Abstract
Nicotinic acetylcholine receptors (nAChRs) are highly conserved between humans and non-human primates. Conservation exists at the level of genomic structure, protein structure and epigenetics. Overall homology of nAChRs at the protein level is 98% in macaques versus 89% in mice, which is highly relevant for evaluating subtype-specific ligands that have different affinities in humans versus rodents. In addition to conservation at the protein level, there is high conservation of genomic structure in terms of intron and exon size and placement of CpG sites that play a key role in epigenetic regulation. Analysis of single nucleotide polymorphisms (SNPs) shows that while the majority of SNPs are not conserved between humans and macaques, some functional polymorphisms are. Most significantly, cynomolgus monkeys express a similar α5 nAChR Asp398Asn polymorphism to the human α5 Asp398Asn polymorphism that has been linked to greater nicotine addiction and smoking related disease. Monkeys can be trained to readily self-administer nicotine, and in an initial study we have demonstrated that cynomolgus monkeys bearing the α5 D398N polymorphism show a reduced behavioral sensitivity to oral nicotine and tend to consume it in a different pattern when compared to wild-type monkeys. Thus the combination of highly homologous nAChR, higher cortical functions and capacity for complex training makes non-human primates a unique model to study in vivo functions of nicotinic receptors. In particular, primate studies on nicotine addiction and evaluation of therapies to prevent or overcome nicotine addiction are likely to be highly predictive of treatment outcomes in humans.
Keywords: nicotinic receptor, nicotine, non-human primate, rhesus, cynomolgus, macaque
1. Introduction
Given the evolutionary closeness of non-human primates (NHP) to humans it is not surprising that there is strong structural conservation between human and non-human nicotinic acetylcholine receptors (nAChRs). The combination of conservation of nAChR structure and conservation of complex behaviors between non-human primates and humans makes non-human primates a valuable model for studies of the roles of nAChR in a variety of behaviors and for evaluation of potential therapeutics. Indeed, advancing neuro-therapeutics to market has proven problematic with drugs targeting the CNS due to longer development times and higher clinical trial failure rates when compared to other drug categories (Klein et al., 2011; Kaitin and DiMasi, 2011; Pritchard, 2008). One aspect of the challenge is the greater functional difference between rodent and human brain than exists for other systems or organs such as cardiovascular or renal. This reflects both differences in anatomy and specificity and distribution of receptors for ligands. For example, the prefrontal cortex, which plays a key regulatory role in addictive behavior, is significantly underdeveloped in rodents compared to primates (Wallis, 2012). In terms of ligands, varenicline is significantly more potent against human than rat α3β4, while cytisine is significantly less potent against human than rat α3β4 (Stokes and Papke, 2012). Similarly the most specific and potent antagonist for the α9α10 nAChR, α-conotoxin (α-CTx) RgIA, is 300-fold less potent against human than rat α9α10 (Azam and McIntosh, 2012) and this difference in potency derives from a single amino acid change (T56I). Thus non-human primate (NHP) models, in which neuroanatomy and nAChR receptors are more homologous to humans, will likely be essential for the development of the next generation of smoking cessation therapies.
The advent of low cost, whole genome sequencing has greatly expanded available information about NHP nAChRs. Currently available NHP genome assemblies in public databases include baboon, bonobo, bushbaby, chimpanzee, cynomolgus monkey, gibbons, guerilla, lemurs, marmoset, orangutan, rhesus monkey, squirrel monkey, tarser, vervet (http://www.ncbi.nlm.nih.gov/genome/browse/) and more are being added all the time. The sequencing of large pedigreed NHP colonies further opens up possibilities for studying the roles of nAChR in non-human primates particularly linking gene function and polymorphisms to behaviors and quantitative traits. In addition the conservation of key polymorphisms between monkeys and humans, such as in the serotonin transporter and monamine oxidase further emphasizes species similarities and the potential for research discoveries (Wendland et al., 2006; Barr et al., 2003).
2. Non-human primate nicotinic receptor structure and organization
2.1. Genomic Structure
The nAChR subunit genes are divided into subfamilies and tribes based on sequence similarity (Corringer et al., 2000). The α9 and α10 subunits, each encoded by 5 exons, are the most recently cloned nAChR subunits and comprise neuronal nAChR subfamily I. The α7 subunit (neuronal subfamily II) has the most dissimilar genomic structure of all mammalian nAChR genes and is composed of 10 exons, 9 of which are smaller than 200 bp. This subunit more closely resembles the muscle type nAChRs and is likely an ancestral subunit that existed prior to the divergence of insects and vertebrates (Le and Changeux, 1995).
We conducted a comparative analysis of the genomic structures of nAChR subunits between the rhesus and human genomes. We retrieved the genomic sequence of human nAChR subunits using the UCSC Genome Bioinformatics Site (Kuhn et al., 2013). The UCSC Table Browser was used to extract the sequence and structural information of nAChR subunit genes (i.e. chromosomal position, exon start and end positions) from the human hg19 genome build. For rhesus macaque, we used the RefSeq gene annotations when available from the rheMac2 genome to determine the sequence, size and position of coding exons and to calculate length of exonic and intronic sequences. The intron and UTR sizes in rhesus macaque were approximated for some nAChR subunits as the available draft assemblies (rheMac2 and rheMac3) contain gaps, denoted by “ ..NNN…”, that are of unknown true length. The Basic Local Alignment Search Tool (BLAST) was then used to align the extracted human and rhesus nucleotide sequences, optimized for highly similar sequences (megablast). Using this approach, we observed high homology between human and rhesus nAChR exon regions. In instances where RefSeq annotations were unavailable for rhesus genomes we manually compared regions of human sequence to equivalent regions of rhesus genomes on a base-by-base level. Overall, the genomic structure of nAChR subunits was highly conserved between rhesus and human as shown in Table 1.
Table 1.
Genomic structure of nAChR subunit genes in human (hg19) compared with macaca mulatta (rheMac2/3)
| Gene ID | Assembly | RefSeq ID | chr. | Ex.1 5'UTR/CDS |
Intron | Ex. 2 | Intron | Ex. 3 | Intron | Ex. 4 | Intron | Ex. 5 | Intron | Ex.6 (5) CDS/3'UTR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHRNA2 | hg19 | NM_000742 | chr08 | 609a/73 | 1004 | 221 | 381 | 45 | 1996 | 110 | 3235 | 1015 | 1224 | 126/1868 |
| rheMac2 | XM_002805289.1 | chr08 | 536a/73 | 1009 | 221 | 446 | 45 | 1982 | 110 | 3204 | 1015 | 1728 | 126/502 | |
| CHRNA3 | hg19 | NM_000743 | chr15 | 501/82 | 1797 | 140 | 114 | 45 | 1483 | 110 | 14759 | 1012 | 4520 | 129/1299 |
| rheMac2/3 | not annotatedb | chr7 | 472/64 | 1809 | 140 | 114 | 45 | 1468 | 110 | 8605 | 1012 | 11950 | 129/1306 | |
| CHRNA4 | hg19 | NM_000744 | chr20 | 231/76 | 1390 | 152 | 3132 | 45 | 286 | 110 | 4947 | 1375 | 2789 | 126/3428 |
| rheMac2 | XM_001114265.2 | chr10 | ?c | ?c | 152 | 3856 | 45 | 320 | 110 | 3859 | 1372 | 3408 | 126/24 | |
| CHRNA5 | hg19 | NM_000745 | chr15 | 200/106 | 14985 | 152 | 5682 | 45 | 1624 | 110 | 1381 | 832 | 2455 | 162/2016 |
| rheMac2 | XM_001108279.2 | chr07 | 163/106 | 15821 | 152 | 3916 | 45d | ?d | 74d | 1383 | 832 | 2752 | 162/446 | |
| CHRNA6 | hg19 | NM_004198 | chr08 | 356/79 | 3147 | 140 | 5951 | 45 | 2031 | 110 | 103 | 979 | 2535 | 132/542 |
| rheMac2 | XM_001099152.2 | chr08 | 202/79 | 3171 | 140 | 5906 | 45 | 1720 | 110 | 103 | 979 | 2559 | 132/537 | |
| CHRNA9 | hg19 | NM_017581 | chr04 | 139/64 | 295 | 146 | 1237 | 155 | 11517 | 533 | 4564 | – | (542/697) | |
| rheMac2 | XM_001094288.2 | chr05 | 95/64 | 293 | 146 | 1256 | 155 | 11464 | 533 | 4562 | – | (542/434) | ||
| CHRNA10 | hg19 | NM_020402 | chr11 | 72/61 | 1310 | 146 | 445 | 155 | 1431 | 533 | 667 | – | (458/520) | |
| rheMac2 | XM_001113644.1 | chr14 | 72/61 | 1268 | 146 | 432 | 155 | 1417 | 533 | 662 | – | (458/523) | ||
| CHRNB2 | hg19 | NM_000748 | chr01 | 264/64 | 1353 | 146 | 184 | 45 | 421 | 110 | 821 | 973 | 3600 | 171/3945 |
| rheMac2 | XM_001114439.2 | chr01 | 264/64 | 1348 | 146 | 184 | 45 | 421 | 110 | 821 | 973 | 3609 | 171/1161 | |
| CHRNB3 | hg19 | NM_000749 | chr08 | 128/52 | 11118 | 152 | 1521 | 45 | 20159 | 110 | 963 | 883 | 3934 | 135/448 |
| rheMac3 | not annotatedb | chr08 | 128/52 | 12199 | 152 | 1503 | 45 | 15070 | 110 | 694 | 883 | 2697 | 135/444 | |
| CHRNB4 | hg19 | NM_000750 | chr15 | 112/55 | 5491 | 149 | 4106 | 45 | 102 | 110 | 1130 | 979 | 3675 | 159/839 |
| rheMac3 | not annotatedb | chr7 | 111/55 | 5220 | 149 | 4029 | 45 | 102 | 110 | 1148 | 961 | 3710 | 159/419 | |
| CHRNA7 | hg19 | NM_000746 | chr15 | 112/55 | 248 | 140 | 70265 | 45 | 10440 | 110 | 42007 | 80 | 3621 | |
| rheMac2 | NM 001032883.1 | chr07 | 105/55 | 222 | 140 | 70068 | 45 | 10074 | 110 | 44083 | 80 | 3662 | ||
| Ex. 6 | Intron | Ex. 7 | Intron | Ex. 8 | Intron | Ex. 9 | Intron | Ex.10 CDS/3'UTR |
||||||
| hg 19 CHRNA7 cont | 168 | 636 | 195 | 962 | 87 | 3570 | 110 | 4604 | 519/1725 | |||||
| rheMac2 CHRNA7 cont | 168 | 632 | 195 | 947 | 87 | 3640 | 110 | 4532 | 519/471 | |||||
The genomic structure of human nAChR subunit genes was compared to the rhesus macaque. The size and distribution of nAChR subunit exons is highly conserved across nAChR subfamilies and tribes, and across species. Size of introns and exons given in base pairs, bold text is used when human and rhesus values are equivalent.
The 5'UTR of CHRNA2 is split by an intron into two fragments (473 and 136bp in hg19; 421 and 115bp in rheMac2).
There were no RefSeq annotations found in rheMac2 or rheMac3 for these subunits. Exon and intron sizes were determined from manual alignment of human sequences to rheMac2/3 as described in the text.
rheMac2 RefSeq annotation XM_001114265.2 predicted 2 exons upstream of Exon 2. Manual base-checking suggests that neither of these predicted exons align to hsCHRNA4.
rheMac2 RefSeq annotation XM_001108279.2 predicted Exon 3 to be 75 bp. Manual base-checking demonstrates that the last 30 bp align to hsCHRNA5 intron 3 and reveals a gap at the next intron/exon boundary in rmCHRNA4.
In humans, there exists a partial duplication of the gene encoding the α7 subunit (CHRNA7), known as CHRFAM7, which functions as a dominant negative regulator of α7 nAChR (Araud et al., 2011). This fusion gene is comprised of 1 exon of unknown origin, 3 exons of ULK4, and exons 5 through 10 of CHRNA7 (Araud et al., 2011). This duplication event has been proposed to be human specific based on southern blot analysis in chimpanzee and gorilla (Locke et al., 2003) and the high level of homology with human CHRNA7. We used in silico BLAT analysis (Kent, 2002; Kuhn et al., 2013) to look for similar duplication events in the available primate genomes. As proof of principle, we first queried the genomic sequence of human CHRNA7 exons 5–10 (14548 bp) against hg19 which resulted in two alignments of similar size and identity: to CHRNA7 with 100% identity and to CHRFAM7 with 99.8% identity. Using the same approach, we confirmed only single high-homology alignments (to CHRNA7) in all primate genomes available in the BLAT tool (baboon, chimp, bushbaby, gibbon, orangutan, squirrel monkey) except gorilla. Here we saw substantial alignment (98.4% and 98.2%) of the human query at two positions on the gorGor3 genome (chr15minus:8,824,749-8,835,892 and chr15plus:10,541,866-10,552,122) spanning 13528 and 14053 bp, respectively, indicating that there may be a partial duplication event in gorilla as well. Further suggesting that gorillas may well express CHRFAM7, a portion of the ULK4 gene appears to align next to the duplicate copy of CHRNA7. However, the relatively poor quality of the Gor3 genome assembly prevents a definitive statement regarding the existence of CHRFAM7A in gorillas at this time.
2.2. Protein Structure
Nicotinic acetylcholine receptors are members of the Cys-loop superfamily of ligand-gated ion channels. Characteristic of this class, nAChRs are composed of five transmembrane protein subunits arranged in a ring to form a central ion pore (Thompson et al., 2010). These protein subunits are classified as either alpha or beta subunits based on the presence of two adjacent cysteine residues in alpha subunits in the N-terminal extracellular domain (Lukas et al., 1999). Presently, 8 alpha subunits (α2, α3, α4, α5, α6, α7, α9, α10) and 3 beta subunits have been identified in the mammalian central nervous system (Albuquerque et al., 2009). These nAChR subunit proteins range in length from 458 to 627 amino acids (human) with a well-conserved order of structural domains that define their placement within the cellular membrane. Each subunit begins with an N-terminal extracellular domain, followed by three transmembrane domains, a large intracellular loop, a fourth transmembrane domain, and finally a short C-terminal extracellular domain. The transmembrane domains are highly conserved while the large intracellular loop is the most variable region between subunits (Papke, 2014). Functional heteromeric receptors contain at least two alpha subunits and the subunit composition and stoichiometry of nAChRs determines ligand binding affinity and specificity as well as ion channel permeability (Albuquerque et al., 2009). The α7 subunits form primarily homopentamers while a diverse array of functional heteromers can be formed by various combinations of neuronal subfamily III.1 subunits (α2, α3, α4, and α6) with subfamily III.2 subunits (β2, β4) that may also include subfamily III.3 subunits (α5 and β3). The α5 and β3 subunits are unique in that they are structural or “accessory” subunits, and do not contribute to ligand binding but can greatly modulate receptor pharmacology (Kuryatov et al., 2008). Because the true composition and stoichiometry of receptor subunits in vivo is often not known the presence of additional “wild-card” subunits is denoted by an asterisk (i.e. α4β2*). Lastly, the α9 subunit can form functional receptors in recombinant expression systems, but is believed to form heteromers with the closely related α10 subunit, which appears to play a structural role (Plazas et al., 2005).
2.3. Homology of primate nAChRs to human nAChRs
We assessed the homology of primate nAChRs to human using CDS Fasta alignment sequences retrieved from the UCSC table browser (https://genome.ucsc.edu/). The current CDS FastA alignment data set includes the following primate reference genomes: chimpanzee (panTro4), gorilla (gorGor3), orangutan (ponAbe2), gibbon (nomLeu3), rhesus macaque (rheMac3), cynomolgus or crab-eating macaque (macFas5), baboon (papHam1), green monkey (chlSab1), marmoset (calJac3), squirrel monkey (saiBol1), and bushbaby (otoGar3). We calculated percent similarity of non-human primate nAChRs with human using MegAlign ClustalW alignment of coding sequences. One limitation of this data is that a number of gaps exist in the publically available primate reference sequences. Therefore we removed gap penalties in our alignment settings to demonstrate homology only in non-gap coding regions and inserted our own sequencing data when available to fill gaps. Otherwise, the default Multiple-Alignment Parameter settings were selected with Gonnet Series weighting. Averaging across all neuronal nAChRs, conservation of great apes, old world monkeys, and new world monkeys with humans is 98.9%, 97.5%, and 94.2%, respectively at the protein level. This is in contrast to conservation of 88.9% for mice with humans. The primate α7 (98.9%) and β2 (98.7%) had the highest homology with human subunit coding sequence while α4 (95.5%) and β4 (95.5%) had the lowest, as shown in Table 2. Within the nAChR subunit genes we most commonly observed small gaps in the N-terminal region that encodes the signal peptide. These regions are often CpG rich which are resistant to standard high throughput sequencing procedures due to secondary structure formation. The α4 subunit sequence exhibited the highest number of genome assembly sequence gaps with incomplete coding sequence for orangutan, chimpanzee, gorilla, gibbon, rhesus, bushbaby, and marmoset. Given that the α4β2 containing receptors are the most predominant nAChR subtypes in mammalian brain and that these and additional nAChR subunits contribute to the behavioral effects of nicotine it is clearly important to fill these sequencing gaps.
Table 2.
Homology of non-human primate nAChR coding sequences with human (hg19).
| Gene ID | Amino Acids |
great apes | old world | new world | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chimp (pan- Tro4) |
Gorilla (gor- Gor3) |
Orangutan (ponAbe2) |
Gibbon (nom- Leu3) |
Cyno (mac- Fas5) |
Rhesus (rhe- Mac3) |
Baboon (pap- Ham1) |
Green monkey (chlSab1) |
Marmoset (calJac3) |
Squirrel monkey (saiBol1) |
Bushbaby (otoGar3) |
Mouse (mm10) |
||
| CHRNA2 | 529 | 98.9 | 98.5 | 97.5 | 97.9 | 97.2 | 96.8 | 97.0 | 97.4 | 95.8 | 95.5 | 86.6 | 83.6 |
| CHRNA3 | 505 | 99.6 | 99.4 | 99.8 | 99.6 | 98.8 | 98.4 | 98.8 | 98.8 | 98.6 | 96.3 | 94.8 | 92.9 |
| CHRNA4 | 627 | 98.3 | 98.7 | 97.7 | 97.0 | 96.5 | 96.2 | 96.6 | 92.7 | 96.7 | 87.3 | 92.6 | 85.3 |
| CHRNA5 | 468 | 99.1 | 99.6 | 98.7 | 98.7 | 98.5 | 98.8 | 99.2 | 97.9 | 96.4 | 95.8 | 94.4 | 89.3 |
| CHRNA6 | 494 | 99.6 | 99.1 | 97.6 | 97.8 | 96.4 | 96.4 | 96.0 | 97.2 | 97.0 | 96.9 | 88.3 | 85.6 |
| CHRNA7 | 502 | 100.0 | 99.8 | 98.8 | 99.2 | 99.0 | 99.2 | 98.6 | 99.0 | 98.4 | 99.0 | 96.8 | 93.6 |
| CHRNA9 | 479 | 99.2 | 98.5 | 99.6 | 98.7 | 98.5 | 98.5 | 98.3 | 98.1 | 98.3 | 97.9 | 93.7 | 91.0 |
| CHRNA10 | 466 | 99.1 | 99.1 | 98.9 | 98.7 | 96.7 | 96.7 | 96.9 | 96.9 | 96.7 | 96.0 | 93.8 | 92.6 |
| CHRNB2 | 502 | 100.0 | 100.0 | 99.0 | 99.4 | 99.2 | 99.0 | 99.4 | 99.6 | 98.2 | 98.2 | 94.0 | 94.4 |
| CHRNB3 | 458 | 99.8 | 99.6 | 98.3 | 99.1 | 98.4 | 99.1 | 98.4 | 97.6 | 98.0 | 98.7 | 92.3 | 87.0 |
| CHRNB4 | 498 | 98.1 | 99.0 | 97.5 | 98.2 | 95.7 | 96.1 | 95.3 | 95.9 | 92.0 | 93.3 | 89.5 | 83.1 |
| Average similarity | 99.2 | 99.2 | 98.5 | 98.6 | 97.7 | 97.7 | 97.7 | 97.4 | 96.9 | 95.9 | 92.4 | 88.9 | |
ClustalW alignment of human and non-human primate nAChR coding sequences demonstrates greater than 95% homology with human for the majority of NHP species assessed, compared to an average of 88.9% homology for human with mouse. CHRNB2 and the ancestral subunit, CHRNA7, displayed the greatest homology to human across NHP families. The cynomolgus and rhesus macaques, commonly used for biomedical research, were 99.6% homologous to each other over all subunits (data not shown) and 97.7% homologous to human across nAChR coding sequences. Genomic assembly used shown in parentheses.
2.4. Conservation of polymorphisms between NHP and humans
While primates share a common genetic structure on the macro-scale, advances in whole-genome sequencing technologies have allowed for the study of population diversity at single nucleotide resolution. The subject of genetic diversity within humans is highly significant as single nucleotide polymorphisms (SNPs) provide key insights into gene function, pathophysiology, and can greatly influence individual responses to therapeutics. The macaques of the Cercopithecidae family of Old World monkeys have been used extensively as translational models with which to study drug pharmacokinetics, safety, and efficacy (Benwell et al., 1988). The most commonly used model species of this family are the cynomolgus macaques, originating from Southeast Asia, and the rhesus macaques whose territory ranges from Eastern China through Western India and Pakistan (Street et al., 2007). Genetic diversity across these model species influences their behavior, physiology, and phenotypic differences in response to experimental protocols (Champoux et al., 1997; Clarke and O'Neil, 1999). Greater genetic diversity has been observed in populations of rhesus macaques from China versus those originating from India (Ferguson et al., 2007; Yuan et al., 2012). One study estimated that rhesus and cynomolgus macaques share approximately half (52%) of SNPs measured in both species (Street et al., 2007). In both the rhesus and cynomolgus macaques, the ratio of non-synonymous/synonymous coding SNPs is significantly lower than in humans (Higashino et al., 2012). Therefore, despite having greater genetic diversity, the number of damaging non-synonymous polymorphisms in rhesus macaque is estimated to be similar to the number in humans (Yuan et al., 2012).
2.5. Conservation of single nucleotide polymorphisms in CHRNA5/A3/B4
Genome-wide association studies (GWAS) and twin studies have indicated a key role for genetics in smoking related phenotypes including cigarettes smoked per day (Tobacco and Genetics Consortium, 2010), response to smoking cessation therapy (Bergen et al., 2013), and smoking related diseases including COPD (Gabrielsen et al., 2013) and risk of lung cancer (Amos et al., 2008). In particular, a number of studies have shown allelic variation in the α5-α3-β4 subunit gene cluster located at human chromosome region 15q25 to be significantly associated with nicotine dependency and smoking-related diseases (Saccone et al., 2010). One particular single nucleotide polymorphism (SNP) in the α5 subunit gene (rs16969968), in which the amino acid 398 is changed from aspartic acid to asparagine (α5D398N), has been independently validated as a factor in multiple nicotine dependent phenotypes (Bierut, 2010). For example, one study showed that carriers of one copy of rs16969968 smoke on average one additional cigarette per day (CPD) than non-carriers while carriers of two risk alleles smoke on average an additional two CPD, based on objective measures of plasma cotinine (Munafo et al., 2012). Another study by Lips et al., measured an increased consumption of 0.3 CPD in carriers of one minor allele and an increase of 1.2 CPD in carriers of two risk alleles based on self-reporting of consumption by 17,000 research subjects (Lips et al., 2010). This amount may be less than predicted by Manufo et al., due to underreporting of CPD. It is also possible that the association of cotinine levels with α5 genotype is stronger than the association with reported number of cigarettes per day due to genotype effects on smoking topography (Macqueen et al., 2014). Furthermore, rs16969968 is associated with increased risk of lung cancer but the question remains whether the small increase in smoking associated with this polymorphism is sufficient to drive a higher lung cancer incidence.
As the role of increased nicotine addiction versus direct effects of nicotine on cell signaling in the periphery cannot be teased apart in clinical studies, animal models are crucial in order to understand the role of nAChRs in both the brain and periphery. Genetic mouse knock-out models exist for a number of nAChR subunits and have proven invaluable in determining a role for these subunits in nicotine associated behaviors such as dependency, aversion, and withdrawal (for review see (Stoker and Markou, 2013; Fowler et al., 2008)). However, homology of mouse nAChR subunits with humans at the protein level is significantly less than NHP model species (88.9% across all subunits for mouse, versus 97.7% for rhesus or cynomolgus macaques). Additionally, complete ablation of a receptor subunit may result in compensation by other subunits and a much different phenotype than a SNP. Finally, in an ideal animal model with which to study the effects of nicotine on brain signaling and behavior, the animal should have similar neuroanatomy, voluntarily self-administer nicotine and metabolize nicotine similarly to humans. Macaque monkeys meet these requirements and have been used extensively by our group and others for study of ethanol, nicotine and other drugs (Grant et al., 2008; McCallum et al., 2006b; Ferguson et al., 2013).
To assess the potential of macaque primates for characterizing mechanisms underlying how nAChR polymorphisms affect nicotine dependence and nicotine-mediated pathophysiology, we screened macaque primates for SNPs for comparison with human polymorphisms with particular emphasis on identifying common SNPs with known association to nicotine dependency. In 12 rhesus macaque monkeys (6 of Indian origin, 6 of Chinese origin (Ferguson et al., 2007)), we observed 869 SNPs in the α5-α3-β4 cluster (RheMac3 chr7:57,587,673-57,704,684). This region is equivalent in humans to hg19 chr15:78,845,491-78,945,185 which contains 510 SNPs (with an allele frequency ≥1%) according to the UCSC databases “Common SNPs (build138)”, respectively. One caveat of this comparison is that the SNP detection pipeline used for our rhesus sequence data was not also used to call SNPs in human sequence data. However, the relatively greater number of SNPs in rhesus macaque vs human at this locus is supported by a previous study which sequenced the 3’ end of 94 genes across the rhesus genome and found an average of 1 SNP/107 bp in rhesus compared with 1 SNP/179 bp in human (Street et al., 2007). Across this nAChR gene locus 29 SNP positions are conserved between rhesus and human, however only 10 of these positions share the same reference and variant nucleotide between the two species. Of these 10 conserved SNPs, 1 is in the 5’ UTR of CHRNA5 (human SNP tag rs201545082), 8 are intronic (rs147796977, rs62008248, rs112973744, rs185308922, rs189302121, rs76832402, rs78319611, rs79105076), and 1 is a synonymous SNP in exon 3 of CHRNA5 (rs75600623). Thus, relatively few rhesus SNPs in this cluster aligned with common human SNPs.
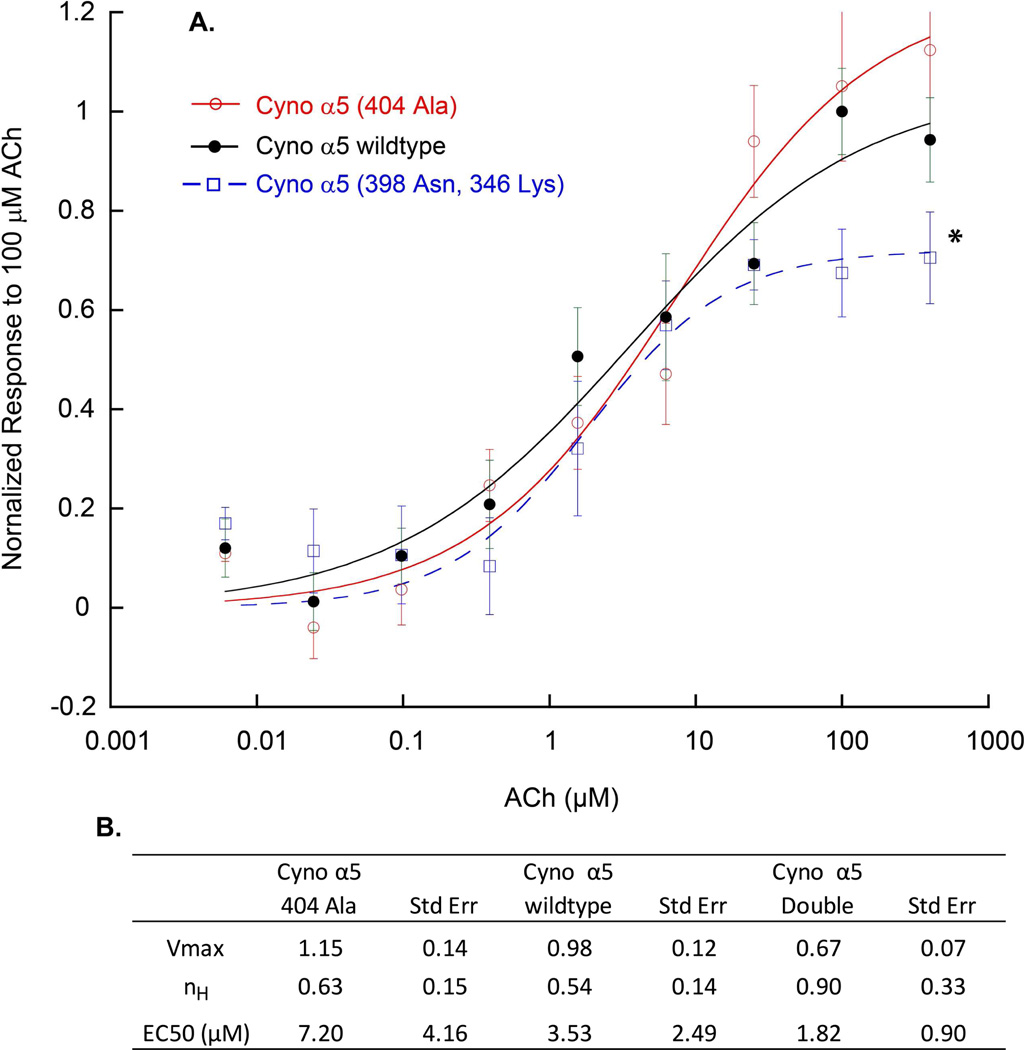
We next assessed the cynomolgus macaque for polymorphisms that may be relevant to nicotine dependent phenotypes. Currently, complete SNP data across the α5-α3-β4 locus for cynomolgus macaques is unavailable. We therefore performed traditional Sanger sequencing across the coding regions of CHRNA5 in 41 cynomolgus macaques. Primer sequences were designed in Primer 3 against Macaca mulatta to sequence across CHRNA5 exons. For PCR amplification we used the NEB Flex polymerase system with the following cycling parameters: 92° - 120 seconds; 34 cycles (92° - 30s, 61° −30s, 72° - 60s); 72° - 600s. PCR products were treated with Exonuclease I and Antarctic Phosphatase (37° - 30m, 95° - 5m) prior to sequencing on an Applied Biosystems 3730xl DNA Sequencer. Sequencing results were analyzed in Sequencher software by assembling to the corresponding human exon reference sequence and manually scanning chromatograms for secondary peaks. We identified 6 SNPs in cynomolgus macaque CHRNA5 exons, 4 of which were non-synonymous SNPs with minor allele frequencies (MAF) ranging between 0.237 and 0.333. Notably cynomolgus macaques, though not rhesus, expressed the same α5 D398N polymorphism that is associated with nicotine dependency and smoking-related diseases in humans (rs16969968). Therefore, using a custom TaqMan allelic discrimination assay we screened an additional 100 animals for this SNP and calculated a minor allele frequency of 0.374 which is similar to the corresponding allele frequency in a population of Utah Residents with Northern and Western European Ancestry (0.385) (HapMap CEU), although neither frequency measure can be considered to be representative of either species as a complete population. However this finding suggests that the polymorphism is relatively common in specific populations of cynomolgus macaques, which makes them a potentially useful model for studies of nicotine dependency and cessation therapies. A second α5 SNP was found in cynomolgus macaques, N346K, which was in linkage disequilibrium with the α5 D398N polymorphism with equivalent major and minor allele frequencies at both loci such that all animals polymorphic for α5 D398N were also polymorphic for α5 N346K. In addition to the α5 D398N polymorphism, a relatively frequent (23% allele frequency) polymorphism in α5 of Thr404Ala was also found in cynomolgus monkeys. This may affect a potential phosphorylation site in cynomolgus α5. Expression of the cynomolgus α5 D398N polymorphism in xenopus oocytes and HEK293 cells together with α4β2 subunits showed that the monkey equivalent of the human α5D98N polymorphism produced smaller currents than the wild type cynomolgus α5 similar to the results previously reported for the human polymorphism (Kuryatov et al., 2011; Bierut et al., 2008) (Figure 1). In preliminary studies, no significant differences between cynomolgus α5 404Ala and wild-type α5 were seen, though a slight trend towards increased maximal response was observed.
Figure 1. Expression of (α4β2)2α5 nAChRs in a human α4β2 concatamer HEK cell line transiently transfected with cynomolgus α5 variants.
The figure represents the combined results of three different transfections. For transient transfections, 60-mm dishes of a 50% confluent human β2QAPα4 cell line were transfected with 2µg of α5 cDNAs using the FuGENE 6 DNA transfection kit. After 24 h, cells were plated onto black 96-well plates for the FLEXstation. Three days after transfection nAChR function was determined in the cell lines using a FLEXstation (Molecular Devices, Sunnyvale, CA) bench-top scanning fluorometer and a membrane potential-sensitive fluorescent indicator as described by Kuryatov et al. (Kuryatov et al., 2005). Responses were normalized to the wild type response to 100µM ACh. Results showed that the α5 404-Ala mutant had essentially similar functional properties as the wild type. * Responses of the α5 398-Asn, 346-Lys mutant to 100 µM were statistically different from wild type (Student test p <0.05). (Cyno = cynomolgus monkey)
For the studies shown in figure 1, the HEK tsA201 cells were maintained in Dulbecco Modified Eagle Medium (DMEM, high glucose) (Life Technologies, Inc.) supplemented with 10% fetal bovine serum (Hyclone), 100 units/ml penicillin, 100 µg/ml streptomycin, and 2 mM L-glutamine (Life Technologies, Inc.) at 37° C, 5% CO2. 60mm dishes of 50% confluent α4β2 cells were transfected with α5 variants cDNAs using the FuGENE 6 DNA transfection kit (Roche Diagnostics, Indianapolis, IN). The transfected cells were passed onto blackwall/clear-bottom 96-well plate (Costar Corning Incorporated, Corning, NY) the day before testing. Functional cellular assays were conducted on FLEXstation (Molecular Devices, Sunnyvale, CA) bench-top scanning fluorometer similar to Kuryatov et al., (Kuryatov et al., 2005). Membrane potential assay kit (Molecular Devices) was used according to manufacturer’s protocol. The Hill equation was fitted to the concentration-response relationship using a nonlinear least-squares error curve-fit method (Kaleidagraph, Synergy Software, Reading, PA): I(x) = Imax[xn/(xn+EC50n)], where I(x) is the current measured at the agonist concentration x, Imax is the maximal concentration for the half-maximal response, and n is the Hill coefficient.
2.6. CpG conservation between human and NHP models
In addition to polymorphisms in nAChR subunit genes that may modify nicotine dependent phenotypes, differences in epigenetic regulation between model species are likely to modify complex behaviors and diseases. The field of epigenomics is rapidly expanding as next generation sequencing and high throughput technologies have been adapted to measure covalent modifications of DNA and histones. Both gene-specific and genome-wide approaches (Bell et al., 2012) have been utilized in non-human primate models including differences between genetically similar primate species (i.e. human and chimpanzee or human and macaque) that may help explain species-specific traits and diseases. One common approach is to measure methylation of CpG islands, as these CpG dense clusters of DNA often co-locate with the promoter regions of genes and are thought to be associated with differential gene expression. DNA methylation of promoter CpG islands has been measured in only a few nAChR subunit genes.
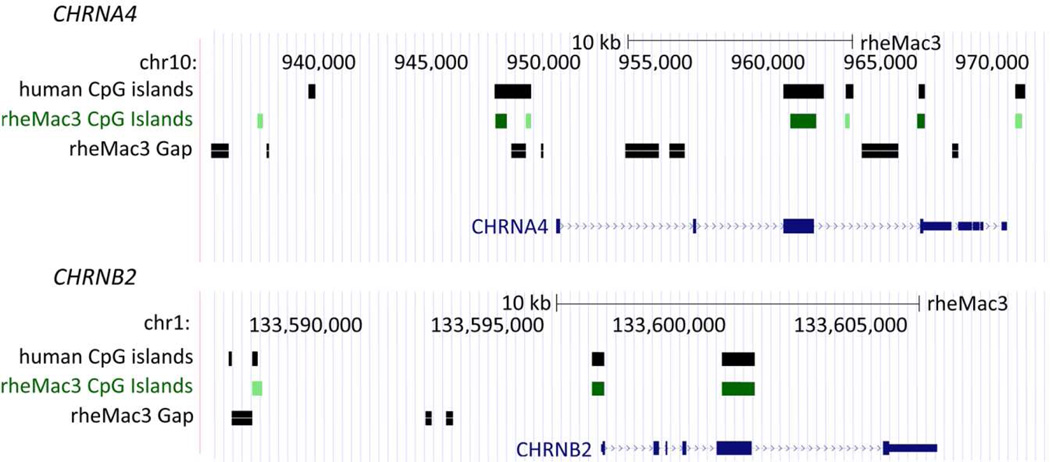
We therefore performed an analysis of CpG island conservation between humans and rhesus macaque at nAChR subunit genes. In brief, we performed base-wise intersection of human CpG islands with CpG islands annotated for the publically available rhesus genome drafts (rheMac2 or rheMac3) using UCSC defined CpG islands (GC content >50%, span greater than 200 bp, and a greater ratio of CG dinucleotides in the segment than expected based on C and G content). In human (hg19) we observed a total of 19 CpG islands within 20kb of any nAChR subunit gene (6 at CHRNA4; 4 at CHRNB2; 2 each at CHRNA2, CHRNA3, and CHRNA7; and 1 each at CHRNB4, CHRNA10, and CHRNA5). We observed conservation or partial conservation in 15/19 human nAChR CpG islands. However, neither rheMac3 (Chinese-origin) or rheMac2 (Indian-origin) drafts contains complete information for the 15 rhesus conserved CpGs due to gaps in these assemblies. Therefore, until a more complete genome for rhesus, and similarly other NHP models, is available caution must be applied when interpreting these results (Zhang et al., 2012). However, the similar structure of CpG islands in and around these genes between primate species is supported by the high level of DNA sequence homology described above, especially in the coding and promoter regions. An example of this analysis shows the CpG homology between human and rhesus α4 and β2 genes.
To date differential methylation has been observed within the nAChR 15q25 susceptibility locus in human lung cancer (Scherf et al., 2012; Paliwal et al., 2010), in CHRNA4 in mouse tissues relative to brain, and at CHRNA5 in association with childhood adversity and its interaction with nicotine and alcohol dependency (Xie et al., 2012; Zhang et al., 2013). The levels of methylation in nAChR subunit genes in non-human primate models has yet to be described although this epigenetic modification is expected to account for at least 12–18% of differential gene expression between primate species (Hernando-Herraez et al., 2013).
3. Nicotinic receptor expression in non-human primate brain
Our knowledge of the distribution of nAChRs in the central nervous system has primarily come from studies using in situ hybridization (ISH) and autoradiography. ISH has the advantage of specificity to detect individual mRNA subunits but does not provide information at the protein level. Conversely, autoradiography allows for detection of functional receptor proteins but can lack specificity as many of the available radio-labeled ligands for autoradiography bind to multiple receptor subtypes, though this problem has been alleviated with the identification of conotoxin ligands with increased specificity. The next two sections will review what is known about distribution of nAChR subunit mRNA and functional protein receptors in non-human primates.
3.1. Non-human primate nAChR mRNA distribution
In 2000, the Changeux lab conducted a systematic survey of nAChR subunit mRNAs in the brain of adult Macaca mulatta (Rhesus macaque) as measured by in situ hybridization (Han et al., 2000). Signal was detected in multiple brain areas for all of the subunits tested which included α2-α7 and β2-β4. Across all brain regions measured, the signal intensity was greatest for β2 followed by α4 and α2. These three subunits were detected in the majority of areas surveyed while α3 and β4 were detected only in hippocampus and epithalamus. This is in partial agreement with an earlier study which examined α3 mRNA distribution in Macaca fascicularis (cynomolgus macaque) and demonstrated strongest α3 signal in the dentate gyrus, pineal gland, dorsal lateral geniculate, and medial habenula (Cimino et al., 1992). In rhesus macaque, the α7 subunit was barely to moderately detectable in isocortex, in parts of the mesencephalon and epithalamus, and in nuclei of the thalamus and was strongly expressed in the dentate gyrus of the hippocampal formation and the supraoptic nucleus of the hypothalamus and in the lateral habenula. Localization of the α5 subunit was similar to α7 with the exception of being undetected in the hippocampal formation or epithalamus. Lastly, β3 was strongly detected in substantia nigra, pars compacta and weakly evident in the ventral tegmental area and hippocampal formation (Han et al., 2000). Curiously, α5 was not detected in rhesus habenula, but this may have reflected technical issues with the probe used for hybridization.
A similar study on the distribution of α4, α6, α7 and β2-β4 nAChR subunit mRNAs in adult male squirrel monkeys (Saimiri sciureus) was also published by Quik et. al in 2000 (Quik et al., 2000). Expression and localization of the α6, α7 and β3 mRNA subunits was similar between the two studies with strongest expression of both subunits in the medial habenula and substantia nigra. The α4 subunit was widely distributed in the brains of both species however the level of expression as measured by signal intensity following in situ hybridization was not consistent between the two studies. In squirrel monkeys the strongest signal for α4 was in sub-regions of telencephalon while the signal in this area in macaque was weak and vice versa for nuclei of the thalamus. A further discrepancy is the widely distributed and high expression of β4 subunits in squirrel monkey with expression in macaque limited to the medial habenula and pineal gland. Lastly, the expression of β2 subunits was much stronger in macaque relative to squirrel monkey. Possible explanations for these discrepancies include true differential expression across these two species, age or gender differences of the animals studied, as well as artifactual differences due to probe design, hybridization strength, and detection methods used. Though there were differences between the two studies, importantly as pointed out by Quik et al (Quik et al., 2000), overall the distribution of nAChR expression in NHP brain was significantly more similar to that of human expression than was rodent. Analysis of nAChR expression as measured by radioligand binding is discussed below.
3.2 Analysis of nAChR expression by positron emission tomography (PET)
The localization and pharmacokinetics of nAChR in primate brain is a subject of intense study with the emergence of new PET radioligands that bind with subunit specificity. While several radioligands targeting α4β2* and α7 receptors have been developed and tested in non-human primates, they often demonstrate kinetic profiles that are still not ideal for widespread diagnostic use in humans (Horti et al., 2013). For α4β2* receptors, the greatest level of radioligand uptake and binding occurred in the anteroventral thalamus, lateral geniculate nucleus and midbrain tegmental nuclei; other brain regions expressing significant binding included cortex (insula, temporal, cingulate, frontal), subiculum and striatum (Hillmer et al., 2011; Horti et al., 2013). For α7 receptors, distribution was highly concentrated in thalamus, insular cortex, hippocampus and cerebellum, with binding also reported in other cortical regions (occipital, temporal, frontal) and the striatum (Hashimoto et al., 2008; Horti et al., 2014). PET scans largely mirrored earlier work characterizing the regional distribution of α4β2* and α7 receptors in monkey brain via receptor autoradiography (Kulak et al., 2006; Han et al., 2003).
4. Nicotine administration in non-human primate models
4.1 Changes in CNS nAChR expression in response to nicotine
Over the past several years Quik and colleagues have developed and investigated a model of chronic nicotine treatment in squirrel monkeys that involves introduction of nicotine (up to a final concentration 650 µg/ml) in sweetened drinking water. This exposure paradigm is designed to mimic the intermittent and chronic delivery of nicotine as occurs with smoking, and is reported to yield plasma nicotine levels on the lower end of those observed in human smokers (60 – 90 nM) (Matta et al., 2007). Although nicotine intake is involuntary, this model has proven fruitful in characterizing nAChR binding sites and function in primate brain following chronic treatment, particularly in regions associated with nicotine addiction. Some of the earliest reports using this model demonstrated both an enhanced nicotine-evoked dopamine release from synaptosomes and an increased number of α4β2* nAChR binding sites in the nucleus accumbens and striatal subregions following chronic nicotine exposure (McCallum et al., 2006a; McCallum et al., 2006b). This enhanced binding is consistent with the upregulation of α4β2* receptors reported in the postmortem cortex and other brain regions of smokers (Benwell et al., 1988; Breese et al., 1997) and in a comparison of smokers versus non-smokers using a subtype-specific PET radioligand (Mukhin et al., 2008). Notably, chronic nicotine treatment exhibits at least some selectivity in upregulating α4β2* bindings sites, as either no change or a decrease in α3/α6β2* sites were detected (McCallum et al., 2006a; McCallum et al., 2006b). This selectivity likely reflects the high occupancy rates and desensitization of α4β2* receptors that is observed in cigarette smokers (Brody et al., 2006).
4.2 A non-human primate model of nicotine self-administration
Studies to date in non-human primates have focused on changes in nAChR binding sites consequent to nicotine exposure. Our identification of a non-synonymous SNP in the α5 receptor subunit in cynomolgus monkeys that is homologous to the D398N mutation in humans (rs16969968) affords the unique opportunity to study the influence of α5 subunit on drug taking behavior in a primate model. Individuals homozygous for α5N have an increased risk for nicotine addiction, smoke more cigarettes per day, exhibit an earlier age of onset for tobacco use and have greater difficulty in quitting (Berrettini and Doyle, 2012; Bierut et al., 2008; Freathy et al., 2009; Weiss et al., 2008; Munafo et al., 2012). We hypothesized that cynomolgus monkeys with this mutation would exhibit a differential behavioral sensitivity to nicotine across a range of drug concentrations and would self-administer greater amounts of nicotine under a chronic, open access condition when compared to wild-type animals. To address this question, we developed a two-bottle choice self-administration procedure with oral nicotine that was based largely on earlier work in the development of non-human primate models of ethanol self-administration (Vivian et al., 2001; Grant et al., 2008).
In brief, animals were acclimated to a 14-hr/10-hr light/dark schedule (light on at 0800 hrs) in cages with an operant panel over a 3-month period. During this time all subjects were trained to consume water from two drinking spouts, obtain 1-g banana-flavored pellets from a food receptacle, and present their leg for awake venipuncture to measure plasma nicotine as previously described (Vivian et al., 2001; Grant et al., 2008). For the duration of the study, single daily 22-hour sessions were conducted 6 days/week. All sessions began at 12 noon and ended at 10am the following morning. Within each session, a set food ration was administered over three meals that were scheduled during the first, third, and fifth hours. Meal times were signaled by red stimulus lights (3-light array positioned above each spout), and subjects were required to pull a dowel and finger poke for the delivery of each food pellet. Drinking spouts were accessible anytime during the session (fluid availability was not linked to pellet delivery) and were controlled by solenoid valves. Both valves were opened following a dowel pull (and activation of green stimulus light), but once fluid was chosen from one spout the opposite spout was inactivated until the dowel was pulled again to provide a subsequent choice between drinking spouts. Fluid was continuously available from an activated spout as long as the dowel remained pulled. Sessions were run Monday thru Saturday (i.e., 6 days/week). On Sunday, water was the only fluid available, and food was delivered directly to the animal by technical staff. Cumulative records of fluid intake from each spout were monitored by weight displacement from reservoirs via a computer interface using National Instruments hardware and LabView software (accuracy to 0.1 ml and 0.1 second, over the entire 22-hour session). From this record, drinking within a given increment of time (e.g., hourly) could be derived.
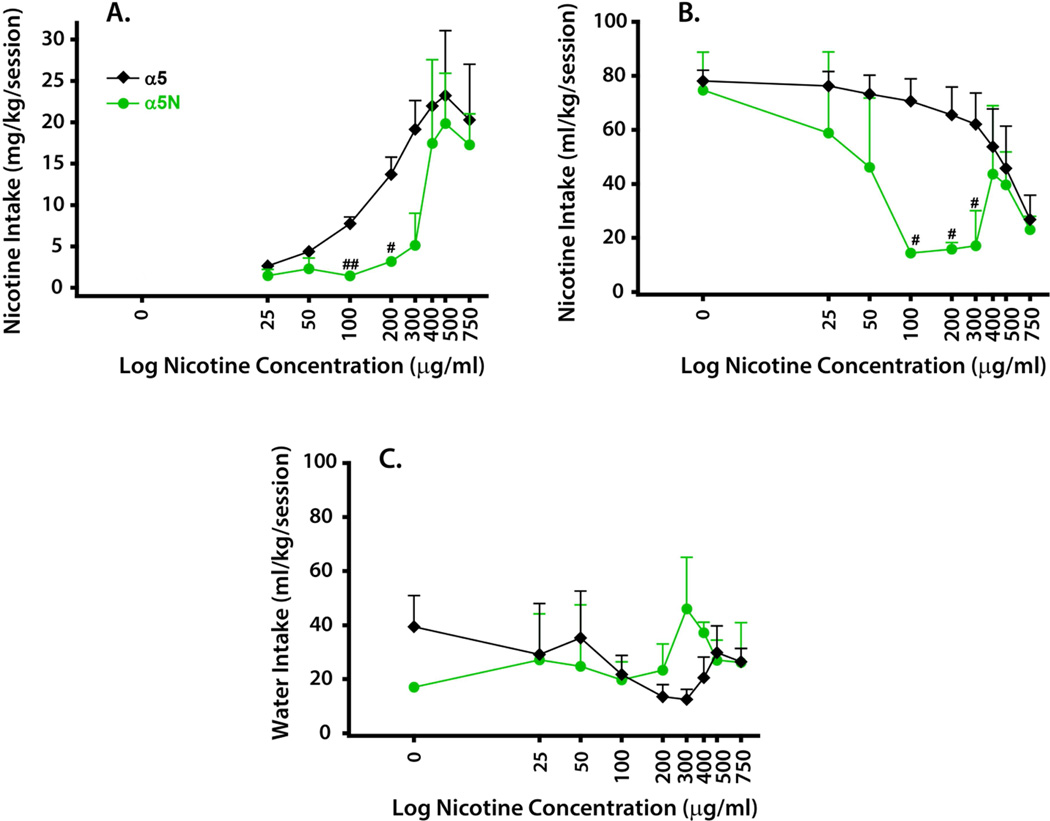
Genotyping of male cynomolgus monkeys (5–6 years old) yielded five subjects that were homozygous or heterozygous wild-type (collapsed as ‘α5’ group) and two subjects homozygous for α5 398N (‘α5N’ group). Following acclimation and panel training, all subjects were provided a choice between a nicotine solution (sweetened with 10% w/v maltose/dextrin; M/D) and water. The nicotine solution was always presented on the same spout for a given subject, but its placement was counterbalanced between left and right sides across subjects. In experimental Phase I, the nicotine spout contained M/D alone for 6 days (Monday-Saturday), and then M/D plus nicotine in incrementally greater concentrations (25- to 750 µg/ml) for 6 days each. The seventh day of each week (Sunday) was a “washout” day with access to water only. Thus, each nicotine concentration was available continuously for 1 week (excluding Sunday), and the next highest concentration was started each Monday. During this experimental phase a 500-ml cutoff for the nicotine solution was instituted to prevent intake of hazardous levels of nicotine and the conditioning of a nicotine aversion. The nicotine solution was available continuously for 22-hours, or until the cutoff was reached, whichever came first. Phase I of the study lasted 9 weeks, until each concentration had been tested for six consecutive days. Subjects exhibited a prototypical inverted-U concentration-response curve with maximal mg/kg nicotine intake occurring with access to the 500 µg/ml solution (Figure 3A). A two-way repeated measures (RM) analysis of variance (ANOVA) detected a significant main effect of nicotine concentration on mg/kg intake [F(7,35) = 7.19; p < 0.001], but neither a main effect of genotype nor a concentration x genotype interaction were apparent. Based on our a priori hypothesis that α5 receptor genotype would alter the behavioral sensitivity to nicotine in a concentration-dependent manner, genotype comparisons at each nicotine concentration were subsequently conducted with separate t-tests. In general, α5N subjects consumed low doses of oral nicotine (≈ 5 mg/kg) when concentrations of 300 µg/ml and less were offered, but consumed comparable doses as α5 wild-type subjects with concentrations of 400 µg/ml and greater. When analyses were run for each concentration, t-tests revealed that α5N subjects consumed significantly less nicotine than wild-types when the 100 µg/ml (t = 4.05; p < 0.01) and 200 µg/ml (t = 2.82; p < 0.05) concentrations were offered (Figure 3A).
Figure 3. Nicotine concentration-dependent nicotine self-administration.
Nicotine intake is reported as dose (mg/kg/session; panel A) or volume (ml/kg/session; panel B) and concurrent water intake as volume (ml/kg/session; panel C). In each case, data points depict mean ± SEM of 6-session averages for wild-type (α5, n = 5) and homozygous 398N mutant (α5N, n = 2) subjects. #p ≤ 0.05 and ##p ≤ 0.01 versus wild-type.
In order to ascertain whether genotype influenced the intake of M/D vehicle (i.e., 0 µg/ml nicotine) and to compare nicotine and water intakes, the volume consumed at each spout was examined. The volume of M/D vehicle consumed did not differ as a function of genotype (Figure 3B). A two-way RM ANOVA determined a main effect of nicotine concentration [F(8,40) = 7.56; p < 0.001] as well as a genotype x concentration interaction [F(8,40) = 3.30; p < 0.01] for the volume of nicotine solution self-administered. Consistent with the mg/kg nicotine intake findings, nicotine solution volumes consumed at the 100, 200 and 300 µg/ml concentrations were significantly decreased in the α5N subjects (all p’s < 0.05) when compared to wild-type animals (Figure 3B), as determined by the Holm-Sidak multiple comparisons procedure. In contrast, a two-way RM ANOVA revealed no effect of nicotine concentration, genotype or their interaction on the volume of water consumed (Figure 3C). These concentration-response relationships indicate that α5N mutants likely express a reduced behavioral sensitivity to oral nicotine.
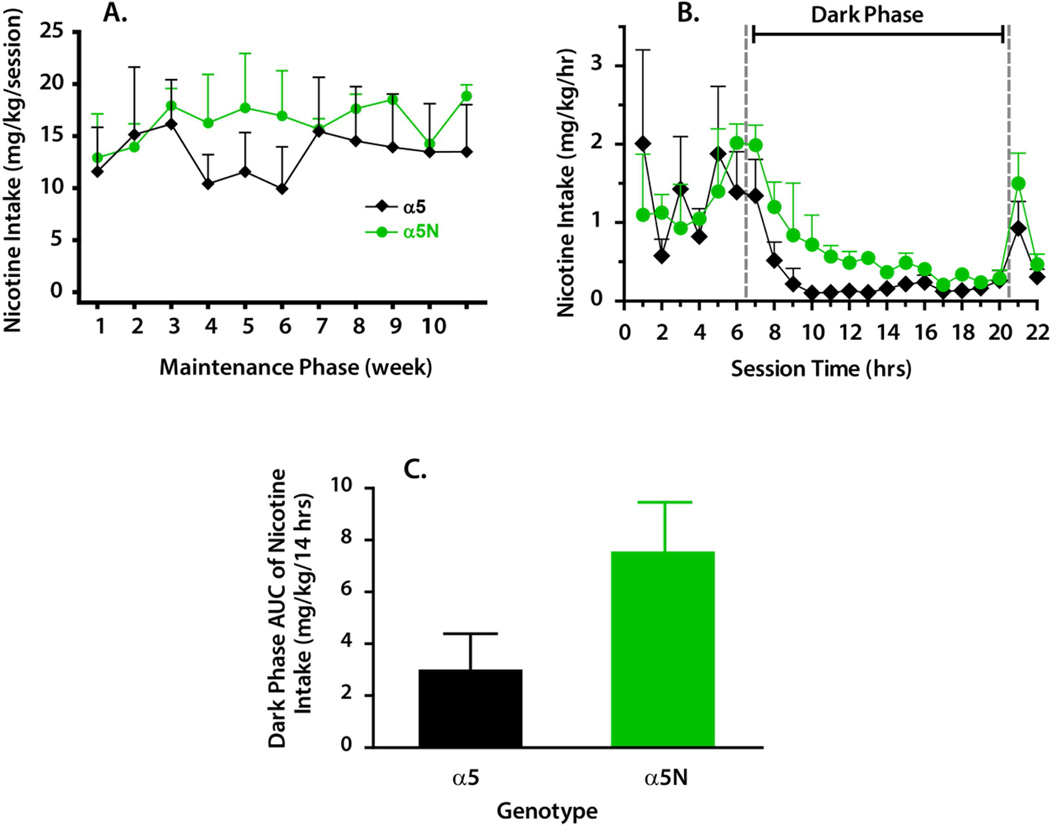
Immediately following the presentation of the 750 µg/ml nicotine solution and a single washout day, experimental Phase II was initiated. During this phase the volume cutoff for the nicotine solution was removed and animals were maintained on 400 µg/ml nicotine over an additional 11-week period. Levels of plasma nicotine measured every other week throughout Phase II ranged from 100 – 600 nM and were highly correlated with mg/kg nicotine intakes that occurred between the session start and blood sampling 3-hrs into the session (r = 0.838, p < 0.0001, n = 31). These plasma nicotine levels are consistent with the peak blood levels of 200–400 nM observed in humans during active smoking (Matta et al., 2007; Russell et al., 1980; Benowitz et al., 1982). Although the α5N animals consumed on average 25% greater mg/kg nicotine per 22-hr session than wild-type animals throughout these 11 weeks, a two-way RM ANOVA showed no significant effect of week, genotype or factorial interaction for mg/kg nicotine intake per session (Figure 4A). In order to determine whether genotype influenced the diurnal pattern of nicotine self-administration hourly nicotine intakes across the 22-hr sessions were plotted (Figure 4B). A 2-way RM ANOVA detected only a main effect of hour [F(21,105) = 3.12; p < 0.001]. A couple observations regarding pattern were worth noting. First, α5 wild-type subjects consumed the vast majority of their daily nicotine dose in conjunction with meals scheduled during the 1st, 3rd and 5th hours of the drinking session whereas the α5N animals self-administered nicotine in a pattern independent of meals. Second, α5N animals consistently consumed 0.5 mg/kg nicotine or more per hour throughout the first half of the dark phase whereas intake by the α5 wild-type subjects rarely occurred beyond the initial two hours of lights out.
Figure 4.
Chronic nicotine self-administration (phase II). In panel A, nicotine intake (mg/kg/session) with open access to a 400 µg/ml nicotine solution across an 11-week maintenance period is depicted. Each data point represents a 6-day average of intake for the 22-hr sessions conducted per week. In panel B, the diurnal pattern of nicotine intakes (mg/kg/hr) across the 22-hr sessions is illustrated. Each data point illustrates the average hourly intake that occurred throughout all sessions run in weeks 8, 9 and 11 (week 10 data omitted due to ketamine dosing of all subjects for an MRI procedure). Session time between the two dashed vertical lines indicates the dark phase of the light cycle. In panel C, an area under the curve (AUC) analysis of dark phase nicotine intake (same values as hrs 7–20 in panel B) is shown. All data points or bars represent the mean ± SEM of wild-type (α5, n = 5) and homozygous 398N mutant (α5N, n = 2) subjects.
A separate ANOVA of area under the curve (AUC) for nicotine intake during session hrs 7–20 (Figure 4C) revealed a trend towards increased dark phase consumption in α5N compared to α5 wild-type animals [F(1,5) = 3.84; p = 0.10]. This latter finding is consistent with observations that a greater degree of nicotine dependence is associated with shorter sleep duration in humans (Cohrs et al., 2012) and that nicotine dependence is associated with a reduction in the disparity between light and dark phase rates of self-administration in rodents (O'Dell et al., 2007).
In summary, our data to date suggest that α5N animals may express a reduced behavioral sensitivity to oral nicotine solutions. This interpretation is consistent with earlier in vitro studies showing that the D398N mutation is associated with a hypo-functional α5 receptor; including a decreased nicotinic agonist-evoked intracellular calcium response (Tammimaki et al., 2012) as well as a reduced calcium permeability and heightened desensitization when the α5 subunit is an accessory to α4β2* receptors (Kuryatov et al., 2011). Changes in α5 receptor function due to gene variants are particularly relevant for nicotine self-administration given the recently described role for habenular α5 nAChRs in mediating nicotine reinforcement and aversion (Fowler et al., 2011; Frahm et al., 2011). There are a few possible explanations for the absence of significant genotype effects on the maintenance of nicotine self-administration in experimental phase II. First, the 400 µg/ml concentration of nicotine was selected because it represented the inflection point in which α5N animals self-administered comparable amounts of nicotine as the α5 wildtype subjects (Figure 3A). It is possible that selection of a higher (or more aversive) nicotine concentration would have unveiled a greater influence of the D398N mutation. Second, the duration of chronic nicotine self-administration was likely insufficient to capture the full development of a nicotine dependence. Subtle, yet incremental, shifts in the nicotine consumption patterns of α5N animals across weeks (e.g., number of intake bouts per session) was indicative of a drinking phenotype that continued to evolve (data not shown). Lastly, inclusion of additional α5N subjects would have provided greater statistical power for the detection of genotype effects for this complex behavior. Overall this data, combined with the high homology of NHP nAChR to human nAChR supports the utility of non-human primates in evaluating the pathophysiology of nAChR and particularly the utility of NHP in the development of more effective drugs for smoking cessation.
5. Conclusions
The high homology of non-human primate nicotinic receptors to human nicotinic receptors combined with the high degree of neuroanatomic homology between humans and monkeys provides unique advantages of monkey models for studying roles of nicotinic receptors in complex behaviors. In addition, the high degree of structural homology between monkey and human nAChR suggests that evaluation of efficacy of drugs for which primate models exist such as Parkinson’s disease, and nicotine or alcohol self-administration is likely to be predictive of treatment outcomes in humans. Conservation of genomic characteristics such as CpG islands and polymorphisms also demonstrates the utility of NHP models for human personalized medicine treatment developments. These advantages have to be balanced by expense and complexity of non-human primate models. Other disadvantages of NHP include lack of genetic models, though recent advantages in gene transfer and gene modification technologies are beginning to mitigate these limitations.
Figure 2. CpG island conservation between human and rhesus macaque nAChR subunits.
Human CpG islands are partially conserved across nAChR subunit genes in rhesus macaque. For example in CHRNA4, rhesus CpG islands aligned with 5 of 6 human CpG clusters and in CHRNB2, rhesus CpG islands overlapped 3 of 4 human CpG islands.
Highlights.
Genomic structure of macaque nAChRs are compared to human.
Non-human primate nAChR coding sequences are compared to human.
Cynomolgus monkeys are shown to express an α5 Asp398Asn polymorphism like humans.
Monkeys can be trained to self-administer nicotine in multiple paradigms.
The α5 Asp398Asn polymorphism affects nicotine self-administration in monkeys.
Acknowledgements
We wish to thank Yibing Jia and the ONPRC molecular and cellular biology service core for assistance with sequencing nicotinic receptors and the ONPRC Primate Genetics Program for bioinformatics support.
Supported by NIH grants P51 OD011092, U01 AA013510, T32 DA007262
Abbreviations
- α-CTx
α-conotoxin
- GWAS
genome wide association study
- NHP
non-human primate
- nAChR
nicotinic acetylcholine receptor
- SNP
single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Lyndsey E. Shorey-Kendrick, Email: shorey@ohsu.edu.
Matthew M. Ford, Email: fordma@ohsu.edu.
Daicia C. Allen, Email: allendai@ohsu.edu.
Alexander Kuryatov, Email: kouriato@mail.med.upenn.edu.
Jon Lindstrom, Email: jslkk@mail.med.upenn.edu.
Larry Wilhelm, Email: wilhella@ohsu.edu.
Kathleen A. Grant, Email: grantka@ohsu.edu.
Eliot R. Spindel, Email: spindele@ohsu.edu.
References
- Albuquerque EX, Pereira EF, Alkondon M, Rogers SW. Mammalian Nicotinic Acetylcholine Receptors: From Structure to Function. Physiol Rev. 2009;89:73–120. doi: 10.1152/physrev.00015.2008. PM:19126755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, Houlston RS. Genome-Wide Association Scan of Tag SNPs Identifies a Susceptibility Locus for Lung Cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. PM:18385676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araud T, Graw S, Berger R, Lee M, Neveu E, Bertrand D, Leonard S. The Chimeric Gene CHRFAM7A, a Partial Duplication of the CHRNA7 Gene, Is a Dominant Negative Regulator of Alpha7*NAChR Function. Biochem Pharmacol. 2011;82:904–914. doi: 10.1016/j.bcp.2011.06.018. PM:21718690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam L, McIntosh JM. Molecular Basis for the Differential Sensitivity of Rat and Human Alpha9alpha10 NAChRs to Alpha-Conotoxin RgIA. J Neurochem. 2012;122:1137–1144. doi: 10.1111/j.1471-4159.2012.07867.x. PM:22774872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr CS, Newman TK, Becker ML, Parker CC, Champoux M, Lesch KP, Goldman D, Suomi SJ, Higley JD. The Utility of the Non-Human Primate; Model for Studying Gene by Environment Interactions in Behavioral Research. Genes Brain Behav. 2003;2:336–340. doi: 10.1046/j.1601-1848.2003.00051.x. PM:14653305. [DOI] [PubMed] [Google Scholar]
- Bell CG, Wilson GA, Butcher LM, Roos C, Walter L, Beck S. Human-Specific CpG "Beacons" Identify Loci Associated With Human-Specific Traits and Disease. Epigenetics. 2012;7:1188–1199. doi: 10.4161/epi.22127. PM:22968434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz NL, Kuyt F, Jacob P. Circadian Blood Nicotine Concentrations During Cigarette Smoking. Clin Pharmacol Ther. 1982;32:758–764. doi: 10.1038/clpt.1982.233. PM:7140139. [DOI] [PubMed] [Google Scholar]
- Benwell ME, Balfour DJ, Anderson JM. Evidence That Tobacco Smoking Increases the Density of (−)-[3H]Nicotine Binding Sites in Human Brain. J Neurochem. 1988;50:1243–1247. doi: 10.1111/j.1471-4159.1988.tb10600.x. PM:3346676. [DOI] [PubMed] [Google Scholar]
- Bergen AW, Javitz HS, Krasnow R, Nishita D, Michel M, Conti DV, Liu J, Lee W, Edlund CK, Hall S, Kwok PY, Benowitz NL, Baker TB, Tyndale RF, Lerman C, Swan GE. Nicotinic Acetylcholine Receptor Variation and Response to Smoking Cessation Therapies. Pharmacogenet Genomics. 2013;23:94–103. doi: 10.1097/FPC.0b013e32835cdabd. PM:23249876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrettini WH, Doyle GA. The CHRNA5-A3-B4 Gene Cluster in Nicotine Addiction. Mol Psychiatry. 2012;17:856–866. doi: 10.1038/mp.2011.122. PM:21968931. [DOI] [PubMed] [Google Scholar]
- Bierut LJ. Convergence of Genetic Findings for Nicotine Dependence and Smoking Related Diseases With Chromosome 15q24-25. Trends Pharmacol Sci. 2010;31:46–51. doi: 10.1016/j.tips.2009.10.004. PM:19896728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, Saccone NL, Saccone SF, Bertelsen S, Fox L, Horton WJ, Breslau N, Budde J, Cloninger CR, Dick DM, Foroud T, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Kuperman S, Madden PA, Mayo K, Nurnberger J, Jr., Pomerleau O, Porjesz B, Reyes O, Schuckit M, Swan G, Tischfield JA, Edenberg HJ, Rice JP, Goate AM. Variants in Nicotinic Receptors and Risk for Nicotine Dependence. Am J Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. PM:18519524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breese CR, Marks MJ, Logel J, Adams CE, Sullivan B, Collins AC, Leonard S. Effect of Smoking History on [3H]Nicotine Binding in Human Postmortem Brain. J Pharmacol Exp Ther. 1997;282:7–13. PM:9223534. [PubMed] [Google Scholar]
- Brody AL, Mandelkern MA, London ED, Olmstead RE, Farahi J, Scheibal D, Jou J, Allen V, Tiongson E, Chefer SI, Koren AO, Mukhin AG. Cigarette Smoking Saturates Brain Alpha 4 Beta 2 Nicotinic Acetylcholine Receptors. Arch Gen Psychiatry. 2006;63:907–915. doi: 10.1001/archpsyc.63.8.907. PM:16894067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux M, Higley JD, Suomi SJ. Behavioral and Physiological Characteristics of Indian and Chinese-Indian Hybrid Rhesus Macaque Infants. Dev Psychobiol. 1997;31:49–63. PM:9222116. [PubMed] [Google Scholar]
- Cimino M, Marini P, Fornasari D, Cattabeni F, Clementi F. Distribution of Nicotinic Receptors in Cynomolgus Monkey Brain and Ganglia: Localization of Alpha 3 Subunit MRNA, Alpha-Bungarotoxin and Nicotine Binding Sites. Neurosci. 1992;51:77–86. doi: 10.1016/0306-4522(92)90472-e. PM:1465189. [DOI] [PubMed] [Google Scholar]
- Clarke MR, O'Neil JA. Morphometric Comparison of Chinese-Origin and Indian-Derived Rhesus Monkeys (Macaca Mulatta) Am J Primatol. 1999;47:335–346. doi: 10.1002/(SICI)1098-2345(1999)47:4<335::AID-AJP5>3.0.CO;2-Y. PM:10206210. [DOI] [PubMed] [Google Scholar]
- Cohrs S, Rodenbeck A, Riemann D, Szagun B, Jaehne A, Brinkmeyer J, Grunder G, Wienker T, Diaz-Lacava A, Mobascher A, Dahmen N, Thuerauf N, Kornhuber J, Kiefer F, Gallinat J, Wagner M, Kunz D, Grittner U, Winterer G. Impaired Sleep Quality and Sleep Duration in Smokers-Results From the German Multicenter Study on Nicotine Dependence. Addict Biol. 2012 doi: 10.1111/j.1369-1600.2012.00487.x. PM:22913370. [DOI] [PubMed] [Google Scholar]
- Corringer PJ, Le Novere N, Changeux JP. Nicotinic Receptors at the Amino Acid Level. Annu Rev Pharmacol Toxicol. 2000;40:431–458. doi: 10.1146/annurev.pharmtox.40.1.431. PM:10836143. [DOI] [PubMed] [Google Scholar]
- Ferguson B, Street SL, Wright H, Pearson C, Jia Y, Thompson SL, Allibone P, Dubay CJ, Spindel E, Norgren RB., Jr. Single Nucleotide Polymorphisms (SNPs) Distinguish Indian-Origin and Chinese-Origin Rhesus Macaques (Macaca Mulatta) BMC Genomics. 2007;8:43. doi: 10.1186/1471-2164-8-43. PM:17286860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson CS, Miksys S, Palmour RM, Tyndale RF. Ethanol Self-Administration and Nicotine Treatment Induce Brain Levels of CYP2B6 and CYP2E1 in African Green Monkeys. Neuropharmacology. 2013;72C:74–81. doi: 10.1016/j.neuropharm.2013.04.022. PM:23639433. [DOI] [PubMed] [Google Scholar]
- Fowler CD, Arends MA, Kenny PJ. Subtypes of Nicotinic Acetylcholine Receptors in Nicotine Reward, Dependence, and Withdrawal: Evidence From Genetically Modified Mice. Behav Pharmacol. 2008;19:461–484. doi: 10.1097/FBP.0b013e32830c360e. PM:18690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler CD, Lu Q, Johnson PM, Marks MJ, Kenny PJ. Habenular Alpha5 Nicotinic Receptor Subunit Signalling Controls Nicotine Intake. Nature. 2011 doi: 10.1038/nature09797. PM:21278726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frahm S, Slimak MA, Ferrarese L, Santos-Torres J, Antolin-Fontes B, Auer S, Filkin S, Pons S, Fontaine JF, Tsetlin V, Maskos U, Ibanez-Tallon I. Aversion to Nicotine Is Regulated by the Balanced Activity of Beta4 and Alpha5 Nicotinic Receptor Subunits in the Medial Habenula. Neuron. 2011;70:522–535. doi: 10.1016/j.neuron.2011.04.013. PM:21555077. [DOI] [PubMed] [Google Scholar]
- Freathy RM, Ring SM, Shields B, Galobardes B, Knight B, Weedon MN, Smith GD, Frayling TM, Hattersley AT. A Common Genetic Variant in the 15q24 Nicotinic Acetylcholine Receptor Gene Cluster (CHRNA5-CHRNA3-CHRNB4) Is Associated With a Reduced Ability of Women to Quit Smoking in Pregnancy. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp216. PM:19429911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielsen ME, Romundstad P, Langhammer A, Krokan HE, Skorpen F. Association Between a 15q25 Gene Variant, Nicotine-Related Habits, Lung Cancer and COPD Among 56 307 Individuals From the HUNT Study in Norway. Eur J Hum Genet. 2013 doi: 10.1038/ejhg.2013.26. PM:23443019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant KA, Leng X, Green HL, Szeliga KT, Rogers LS, Gonzales SW. Drinking Typography Established by Scheduled Induction Predicts Chronic Heavy Drinking in a Monkey Model of Ethanol Self-Administration. Alcohol Clin Exp Res. 2008;32:1824–1838. doi: 10.1111/j.1530-0277.2008.00765.x. PM:18702645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han ZY, Le NN, Zoli M, Hill JA, Jr., Champtiaux N, Changeux JP. Localization of NAChR Subunit MRNAs in the Brain of Macaca Mulatta. Eur J Neurosci. 2000;12:3664–3674. doi: 10.1046/j.1460-9568.2000.00262.x. PM:11029636. [DOI] [PubMed] [Google Scholar]
- Han ZY, Zoli M, Cardona A, Bourgeois JP, Changeux JP, Le NN. Localization of [3H]Nicotine, [3H]Cytisine, [3H]Epibatidine, and [125I]Alpha-Bungarotoxin Binding Sites in the Brain of Macaca Mulatta. J Comp Neurol. 2003;461:49–60. doi: 10.1002/cne.10659. PM:12722104. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Nishiyama S, Ohba H, Matsuo M, Kobashi T, Takahagi M, Iyo M, Kitashoji T, Tsukada H. [11C]CHIBA-1001 As a Novel PET Ligand for Alpha7 Nicotinic Receptors in the Brain: a PET Study in Conscious Monkeys. PLoS ONE. 2008;3:e3231. doi: 10.1371/journal.pone.0003231. PM:18800169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando-Herraez I, Prado-Martinez J, Garg P, Fernandez-Callejo M, Heyn H, Hvilsom C, Navarro A, Esteller M, Sharp AJ, Marques-Bonet T. Dynamics of DNA Methylation in Recent Human and Great Ape Evolution. PLoS Genet. 2013;9:e1003763. doi: 10.1371/journal.pgen.1003763. PM:24039605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashino A, Sakate R, Kameoka Y, Takahashi I, Hirata M, Tanuma R, Masui T, Yasutomi Y, Osada N. Whole-Genome Sequencing and Analysis of the Malaysian Cynomolgus Macaque (Macaca Fascicularis) Genome. Genome Biol. 2012;13:R58. doi: 10.1186/gb-2012-13-7-r58. PM:22747675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer AT, Wooten DW, Moirano JM, Slesarev M, Barnhart TE, Engle JW, Nickles RJ, Murali D, Schneider ML, Mukherjee J, Christian BT. Specific Alpha4beta2 Nicotinic Acetylcholine Receptor Binding of [F-18]Nifene in the Rhesus Monkey. Synapse. 2011;65:1309–1318. doi: 10.1002/syn.20965. PM:21674627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horti AG, Gao Y, Kuwabara H, Wang Y, Abazyan S, Yasuda RP, Tran T, Xiao Y, Sahibzada N, Holt DP, Kellar KJ, Pletnikov MV, Pomper MG, Wong DF, Dannals RF. 18F-ASEM, a Radiolabeled Antagonist for Imaging the Alpha7-Nicotinic Acetylcholine Receptor With PET. J Nucl Med. 2014;55:672–677. doi: 10.2967/jnumed.113.132068. PM:24556591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horti AG, Kuwabara H, Holt DP, Dannals RF, Wong DF. Recent PET Radioligands With Optimal Brain Kinetics for Imaging Nicotinic Acetylcholine Receptors. J Labelled Comp Radiopharm. 2013;56:159–166. doi: 10.1002/jlcr.3020. PM:24285321. [DOI] [PubMed] [Google Scholar]
- Kaitin KI, DiMasi JA. Pharmaceutical Innovation in the 21st Century: New Drug Approvals in the First Decade, 2000–2009. Clin Pharmacol Ther. 2011;89:183–188. doi: 10.1038/clpt.2010.286. PM:21191382. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-Like Alignment Tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. PM:11932250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DF, Glick ID, Shader RI. Central Nervous System Drug Development, Basic, and Clinical Research: Thinking Outside the Box. J Clin Psychopharmacol. 2011;31:553–554. doi: 10.1097/JCP.0b013e3182305656. PM:21869701. [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Haussler D, Kent WJ. The UCSC Genome Browser and Associated Tools. Brief Bioinform. 2013;14:144–161. doi: 10.1093/bib/bbs038. PM:22908213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak JM, Carroll FI, Schneider JS. [125I]Iodomethyllycaconitine Binds to Alpha7 Nicotinic Acetylcholine Receptors in Monkey Brain. Eur J Neurosci. 2006;23:2604–2610. doi: 10.1111/j.1460-9568.2006.04804.x. PM:16817863. [DOI] [PubMed] [Google Scholar]
- Kuryatov A, Berrettini W, Lindstrom J. Acetylcholine Receptor (AChR) Alpha5 Subunit Variant Associated With Risk for Nicotine Dependence and Lung Cancer Reduces (Alpha4beta2)Alpha5 AChR Function. Mol Pharmacol. 2011;79:119–125. doi: 10.1124/mol.110.066357. PM:20881005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuryatov A, Luo J, Cooper J, Lindstrom J. Nicotine Acts As a Pharmacological Chaperone to Up-Regulate Human Alpha4beta2 Acetylcholine Receptors. Mol Pharmacol. 2005;68:1839–1851. doi: 10.1124/mol.105.012419. PM:16183856. [DOI] [PubMed] [Google Scholar]
- Kuryatov A, Onksen J, Lindstrom J. Roles of Accessory Subunits in Alpha4beta2(*) Nicotinic Receptors. Mol Pharmacol. 2008;74:132–143. doi: 10.1124/mol.108.046789. PM:18381563. [DOI] [PubMed] [Google Scholar]
- Le NN, Changeux JP. Molecular Evolution of the Nicotinic Acetylcholine Receptor: an Example of Multigene Family in Excitable Cells. J Mol Evol. 1995;40:155–172. doi: 10.1007/BF00167110. PM:7699721. [DOI] [PubMed] [Google Scholar]
- Lips EH, Gaborieau V, McKay JD, Chabrier A, Hung RJ, Boffetta P, Hashibe M, Zaridze D, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Field JK, Liloglou T, Xinarianos G, McLaughlin J, Liu G, Skorpen F, Elvestad MB, Hveem K, Vatten L, Study E, Benhamou S, Lagiou P, Holcatova I, Merletti F, Kjaerheim K, Agudo A, Castellsague X, Macfarlane TV, Barzan L, Canova C, Lowry R, Conway DI, Znaor A, Healy C, Curado MP, Koifman S, Eluf-Neto J, Matos E, Menezes A, Fernandez L, Metspalu A, Heath S, Lathrop M, Brennan P. Association Between a 15q25 Gene Variant, Smoking Quantity and Tobacco-Related Cancers Among 17 000 Individuals. Int J Epidemiol. 2010;39:563–577. doi: 10.1093/ije/dyp288. PM:19776245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Archidiacono N, Misceo D, Cardone MF, Deschamps S, Roe B, Rocchi M, Eichler EE. Refinement of a Chimpanzee Pericentric Inversion Breakpoint to a Segmental Duplication Cluster. Genome Biol. 2003;4:R50. doi: 10.1186/gb-2003-4-8-r50. PM:12914658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas RJ, Changeux JP, le N, Albuquerque EX, Balfour DJ, Berg DK, Bertrand D, Chiappinelli VA, Clarke PB, Collins AC, Dani JA, Grady SR, Kellar KJ, Lindstrom JM, Marks MJ, Quik M, Taylor PW, Wonnacott S. International Union of Pharmacology. XX. Current Status of the Nomenclature for Nicotinic Acetylcholine Receptors and Their Subunits. Pharmacol Rev. 1999;51:397–401. PM:10353988. [PubMed] [Google Scholar]
- Macqueen DA, Heckman BW, Blank MD, Janse Van RK, Park JY, Drobes DJ, Evans DE. Variation in the Alpha 5 Nicotinic Acetylcholine Receptor Subunit Gene Predicts Cigarette Smoking Intensity As a Function of Nicotine Content. Pharmacogenomics J. 2014;14:70–76. doi: 10.1038/tpj.2012.50. PM:23358500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta SG, Balfour DJ, Benowitz NL, Boyd RT, Buccafusco JJ, Caggiula AR, Craig CR, Collins AC, Damaj MI, Donny EC, Gardiner PS, Grady SR, Heberlein U, Leonard SS, Levin ED, Lukas RJ, Markou A, Marks MJ, McCallum SE, Parameswaran N, Perkins KA, Picciotto MR, Quik M, Rose JE, Rothenfluh A, Schafer WR, Stolerman IP, Tyndale RF, Wehner JM, Zirger JM. Guidelines on Nicotine Dose Selection for in Vivo Research. Psychopharmacology (Berl) 2007;190:269–319. doi: 10.1007/s00213-006-0441-0. PM:16896961. [DOI] [PubMed] [Google Scholar]
- McCallum SE, Parameswaran N, Bordia T, Fan H, McIntosh JM, Quik M. Differential Regulation of Mesolimbic Alpha 3/Alpha 6 Beta 2 and Alpha 4 Beta 2 Nicotinic Acetylcholine Receptor Sites and Function After Long-Term Oral Nicotine to Monkeys. J Pharmacol Exp Ther. 2006a;318:381–388. doi: 10.1124/jpet.106.104414. PM:16622038. [DOI] [PubMed] [Google Scholar]
- McCallum SE, Parameswaran N, Bordia T, Fan H, Tyndale RF, Langston JW, McIntosh JM, Quik M. Increases in Alpha4* but Not Alpha3*/Alpha6* Nicotinic Receptor Sites and Function in the Primate Striatum Following Chronic Oral Nicotine Treatment. J Neurochem. 2006b;96:1028–1041. doi: 10.1111/j.1471-4159.2005.03646.x. PM:16412091. [DOI] [PubMed] [Google Scholar]
- Mukhin AG, Kimes AS, Chefer SI, Matochik JA, Contoreggi CS, Horti AG, Vaupel DB, Pavlova O, Stein EA. Greater Nicotinic Acetylcholine Receptor Density in Smokers Than in Nonsmokers: a PET Study With 2-18F-FA-85380. J Nucl Med. 2008;49:1628–1635. doi: 10.2967/jnumed.108.050716. PM:18794265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafo MR, Timofeeva MN, Morris RW, Prieto-Merino D, Sattar N, Brennan P, Johnstone EC, Relton C, Johnson PC, Walther D, Whincup PH, Casas JP, Uhl GR, Vineis P, Padmanabhan S, Jefferis BJ, Amuzu A, Riboli E, Upton MN, Aveyard P, Ebrahim S, Hingorani AD, Watt G, Palmer TM, Timpson NJ, Davey SG. Association Between Genetic Variants on Chromosome 15q25 Locus and Objective Measures of Tobacco Exposure. J Natl Cancer Inst. 2012;104:740–748. doi: 10.1093/jnci/djs191. PM:22534784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dell LE, Chen SA, Smith RT, Specio SE, Balster RL, Paterson NE, Markou A, Zorrilla EP, Koob GF. Extended Access to Nicotine Self-Administration Leads to Dependence: Circadian Measures, Withdrawal Measures, and Extinction Behavior in Rats. J Pharmacol Exp Ther. 2007;320:180–193. doi: 10.1124/jpet.106.105270. PM:17050784. [DOI] [PubMed] [Google Scholar]
- Paliwal A, Vaissiere T, Krais A, Cuenin C, Cros MP, Zaridze D, Moukeria A, Boffetta P, Hainaut P, Brennan P, Herceg Z. Aberrant DNA Methylation Links Cancer Susceptibility Locus 15q25.1 to Apoptotic Regulation and Lung Cancer. Cancer Res. 2010;70:2779–2788. doi: 10.1158/0008-5472.CAN-09-4550. PM:20332232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papke RL. Merging Old and New Perspectives on Nicotinic Acetylcholine Receptors. Biochem Pharmacol. 2014;89:1–11. doi: 10.1016/j.bcp.2014.01.029. PM:24486571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plazas PV, Katz E, Gomez-Casati ME, Bouzat C, Elgoyhen AB. Stoichiometry of the Alpha9alpha10 Nicotinic Cholinergic Receptor. J Neurosci. 2005;25:10905–10912. doi: 10.1523/JNEUROSCI.3805-05.2005. PM:16306403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JF. Risk in CNS Drug Discovery: Focus on Treatment of Alzheimer's Disease. BMC Neurosci. 2008;9(Suppl 3):S1. doi: 10.1186/1471-2202-9-S3-S1. PM:19090998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quik M, Polonskaya Y, Gillespie A, Jakowec M, Lloyd GK, Langston JW. Localization of Nicotinic Receptor Subunit MRNAs in Monkey Brain by in Situ Hybridization. J Comp Neurol. 2000;425:58–69. doi: 10.1002/1096-9861(20000911)425:1<58::aid-cne6>3.0.co;2-x. PM:10940942. [DOI] [PubMed] [Google Scholar]
- Russell MA, Jarvis M, Iyer R, Feyerabend C. Relation of Nicotine Yield of Cigarettes to Blood Nicotine Concentrations in Smokers. Br Med J. 1980;280:972–976. doi: 10.1136/bmj.280.6219.972. PM:7417765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, Giegling I, Han S, Han Y, Keskitalo-Vuokko K, Kong X, Landi MT, Ma JZ, Short SE, Stephens SH, Stevens VL, Sun L, Wang Y, Wenzlaff AS, Aggen SH, Breslau N, Broderick P, Chatterjee N, Chen J, Heath AC, Heliovaara M, Hoft NR, Hunter DJ, Jensen MK, Martin NG, Montgomery GW, Niu T, Payne TJ, Peltonen L, Pergadia ML, Rice JP, Sherva R, Spitz MR, Sun J, Wang JC, Weiss RB, Wheeler W, Witt SH, Yang BZ, Caporaso NE, Ehringer MA, Eisen T, Gapstur SM, Gelernter J, Houlston R, Kaprio J, Kendler KS, Kraft P, Leppert MF, Li MD, Madden PA, Nothen MM, Pillai S, Rietschel M, Rujescu D, Schwartz A, Amos CI, Bierut LJ. Multiple Independent Loci at Chromosome 15q25.1 Affect Smoking Quantity: a Meta-Analysis and Comparison With Lung Cancer and COPD. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001053. PM:20700436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf DB, Sarkisyan N, Jacobsson H, Claus R, Bermejo JL, Peil B, Gu L, Muley T, Meister M, Dienemann H, Plass C, Risch A. Epigenetic Screen Identifies Genotype-Specific Promoter DNA Methylation and Oncogenic Potential of CHRNB4. Onc. 2012 doi: 10.1038/onc.2012.344. PM:22945651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker AK, Markou A. Unraveling the Neurobiology of Nicotine Dependence Using Genetically Engineered Mice. Curr Opin Neurobiol. 2013;23:493–499. doi: 10.1016/j.conb.2013.02.013. PM:23545467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes C, Papke RL. Use of an Alpha3beta4 Nicotinic Acetylcholine Receptor Subunit Concatamer to Characterize Ganglionic Receptor Subtypes With Specific Subunit Composition Reveals Species-Specific Pharmacologic Properties. Neuropharmacology. 2012;63:538–546. doi: 10.1016/j.neuropharm.2012.04.035. PM:22580377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street SL, Kyes RC, Grant R, Ferguson B. Single Nucleotide Polymorphisms (SNPs) Are Highly Conserved in Rhesus (Macaca Mulatta) and Cynomolgus (Macaca Fascicularis) Macaques. BMC Genomics. 2007;8:480. doi: 10.1186/1471-2164-8-480. PM:18166133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammimaki A, Herder P, Li P, Esch C, Laughlin JR, Akk G, Stitzel JA. Impact of Human D398N Single Nucleotide Polymorphism on Intracellular Calcium Response Mediated by Alpha3beta4alpha5 Nicotinic Acetylcholine Receptors. Neuropharmacology. 2012;63:1002–1011. doi: 10.1016/j.neuropharm.2012.07.022. PM:22820273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Lester HA, Lummis SC. The Structural Basis of Function in Cys-Loop Receptors. Q Rev Biophys. 2010;43:449–499. doi: 10.1017/S0033583510000168. PM:20849671. [DOI] [PubMed] [Google Scholar]
- Tobacco and Genetics Consortium. Genome-Wide Meta-Analyses Identify Multiple Loci Associated With Smoking Behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. PM:20418890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian JA, Green HL, Young JE, Majerksy LS, Thomas BW, Shively CA, Tobin JR, Nader MA, Grant KA. Induction and Maintenance of Ethanol Self-Administration in Cynomolgus Monkeys (Macaca Fascicularis): Long-Term Characterization of Sex and Individual Differences. Alcohol Clin Exp Res. 2001;25:1087–1097. PM:11505038. [PubMed] [Google Scholar]
- Wallis JD. Cross-Species Studies of Orbitofrontal Cortex and Value-Based Decision-Making. Nat Neurosci. 2012;15:13–19. doi: 10.1038/nn.2956. PM:22101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RB, Baker TB, Cannon DS, von NA, Dunn DM, Matsunami N, Singh NA, Baird L, Coon H, McMahon WM, Piper ME, Fiore MC, Scholand MB, Connett JE, Kanner RE, Gahring LC, Rogers SW, Hoidal JR, Leppert MF. A Candidate Gene Approach Identifies the CHRNA5-A3-B4 Region As a Risk Factor for Age-Dependent Nicotine Addiction. PLoS Genet. 2008;4:e1000125. doi: 10.1371/journal.pgen.1000125. PM:18618000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland JR, Hampe M, Newman TK, Syagailo Y, Meyer J, Schempp W, Timme A, Suomi SJ, Lesch KP. Structural Variation of the Monoamine Oxidase A Gene Promoter Repeat Polymorphism in Nonhuman Primates. Genes Brain Behav. 2006;5:40–45. doi: 10.1111/j.1601-183X.2005.00130.x. PM:16436187. [DOI] [PubMed] [Google Scholar]
- Xie P, Kranzler HR, Zhang H, Oslin D, Anton RF, Farrer LA, Gelernter J. Childhood Adversity Increases Risk for Nicotine Dependence and Interacts With Alpha5 Nicotinic Acetylcholine Receptor Genotype Specifically in Males. Neuropsychopharmacology. 2012;37:669–676. doi: 10.1038/npp.2011.240. PM:22012472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Zhou Z, Lindell SG, Higley JD, Ferguson B, Thompson RC, Lopez JF, Suomi SJ, Baghal B, Baker M, Mash DC, Barr CS, Goldman D. The Rhesus Macaque Is Three Times As Diverse but More Closely Equivalent in Damaging Coding Variation As Compared to the Human. BMC Genet. 2012;13:52. doi: 10.1186/1471-2156-13-52. PM:22747632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang F, Kranzler HR, Zhao H, Gelernter J. Profiling of Childhood Adversity-Associated DNA Methylation Changes in Alcoholic Patients and Healthy Controls. PLoS ONE. 2013;8:e65648. doi: 10.1371/journal.pone.0065648. PM:23799031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Goodsell J, Norgren RB., Jr. Limitations of the Rhesus Macaque Draft Genome Assembly and Annotation. BMC Genomics. 2012;13:206. doi: 10.1186/1471-2164-13-206. PM:22646658. [DOI] [PMC free article] [PubMed] [Google Scholar]