promoter was associated with increased hippocampal volume. | |
| Choi et al [72] | 2014 | BDNF | Increased BDNF promoter methylation was associated with reduced white matter integrity in patients with major depressive disorder. | |
| Booij et al [73] | 2015 | SLC6A4 | Increased SLC6A4 methylation was associated with childhood trauma and decreased hippocampal volume. | |
| PET | Wang et al [51] | 2012 | SLC6A4 | Increased SLC6A4 promoter methylation was associated with lower 5-HT synthesis in the orbitofrontal cortex. |
| Shumay et al [52] | 2012 | MAOA | Increased methylation near the MAOA promoter was associated with lower MAOA activity in healthy men. |
BDNF = brain derived neurotrophic factor
COMT = catechol-O-methyltransferase
FKBP5 = FK506 binding protein 5
MAOA = monoamine oxidase A
NR3C1 = glucocorticoid receptor
OXTR = oxytocin receptor
PET = positron emission tomography
With regards to the origin of variable methylation, it is generally accepted that epigenetic marks are at least partially heritable [25]. However, animal studies have demonstrated that they can change in response to environmental factors particularly early in development [33]. Correlational studies suggest this may be the case in humans as well. Specifically, it has been shown that methylation patterns, within and near SLC6A4 [34–36], as well as across the genome [37], differ as a function of early or recent life stress or trauma. Perhaps more intriguingly, recent studies in animals [38] and humans [39,40] have raised the possibility that methylation patterns in offspring may partially reflect differences in the experiences of the parents [41]. Finally, as evidenced by studies in monozygotic twin pairs of different ages, ordinary life experiences and normal aging may steadily alter genome-wide methylation patterns in the absence of severe stress or trauma [28].
Taken together, these studies suggest that even if DNA methylation patterns do not reflect personal or parental traumatic experiences, they are likely to at least partially capture the impact of an individual’s unique environment on the functional properties of the genome. Thus, it stands to reason that taking DNA methylation into account will help explain more variability in brain function than DNA sequence-based variation alone. In many ways, explicitly considering the impact of epigenetics is a logical next step in imaging genetics, which has quickly embraced the importance of environmental factors in the emergences of associations between sequence variants and brain function [23]. More tellingly, epigenetic changes have long been postulated to constitute a crucial part of the mechanism through which experience gets “under the skin” [42–44].
As the DNA sequence is the same in every cell within an individual, with rare exceptions [45], there is little question that DNA derived from easily sampled peripheral tissues can serve as a reliable proxy for DNA in the human brain. In contrast, epigenetic marks are expected to be different between cell types and tissues. Thus, it has been questioned whether methylation patterns readily measurable in DNA derived from peripheral tissues such as blood or saliva are veritable proxies for methylation patterns in the brain. Although the methylation status of a number of CpG sites has indeed been shown to differ substantially among tissues [46], some studies report blood-brain correlations in DNA methylation of up to 0.90 [47,48]. Notably, the largest methylation differences occur within or near genes involved in tissue differentiation, including neurogenesis and hematopoiesis [46].
Additional convergence has been reported between methylation patterns in blood and saliva, with some comparisons further suggesting that patterns in DNA derived from saliva rather than blood better map onto variability in brain function [49]. Although much remains unknown about the exact mapping of the entire methylomes of the brain and peripheral tissues (see also Box 3), significant cross-tissue convergence in methylation patterns has been observed for CG-rich promoters across genes [46]. In light of this promising, albeit partial, convergence between blood and brain methylomes, we argue that even in the absence of a full cross-tissue molecular characterization of a particular DNA methylation mark, a careful selection of well-matched epigenetic and phenotypic measures, especially when coupled with replication in independent samples, could reasonably safeguard against false positive associations.
Box 3. Outstanding Questions.
What are the mechanisms through which methylation changes in the brain may be mirrored in the periphery and vice versa? Does this cross-tissue convergence reflect the actions of DNMT1 (Box 2) through multiple cell divisions, ultimately demonstrating the shared developmental origin of cells, or does it at least partially reflect experiences in adulthood?
What is the temporal stability of DNA methylation markers and, perhaps more importantly, the correlation between the brain and the blood methylomes? Does this correlation change as a function of development or experience, and how can such change be leveraged to study the effects of environmental factors on brain function, behavior, and risk for psychopathology?
Which peripheral tissue is the best proxy for DNA methylation in the brain? Most neuroimaging epigenetics studies have assessed peripheral methylation in DNA extracted from blood. However, saliva is easier to collect and research suggests convergence between DNA methylation patterns in blood and saliva [56,104].
How can knowledge of regionally specific DNA methylation in the brain [46] be leveraged to design more specific hypotheses for imaging epigenetics studies?
What is the impact of relatively small-scale inter-individual variability (e.g., 0–5%) in DNA methylation, particularly within otherwise hypomethylated CpG islands, on gene expression across tissues, and are currently available methylation assays capable of accurately capturing variability on such small scale?
Are there epigenetic marks other than DNA methylation (e.g., histone modifications, small non-coding RNAs) that correlate between brain and peripheral tissues and have meaningful effects on brain function?
Despite these signposts, relatively few studies to date have ventured to combine neuroimaging and epigenetics, and those that have, have been rightfully cautious in selecting only DNA methylation marks which have been extensively functionally characterized in concurrent or prior molecular work (Table 1). Given recent advances in our understanding of the brain and blood methylomes, we argue that enough evidence has accumulated to warrant the adoption of less conservative approaches moving forward. This could in turn give the field a much-needed initial push and begin a constructive dialogue of mutual critique and conceptual guidance between the fields of neuroimaging and basic molecular epigenetics.
Challenges and Future Directions
Candidate Genes and Genomic Targets
As in imaging genetics, to ensure solid hypothesis construction, early imaging epigenetics research would be best served by investigating well-understood biological systems (e.g., focus on dopaminergic genes when studying reward-related brain function [50]). By the same token, a focus on genomic regions where the impact of DNA methylation is best understood, such as CG-rich gene promoters, will also benefit the research. Conversely, it would be wise to eschew a focus on markers known to be involved in tissue differentiation and neurodevelopment, which are most likely to be differentially methylated across the brain and peripheral tissues [46]. Employing in vivo molecular imaging such as positron emission tomography (PET), which provides more direct links to brain chemistry [such as [51,52]] or multimodal PET/fMRI may be particularly fruitful by establishing intermediate molecular phenotypes (e.g., differences in serotonin transporter levels) that may bridge DNA methylation and systems-level phenotypes of brain function.
It should be noted that this candidate gene/system approach would be open to the same criticisms and limitations as traditional candidate gene studies [53,54]. However, within this context, suitable candidate genes and systems can also be those identified on the basis of prior GWAS, particularly if buttressed by additional biological evidence. Moreover, an early focus on biological targets of “convenience” may provide important validation of prior genetic associations, as well as hold additional proof-of-concept value for the field of imaging epigenetics, while ultimately setting the stage for epigenome-wide approaches, as the field matures.
Identifying Mechanisms
Whenever possible, additional experiments should be carried out to evaluate the precise molecular mechanisms that may explain any association between a peripheral DNA methylation mark and brain function. Fortunately, this research is inherently interdisciplinary and fosters, if not necessitates, collaboration among labs with unique but complementary expertise. Importantly, although research in neuroimaging epigenetics can and should be informed by basic research in molecular epigenetics, the opposite may be true as well. Given the decreasing cost of DNA methylation assays and the ease with which these can be conducted in DNA derived from blood or saliva, neuroimaging epigenetics studies can be easily implemented in large archival imaging genetics datasets (e.g, [10,55,56]). Biologically plausible correlations between a peripheral DNA methylation mark and a well-defined neural phenotype, especially if replicated across independent cohorts, may then be used as rationale for potentially more costly follow-up molecular experiments. Notably, results from the nascent field of imaging epigenetics have already raised potentially interesting methodological and conceptual questions for the field of basic molecular epigenetics (Box 3).
Mining Public Databases
The need for mechanistic follow-up experiments or independent molecular work can be reduced, though not necessarily obviated, by the increasing availability of shared electronic resources. Whenever possible, information regarding the cross-tissue correlation or functionality of a particular methylation mark can and should be obtained from publically available epigenomics databases. Most notably, the Human Epigenome Roadmap project has recently compiled and analyzed a total of 127 reference human epigenomes [57]. Raw data including basic DNA sequence, DNA methylation, and mRNA levels, assayed in peripheral blood and brain, among other tissue types, can be accessed freely through designated web portals (http://www.roadmapepigenomics.org and http://compbio.mit.edu/roadmap). Additional epigenomic data are available on the website of the International Human Epigenome Consortium (IHEC) (http://epigenomesportal.ca/ihec/index.html). Yet another resource that could be particularly useful in developmental studies is Braincloud (http://braincloud.jhmi.edu), a database containing genomic, epigenomic, and transcriptomic data sampled across stages of development currently available for the prefrontal cortex [50; 51] but with plans to extend the database to additional brain regions.
Longitudinal Studies and Trans-Generational Effects
Given the dynamic nature of the epigenome, neuroimaging epigenetics studies that track concurrent changes in peripheral DNA methylation and brain function, especially in developmental populations, are particularly important. Prospective longitudinal designs in normally developing populations can serve as references for subsequent cross-sectional studies. Longitudinal studies in at-risk populations (e.g., those with a positive family history for disorder) can foster insight into the neuroepigenetic signatures of experience or development associated with specific pathophysiologic mechanisms. The epigenetic correlates of development and experience, as distinct from those associated with sequence variation, would be particularly likely to emerge in prospective longitudinal studies of monozygotic twin pairs who share a common DNA sequence, but accumulate DNA methylation differences over time [28]. Another intriguing, albeit more remote, prospect for the field of imaging epigenetics lies in the possibility to uncover the molecular mechanisms of any potential trans-generational transmission of non-genetic psychiatric disorder risk. Although methodologically challenging, recording epigenetic and phenotypic information from parents and their children, especially in a longitudinal context, may help shed light on some of the epigenetic mechanisms mediating parent-to-offspring transmission of risk. However, extensive follow-up studies in animal models will likely be needed to lend additional empirical support to any emergent findings [58].
Epigenetic Biomarkers
Despite the potential of imaging epigenetics studies to identify molecular mechanisms of risk, it is important to note that the establishment of reliable associations between peripherally assessed DNA methylation marks and neuroimaging phenotypes can be useful even in the absence of explicit knowledge about their precise underlying molecular mechanisms. Just as sequence variation can serve as a readily accessible biomarker of brain function and associated risk, peripherally assessed DNA methylation patterns could be leveraged to index neural endophenotypes. Although the dynamic nature of the epigenome introduces greater potential intra-individual variability in DNA methylation patterns, such state-dependency could also be used to the field’s advantage, especially as the impact of various environmental factors on DNA methylation becomes better understood.
Finally, it is increasingly recognized that individual differences in the vast majority of brain endophenotypes likely reflect the simultaneous impact of multiple sequence variants, as well as the effects of an individual’s unique experiences [23,59]. The polygenic nature of complex biological traits is likely to be mirrored on the level of epigenetics. As new data accumulates, leveraging multilocus genetic and epigenetic analytic strategies to simultaneously capture diverse genetic and environmental contributions to brain function, in a hypothesis-driven or theory-free manner, may ultimately offer a more powerful way to account for variability in disorder risk and perhaps even make predictions about differential vulnerability at the individual level. Furthermore, because DNA methylation patterns, unlike genotypes, can be altered pharmacologically [60], such findings may also give us greater hope for successful individualized treatment and prevention than studies focusing solely on sequence based markers.
Unified Strategy
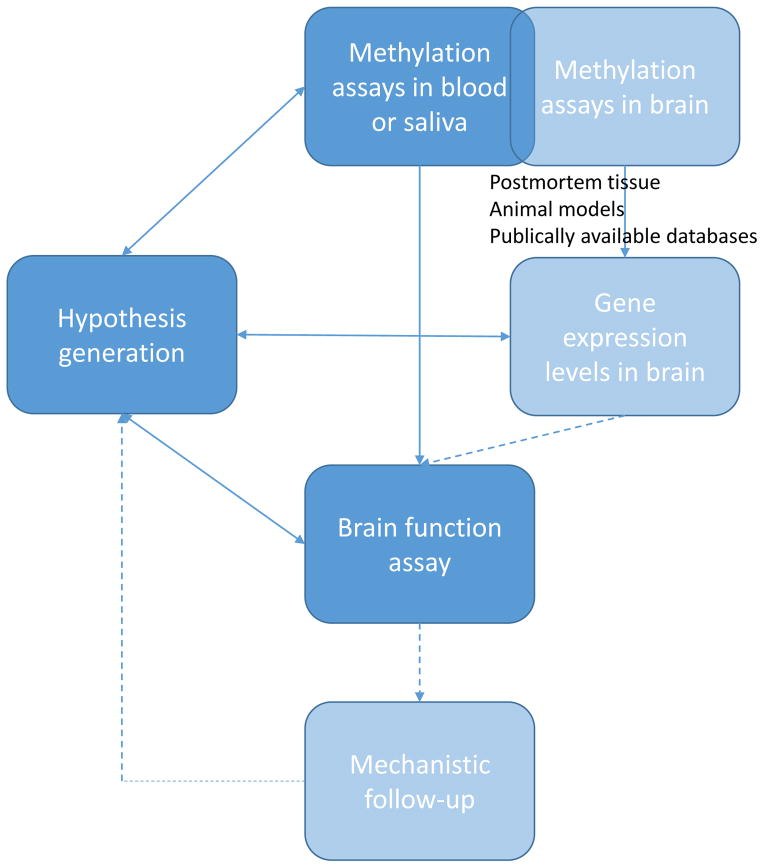
In Figure 1, we outline a possible strategy for the field of imaging epigenetics wherein prior or concurrent cross-tissue molecular characterization of functional impact, carried out through collaboration or obtained via publically available databases, provides biological rationale or validation of a particular imaging epigenetics association. Given the increasing ease with which methylation assays can be conducted in DNA extracted following standard protocols, as well as the accumulation of large and diverse imaging genetics datasets (e.g., [61–63]), imaging epigenetics is poised to capitalize on existing resources at little additional cost. This is particularly important, given that large sample sizes would likely be necessary to adequately capture the potentially larger variability in DNA methylation relative to structural variation, while also properly accounting for the diverse range of environmental experiences and ancestral backgrounds [64] represented in a specific sample and, ultimately, the general population. Results from such imaging epigenetics studies could subsequently fuel additional, and potentially more costly, mechanistic follow-up efforts.
Figure 1.
Proposed strategy for the emergent field of imaging epigenetics. Although concurrent measurements of DNA methylation, gene expression patterns, and brain function are extremely difficult, if not impossible, to obtain in the same individuals, correlations between peripherally assessed DNA methylation and brain function can be corroborated by more extensive cross-tissue functional characterization of methylation-related molecular phenotypes derived from postmortem human brain tissue and animal models, some of which are already summarized in publically available databases. In turn, any biologically plausible associations emerging from imaging epigenetics studies, especially if replicated, could serve as motivation for more costly mechanistic follow-up efforts.
Concluding Remarks
Imaging genetics has established useful links between peripherally accessible genetic markers and variability in brain function associated with individual differences in behavior and relative risk for psychopathology. Going beyond the genome and elucidating the role of epigenetic modifications on these and other associations represents a next frontier. Promising initial studies have already begun to establish links between behaviorally and clinically relevant brain function and peripherally assessed DNA methylation. Despite methodological concerns regarding tissue-specific functional characterization of DNA methylation patterns, we believe enough encouraging initial data has accumulated for the field of neuroimaging epigenetics to move forward with confidence. The establishment of reliable associations between peripherally assessed DNA methylation, varying experiential and environmental factors, and brain function has the capacity to shed additional light on the molecular mechanisms of risk and pave new roads for not only identifying vulnerable individuals but also providing them with precision medicine.
Figure I.
Methylation of CpG islands near gene promoters is typically associated with reduced transcription initiation. Although hypomethylated (A) and hypermethylated (C) CpG islands are generally associated with normal and repressed gene expression, respectively, emergent imaging epigenetic work suggests smaller inter-individual variability in the relative methylation level of otherwise hypomethylated CpG islands may also result in phenotypically relevant differences in gene expression (B).
Highlights.
Gene methylation can be readily assessed in DNA derived from peripheral tissues
Differential DNA methylation predicts brain function
Epigenetic variability in brain function can identify novel mechanisms of risk
Glossary
- Allele
one of several alternative forms or copies of a gene variant or polymorphism; at each polymorphism an individual inherits one allele from their mother and one allele from their father
- BDNF
the gene encoding the human brain derived neurotrophic factor, a protein crucial for the survival, growth, and differentiation of neurons as well as synaptic plasticity supporting learning [74]
- COMT
the gene encoding the human catechol-O-methyltransferase enzyme, which is involved in the degradation of catecholamines such as dopamine, epinephrine, and norepinephrine [75]
- Chromatin
a multi-molecular complex of DNA, RNA and protein, which serves to protect the integrity of genetic information and regulate the dynamics of gene expression
- DNMT1
DNA methyltransferase 1, an enzyme responsible for maintaining DNA methylation patterns during cell division
- Epistatic interactions
interactions between or among several genes or polymorphisms
- FKBP5
the gene encoding the human FK506 binding protein 5, which is involved in HPA axis regulation via its interactions with the glucocorticoid receptor [76]
- Homozygous
individuals carrying two copies of the same allele of a polymorphism
- Monozygotic twins
twins who share 100% of their DNA sequence resulting from the division of a single fertilized egg or zygote
- MAOA
the gene encoding the human enzyme monoamine oxidase A, which is involved in the degradation of monoamines, such as serotonin, dopamine, epinephrine and norepineprhine [77]
- Methylome
the entire set of all methylation marks present across the entire genome; unlike the genome , however, there are likely multiple cell-type specific methylomes
- NR3C1
the gene encoding the human glucocorticoid receptor protein, which is critically implicated in the negative feedback regulation of the stress response mediated by the HPA axis [78]
- Nucleotide
an organic molecule that serves as a subunit or basic building block of DNA or RNA
- OXTR
the gene encoding the human oxytocin receptor, which has been extensively implicated in the regulation of social behavior [79,80]
- Polymorphism
a structural variant in the DNA sequence of a gene
- Proximal promoter
the DNA sequence immediately upstream of the transcription start site of a gene; binding of transcription factors to proximal promoters functions to regulate gene expression
- SLC6A4
the gene encoding the human serotonin transporter protein, which mediates the reuptake of serotonin at serotonergic synapses [81]
- Transcription start site
a DNA sequence marking the beginning of a gene or non-coding RNA transcript
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bookheimer SY, et al. Patterns of brain activation in people at risk for Alzheimer’s disease. N Engl J Med. 2000;343:450–456. doi: 10.1056/NEJM200008173430701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egan MF, et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci U S A. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hariri AR, et al. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297:400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- 4.Hariri AR. The neurobiology of individual differences in complex behavioral traits. Annu Rev Neurosci. 2009;32:225–247. doi: 10.1146/annurev.neuro.051508.135335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hariri AR. Genetic polymorphisms: a cornerstone of translational biobehavioral research. Science translational medicine. 2010;2:18ps16. doi: 10.1126/scitranslmed.3000811. [DOI] [PubMed] [Google Scholar]
- 6.Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nature reviews Neuroscience. 2006;7:818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- 7.Bartova L, et al. Is there a personalized medicine for mood disorders? Eur Arch Psychiatry Clin Neurosci. 2010;260(Suppl 2):S121–126. doi: 10.1007/s00406-010-0152-8. [DOI] [PubMed] [Google Scholar]
- 8.Singh I, Rose N. Biomarkers in psychiatry. Nature. 2009;460:202–207. doi: 10.1038/460202a. [DOI] [PubMed] [Google Scholar]
- 9.Hariri AR, Holmes A. Genetics of emotional regulation: the role of the serotonin transporter in neural function. Trends in cognitive sciences. 2006;10:182–191. doi: 10.1016/j.tics.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Swartz JR, et al. A neural biomarker of psychological vulnerability to future life stress. Neuron. 2015;85:505–511. doi: 10.1016/j.neuron.2014.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fakra E, et al. Effects of HTR1A C(-1019)G on amygdala reactivity and trait anxiety. Arch Gen Psychiatry. 2009;66:33–40. doi: 10.1001/archpsyc.66.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munafo MR, et al. Serotonin transporter (5-HTTLPR) genotype and amygdala activation: a meta-analysis. Biol Psychiatry. 2008;63:852–857. doi: 10.1016/j.biopsych.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caspi A, et al. Genetic sensitivity to the environment: the case of the serotonin transporter gene and its implications for studying complex diseases and traits. A J Psychiatry. 2010;167:509–527. doi: 10.1176/appi.ajp.2010.09101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Risch N, et al. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. JAMA. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caspi A, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 16.Karg K, et al. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: evidence of genetic moderation. Arch Gen Psychiatry. 2011;68:444–454. doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Insel TR, Cuthbert BN. Medicine. Brain disorders? Precisely. Science. 2015;348:499–500. doi: 10.1126/science.aab2358. [DOI] [PubMed] [Google Scholar]
- 18.Murphy SE, et al. The effect of the serotonin transporter polymorphism (5-HTTLPR) on amygdala function: a meta-analysis. Mol Psychiatry. 2013;18:512–520. doi: 10.1038/mp.2012.19. [DOI] [PubMed] [Google Scholar]
- 19.Pezawas L, et al. Evidence of biologic epistasis between BDNF and SLC6A4 and implications for depression. Mol Psychiatry. 2008;13:709–716. doi: 10.1038/mp.2008.32. [DOI] [PubMed] [Google Scholar]
- 20.Drabant EM, et al. Neural mechanisms underlying 5-HTTLPR-related sensitivity to acute stress. A J Psychiatry. 2012;169:397–405. doi: 10.1176/appi.ajp.2011.10111699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemogne C, et al. Cognitive appraisal and life stress moderate the effects of the 5-HTTLPR polymorphism on amygdala reactivity. Hum Brain Mapp. 2011;32:1856–1867. doi: 10.1002/hbm.21150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bigos KL, Weinberger DR. Imaging genetics--days of future past. Neuroimage. 2010;53:804–809. doi: 10.1016/j.neuroimage.2010.01.035. [DOI] [PubMed] [Google Scholar]
- 23.Hyde LW, et al. Understanding risk for psychopathology through imaging gene-environment interactions. Trends in cognitive sciences. 2011;15:417–427. doi: 10.1016/j.tics.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews Genetics. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 25.Bernstein BE, et al. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 26.Brenet F, et al. DNA methylation of the first exon is tightly linked to transcriptional silencing. PloS one. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Dongen J, et al. Epigenetic variation in monozygotic twins: a genome-wide analysis of DNA methylation in buccal cells. Genes. 2014;5:347–365. doi: 10.3390/genes5020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikolova YS, et al. Beyond genotype: serotonin transporter epigenetic modification predicts human brain function. Nat Neurosci. 2014;17:1153–1155. doi: 10.1038/nn.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiers CE. Methylation and the human brain: towards a new discipline of imaging epigenetics. Eur Arch Psychiatry Clin Neurosci. 2012;262:271–273. doi: 10.1007/s00406-011-0261-z. [DOI] [PubMed] [Google Scholar]
- 31.Ursini G, et al. Stress-related methylation of the catechol-O-methyltransferase Val 158 allele predicts human prefrontal cognition and activity. J Neurosci. 2011;31:6692–6698. doi: 10.1523/JNEUROSCI.6631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vukojevic V, et al. Epigenetic modification of the glucocorticoid receptor gene is linked to traumatic memory and post-traumatic stress disorder risk in genocide survivors. J Neurosci. 2014;34:10274–10284. doi: 10.1523/JNEUROSCI.1526-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weaver IC, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 34.Beach SR, et al. Methylation at SLC6A4 is linked to family history of child abuse: an examination of the Iowa Adoptee sample. American journal of medical genetics. Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2010;153B:710–713. doi: 10.1002/ajmg.b.31028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang HJ, et al. Association of SLC6A4 methylation with early adversity, characteristics and outcomes in depression. Prog Neuropsychopharmacol Bol Psychiatry. 2013;44C:23–28. doi: 10.1016/j.pnpbp.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 36.Alasaari JS, et al. Environmental stress affects DNA methylation of a CpG rich promoter region of serotonin transporter gene in a nurse cohort. PloS one. 2012;7:e45813. doi: 10.1371/journal.pone.0045813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehta D, et al. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci. 2014;17:89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee KW, et al. Prenatal Exposure to Maternal Cigarette Smoking and DNA Methylation: Epigenome-Wide Association in a Discovery Sample of Adolescents and Replication in an Independent Cohort at Birth through 17 Years of Age. Environ Health Perspect. 2015;123:193–199. doi: 10.1289/ehp.1408614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oberlander TF, et al. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics: official journal of the DNA Methylation Society. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 41.Szyf M. Nongenetic inheritance and transgenerational epigenetics. Trends in molecular medicine. 2015;21:134–144. doi: 10.1016/j.molmed.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 42.Hyman SE. How adversity gets under the skin. Nat Neurosci. 2009;12:241–243. doi: 10.1038/nn0309-241. [DOI] [PubMed] [Google Scholar]
- 43.McEwen BS. Brain on stress: how the social environment gets under the skin. Proc Natl Acad Sci U S A. 2012;109(Suppl 2):17180–17185. doi: 10.1073/pnas.1121254109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hertzman C, Boyce T. How experience gets under the skin to create gradients in developmental health. Annu Rev Public Health. 2010;31:329–347. 323p. doi: 10.1146/annurev.publhealth.012809.103538. following 347. [DOI] [PubMed] [Google Scholar]
- 45.Vijg J. Somatic mutations, genome mosaicism, cancer and aging. Curr Opin Genet Dev. 2014;26:141–149. doi: 10.1016/j.gde.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies MN, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome biology. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tylee DS, et al. On the outside, looking in: a review and evaluation of the comparability of blood and brain “-omes”. American journal of medical genetics. Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2013;162B:595–603. doi: 10.1002/ajmg.b.32150. [DOI] [PubMed] [Google Scholar]
- 48.Horvath S, et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome biology. 2012;13:R97. doi: 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith AK, et al. DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. American journal of medical genetics. Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2015;168B:36–44. doi: 10.1002/ajmg.b.32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schultz W. Getting formal with dopamine and reward. Neuron. 2002;36:241–263. doi: 10.1016/s0896-6273(02)00967-4. [DOI] [PubMed] [Google Scholar]
- 51.Wang D, et al. Peripheral SLC6A4 DNA methylation is associated with in vivo measures of human brain serotonin synthesis and childhood physical aggression. PloS one. 2012;7:e39501. doi: 10.1371/journal.pone.0039501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shumay E, et al. Evidence that the methylation state of the monoamine oxidase A (MAOA) gene predicts brain activity of MAO A enzyme in healthy men. Epigenetics: official journal of the DNA Methylation Society. 2012;7:1151–1160. doi: 10.4161/epi.21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farrell MS, et al. Evaluating historical candidate genes for schizophrenia. Mol Psychiatry. 2015;20:555–562. doi: 10.1038/mp.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tabor HK, et al. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nature reviews Genetics. 2002;3:391–397. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- 55.Schumann G, et al. The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Mol Psychiatry. 2010;15:1128–1139. doi: 10.1038/mp.2010.4. [DOI] [PubMed] [Google Scholar]
- 56.Nikolova YS, et al. Beyond genotype: serotonin transporter epigenetic modification predicts human brain function. Nat Neurosci. 2014 doi: 10.1038/nn.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roadmap Epigenomics C et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dias BG, et al. Epigenetic mechanisms underlying learning and the inheritance of learned behaviors. Trends Neurosci. 2015;38:96–107. doi: 10.1016/j.tins.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nikolova YS, et al. Multilocus genetic profile for dopamine signaling predicts ventral striatum reactivity. Neuropsychopharmacology. 2011;36:1940–1947. doi: 10.1038/npp.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- 61.Van Essen DC, et al. The Human Connectome Project: a data acquisition perspective. Neuroimage. 2012;62:2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson PM, et al. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014;8:153–182. doi: 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paus T. Population neuroscience: why and how. Hum Brain Mapp. 2010;31:891–903. doi: 10.1002/hbm.21069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barfield RT, et al. Accounting for population stratification in DNA methylation studies. Genet Epidemiol. 2014;38:231–241. doi: 10.1002/gepi.21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jack A, et al. DNA methylation of the oxytocin receptor gene predicts neural response to ambiguous social stimuli. Front Hum Neurosci. 2012;6:280. doi: 10.3389/fnhum.2012.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walton E, et al. MB-COMT promoter DNA methylation is associated with working-memory processing in schizophrenia patients and healthy controls. Epigenetics: official journal of the DNA Methylation Society. 2014;9:1101–1107. doi: 10.4161/epi.29223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Puglia MH, et al. Epigenetic modification of the oxytocin receptor gene influences the perception of anger and fear in the human brain. Proc Natl Acad Sci U S A. 2015;112:3308–3313. doi: 10.1073/pnas.1422096112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frodl T, et al. DNA methylation of the serotonin transporter gene (SLC6A4) is associated with brain function involved in processing emotional stimuli. J Psychiatry Neurosci. 2015;40:140180. doi: 10.1503/jpn.140180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ziegler C, et al. Oxytocin receptor gene methylation: converging multilevel evidence for a role in social anxiety. Neuropsychopharmacology. 2015;40:1528–1538. doi: 10.1038/npp.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klengel T, et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dannlowski U, et al. Serotonin transporter gene methylation is associated with hippocampal gray matter volume. Hum Brain Mapp. 2014;35:5356–5367. doi: 10.1002/hbm.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choi S, et al. Association of brain-derived neurotrophic factor DNA methylation and reduced white matter integrity in the anterior corona radiata in major depression. J Affect Disord. 2014;172C:74–80. doi: 10.1016/j.jad.2014.09.042. [DOI] [PubMed] [Google Scholar]
- 73.Booij L, et al. DNA methylation of the serotonin transporter gene in peripheral cells and stress-related changes in hippocampal volume: a study in depressed patients and healthy controls. PloS one. 2015;10:e0119061. doi: 10.1371/journal.pone.0119061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Binder DK, Scharfman HE. Brain-derived neurotrophic factor. Growth Factors. 2004;22:123–131. doi: 10.1080/08977190410001723308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grossman MH, et al. Chromosomal mapping of the human catechol-O-methyltransferase gene to 22q11.1----q11.2. Genomics. 1992;12:822–825. doi: 10.1016/0888-7543(92)90316-k. [DOI] [PubMed] [Google Scholar]
- 76.Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 77.Hotamisligil GS, Breakefield XO. Human monoamine oxidase A gene determines levels of enzyme activity. Am J Hum Genet. 1991;49:383–392. [PMC free article] [PubMed] [Google Scholar]
- 78.Hollenberg SM, et al. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature. 1985;318:635–641. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gimpl G, Fahrenholz F. The oxytocin receptor system: structure, function, and regulation. Physiol Rev. 2001;81:629–683. doi: 10.1152/physrev.2001.81.2.629. [DOI] [PubMed] [Google Scholar]
- 80.Kumsta R, Heinrichs M. Oxytocin, stress and social behavior: neurogenetics of the human oxytocin system. Curr Opin Neurobiol. 2013;23:11–16. doi: 10.1016/j.conb.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 81.Lesch KP, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 82.Harshman SW, et al. H1 histones: current perspectives and challenges. Nucleic Acids Res. 2013;41:9593–9609. doi: 10.1093/nar/gkt700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 84.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nature structural & molecular biology. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 85.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews Genetics. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 86.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nature reviews Genetics. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nature reviews Neuroscience. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- 88.Sillivan SE, et al. Neuroepigenetic Regulation of Pathogenic Memories. Neuroepigenetics. 2015;1:28–33. doi: 10.1016/j.nepig.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saab BJ, Mansuy IM. Neuroepigenetics of memory formation and impairment: the role of microRNAs. Neuropharmacology. 2014;80:61–69. doi: 10.1016/j.neuropharm.2014.01.026. [DOI] [PubMed] [Google Scholar]
- 90.Koch CM, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Belmont A. Dynamics of chromatin, proteins, and bodies within the cell nucleus. Curr Opin Cell Biol. 2003;15:304–310. doi: 10.1016/s0955-0674(03)00045-0. [DOI] [PubMed] [Google Scholar]
- 92.Branco MR, et al. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nature reviews Genetics. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 93.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet. 1989;83:181–188. doi: 10.1007/BF00286715. [DOI] [PubMed] [Google Scholar]
- 95.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 96.Saxonov S, et al. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- 98.Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baubec T, et al. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell. 2013;153:480–492. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 100.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature reviews Genetics. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 101.Gruenbaum Y, et al. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982;295:620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- 102.Okano M, et al. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 103.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nature reviews Molecular cell biology. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Talens RP, et al. Variation, patterns, and temporal stability of DNA methylation: considerations for epigenetic epidemiology. FASEB J. 2010;24:3135–3144. doi: 10.1096/fj.09-150490. [DOI] [PubMed] [Google Scholar]