Abstract
Seed storage proteins comprise a major part of the protein content of the seed and have an important role on the quality of the seed. These storage proteins are important because they determine the total protein content and have an effect on the nutritional quality and functional properties for food processing. Transgenic plants are being used to develop improved lines for incorporation into plant breeding programs and the nutrient composition of seeds is a major target of molecular breeding programs. Hence, classification of these proteins is crucial for the development of superior varieties with improved nutritional quality. In this study we have applied machine learning algorithms for classification of seed storage proteins. We have presented an algorithm based on nearest neighbor approach for classification of seed storage proteins and compared its performance with decision tree J48, multilayer perceptron neural (MLP) network and support vector machine (SVM) libSVM. The model based on our algorithm has been able to give higher classification accuracy in comparison to the other methods.
Keywords: Classification, Nearest neighbour algorithm, Correlation based feature selection, Machine learning, Seed storage proteins, Bio-informatics
Introduction
A seed storage protein is defined as a protein which is found only in seeds, where they accumulate in large quantities and can be hydrolyzed to release its constituent amino acids. These acids are used as a source for reduced nitrogen by the seedling which is essential for germination and early growth of seedling (Spencer and Boulter 1984). Humans and livestock obtain a major fraction of the total protein from the seeds of crop plants. Composition of storage proteins determines the quality of proteins which is very important from the nutritional aspect. For instance, high quality cereals are characterized by the quality of proteins of the grains which are yet determined by the composition of proteins. These storage proteins determine not only the total protein content of the seed but also its quality for various end uses. In cereals, about 50 % of the total protein in mature grains is comprised of storage proteins and thus have an important role on nutritional quality for humans and livestock and on functional properties in food processing (Shewry and Halford 2002). In the case of wheat, the storage proteins from the gluten fraction are important, whose properties are largely responsible for the ability to use wheat flour to make bread and other products (Shewry and Halford 2002). Osborne (1924) classified the storage proteins into groups on the basis of their extraction and solubility in water (albumins), dilute saline (globulins), alcohol ether mixtures (prolamins), and dilute acid or alkali (glutelins).
A detailed understanding of storage protein structure and diversity is an important prerequisite for attempts to manipulate quality because it indicates the extent to which the composition of the proteins can be maneuvered without affecting their biological properties (Shewry and Halford 2002). Transgenic plants are being used to develop improved lines for incorporation into plant breeding programs. Improving the nutrient composition of seeds is a major target of molecular breeding and much of the recent work on seed storage proteins was performed to provide a basis for improving the nutritional and processing properties of crops using genetic engineering programs (Mandal and Mandal 2000, Kawakatsu et al. 2010). Hence, classification of seed storage proteins is crucial for development of superior varieties with improved nutritional quality. Any effort to characterize and classify these storage proteins would help in augmenting breeding efforts for the development of improved cereal varieties with better nutritional quality (Marla et al. 2010). Traditionally seed storage proteins have been classified on the basis of a few characteristics including solubility (Osborne 1924). However with the advances in laboratory techniques like throughput sequencing, mass spectrometry etc. there has been a surge in the production of protein information. This has necessitated the accurate annotation, classification, characterization and deciphering of the biological function of these sequences.
Accurate classification of proteins into different functional and structural classes is an important task in computational biology. Many methods of function prediction rely on identifying similarity in sequence and/or structure between a protein of unknown function and one or more well-understood proteins while alternative methods include inferring conservation patterns in members of a functionally uncharacterized family for which many sequences and structures are known (Whisstock and Lesk 2003). However, as pointed by them, these inferences are tenuous and provide reasonable guesses at function, but are far from foolproof. This problem can be overcome by predicting the function from features based on protein sequence and structure using classification models. Machine learning approaches have been used for building classification models for protein prediction in a number of studies. They have been found to be useful in protein/enzyme classification, protein structural classification, sub cellular localization of proteins and identification of functionally important sites in proteins. Various machine learning algorithms like neural networks, support vector machines, decision trees and the nearest neighbor based machine learning algorithms have been employed in protein classification studies.
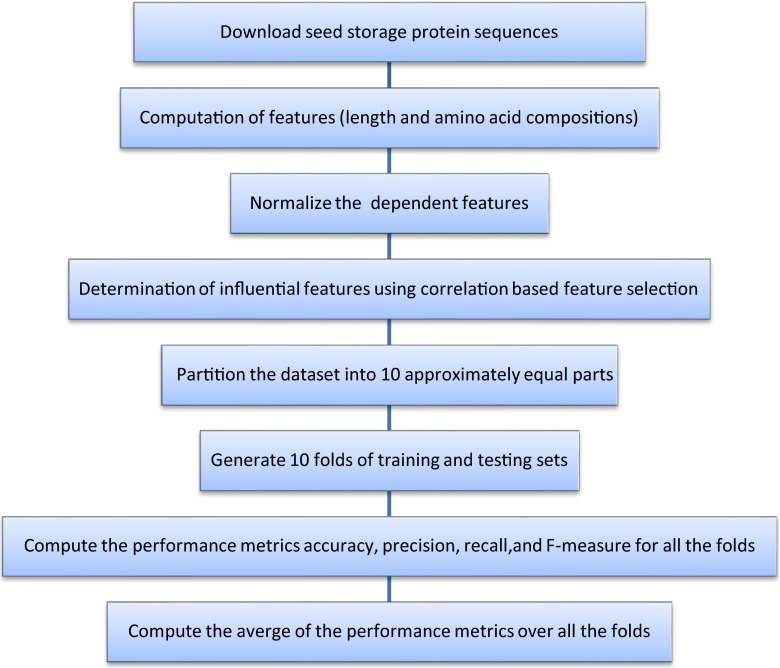
In a recent study, feed forward neural networks using back propagation algorithm have been employed for classification of seed storage proteins in rice into four classes - albumins, globulins, glutelins and prolamins (Marla et al. 2010). The authors have used multi layer perceptron (MLP) neural networks to classify the rice seed storage proteins. In our study we have attempted to classify the seed storage proteins in rice, wheat, maize, castor bean and thale cress using an algorithm (Fig 1.) and compared the performance of this algorithm with the following machine learning algorithms viz. J48 (decision tree) (Quinlan 1993), MLP neural network and LibSVM support vector machine (Chang and Lin 2011). The objectives of our work are as follows:
To identify and extract a set of features from protein sequences, that would help in the classification of the proteins into their respective seed storage classes;
To obtain a reduced set of features that would enable classification of seed storage proteins with better accuracy;
To develop a classification model for predicting the storage class of the proteins with higher accuracy.
Fig. 1.
Flowchart for NM algorithm
In this paper we have presented an algorithm based on correlation based feature selection (CFS) algorithm (Hall 1999) to classify the seed storage proteins.
Material and methods
Development of a new algorithm for the classification of seed storage class
An algorithm has been proposed for the classification of seed storage proteins (Fig. 1). The amino acid compositions and sequence length of the seed storage proteins have been used for representing the features to be input for classification models. The most important features have been ranked based on correlation based feature selection algorithm and a similarity measure based on Euclidean distance has been adopted for developing a classification model.
Source of data and input vector representation
We have conducted the experiments on the seed storage protein datasets of rice (Rice Annotation Project et al. 2007), wheat (Anderson et al. 2013), maize (Schnable et al. 2009; Tenaillon et al. 2001; Hilton and Gaut 1998), castor bean (Rivarola et al. 2011) and thale cress (Swarbreck et al. 2007) available in NCBI database (http://www.ncbi.nlm.nih.gov) (NCBI Resource coordinators 2013). Seed storage protein sequences of rice, wheat, maize, castor bean and thale cress were downloaded in fasta format. A protein sequence is represented as a chain of amino acids. There are 20 amino acids and a protein P is represented as P = X1X2 … Xn, where Xi’s are amino acids. For instance ‘MKIIFFFALLAIAACSASAQFDAVTQVYRQY’ represents a protein sequence where,M, K, I … are the different amino acids.
For any machine learning problem, input vectors presented to a learning algorithm is very crucial for the success of the learning process. In our study we have taken the length of the sequence and amino acid composition of the protein, as the input features for the classification models. Length of the sequence and amino acid composition is computed by the formula given in step 2 of our algorithm (Fig. 1).
The datasets of rice, wheat, maize, castor bean and thale cress consisted of 49, 72, 67, 39 and 196 records for sequences of seed storage proteins respectively. Twenty one features of seed storage proteins including length of sequence and composition of the 20 amino acids have been included as input features while seed storage type has been taken as the class attribute. The classification models were tested on all the five datasets.
Feature selection based on CFS algorithm
Various filter, wrapper and embedded methods have been utilized for selecting the best features which can classify the input sequences accurately. We have used CFS method based on Best-first search (BFS) algorithm for selecting the input features which can best classify the different seed storage proteins. The CFS algorithm selects the features having high correlation with the class but which are un-correlated among themselves.
| NM Algorithm |
| Dataset description Consider the dataset of sequences of seed storage proteins, P = X1X2…Xr, where X1, X 2,…,Xr are amino acids. (1) For each protein P compute the following attributes: Length = Count of all X 'i s in P (2) Let {S(i, j): 1 ≤ i ≤ m, 1 ≤ j ≤ n} denote the set of all proteins, where S (i, j) = jth attribute of the ith protein, where m and n denote the number of proteins and attributes respectively. There are 21independent attributes in our study, hence n = 21. Let S (i, 22) denote the seed storage class of the protein (See Table 5 below for a sample dataset). Algorithm (3) Normalize the dependent attributes using the formulae; (4) Determine the most influential features using correlation based feature selection algorithm in the following manner • The features should be correlated with the class attribute • They should not be correlated among themselves (5) Generate the test and train sets using k-fold cross validation as follows: Define S = UQi as a random partition of S into k approximately equal parts. For i = 1 to k, let Qi be the testing set and the remaining parts be the training set. (6) For each training and testing dataset generated in the above step, classification model is constructed as follows: • For each member of testing set, its distance from the members of training set is calculated using the similarity index as follows: where, “x” is a member of testing set and “y” is a member of training set. • The class of the training instance closest to the given test instance based on the above similarity index, is assigned to the test instance. • Obtain the performance metrics accuracy (ACC), precision (p), recall (r) and F-measure (F), based on the predicted and actual classes of the test instances, using the formulae: where, TP is the number of true positives, TN is the number of true negatives, FP is the number of false positives and FN is the number of false negatives. (7) Compute the average of the performance metrics over all the training and testing data sets. The values of accuracy, precision, recall and F-measure are measures of the goodness of fit of this model to the data. Hence higher measures of accuracy (close to 100 %) and precision, recall and F-measures (close to 1) indicate the suitability of the above model for the classification of seed storage proteins into its respective classes. |
Generation of training and testing sets
The k-fold cross-validation method has been used for generating training and test sets for the classification methods. In k-fold cross-validation, the original sample is randomly partitioned into k subsamples. Of the k subsamples, k − 1 subsamples are used as training data and a single subsample is retained as the validation data for testing the model. The cross-validation process is then repeated k times (the folds), with each of the k subsamples used exactly once as the validation data. The k results from the folds then can be averaged (or otherwise combined) to produce a single estimation. The advantage of this method over repeated random sub-sampling is that all observations are used for both training and validation, and each observation is used for validation exactly once. We have employed 10-fold cross-validation in our study.
Performance evaluation
The performance of the various classification models were measured using the measures of accuracy (ACC), precision (p) and recall (r), and F-measure. Accuracy measures the degree of closeness between observed and true classes. Precision is the fraction of retrieved instances that are relevant, while recall is the fraction of relevant instances that are retrieved. F-measure is the harmonic mean of precision and recall. A high value of F is an indication of higher values of both precision and recall. These measures are defined as follows:
where, TP is the number of true positives, TN is the number of true negatives, FP is the number of false positives and FN is the number of false negatives.
The performance metrics of the models have been tested for statistical significance by Wilcoxon test (Frank 1945).
Experiments and results
We have studied the performance of the classification models achieved by NM on the five datasets of rice, wheat, maize, castor bean and thale cress. The lengths of the protein sequences and their amino acid compositions have been taken as input features. The features identified by the CFS method have been taken for further computations while the other features have been deleted from the data file. The important features selected by CFS have been listed in Table 1. The numbers of features have been reduced from 21 to 8, 7, 8, 15 and 7 in castor bean, maize, rice, thale cress and wheat respectively. There has been a considerable reduction in the number of features after performing feature selection by CFS.
Table 1.
Influential features identified by CFS method
| Data set | # Original features | # Influential features | Features identified | Accuracy (%) |
|---|---|---|---|---|
| Castor bean | 21 | 8 | Length, A, C, E, G, P, R, T | 82.1 |
| Maize | 21 | 7 | Length, C, D, G, P, R, V | 97 |
| Rice | 21 | 8 | Length, A, D, E, L, P, Q, R | 91.8 |
| Thale Cress | 21 | 15 | Length, A, C, E, F, G, H, I, M, Q, R, S, T, V, W | 91.3 |
| Wheat | 21 | 7 | Length, E, G, M, P, Q, R | 98.6 |
The 10-fold cross-validation method has been used for generating training and test sets for the five seed storage datasets. Each dataset was divided into 10 approximately equal parts and used for subsequent calculations.
The classification measures viz. accuracy, precision; recall and F-measures have been computed (Table 2). Highest classification accuracy has been achieved in wheat (98.6 %), followed by maize (97 %), rice (91.8 %), thale cress (91.3 %) and castor bean (82.1 %). The other measures precision, recall and F-measure have also observed to be in the same order in these crops (Table 2).
Table 2.
Performance of NM, J48, MLP and LibSVM on the five seed storage sequence datasets
| Dataset | Measures | NM | J48 | MLP | LibSVM |
|---|---|---|---|---|---|
| Castor bean (39) | Accuracy (%) | 82.1 | 76.9 | 79.5 | 64.1 |
| Precision | 0.825 | 0.771 | 0.824 | 0.557 | |
| Recall | 0.821 | 0.769 | 0.795 | 0.641 | |
| F-measure | 0.821 | 0.769 | 0.807 | 0.578 | |
| Maize (67) | Accuracy (%) | 97 | 85.1 | 92.5 | 74.6 |
| Precision | 0.97 | 0.849 | 0.927 | 0.757 | |
| Recall | 0.97 | 0.851 | 0.925 | 0.746 | |
| F-measure | 0.97 | 0.849 | 0.926 | 0.71 | |
| Rice (49) | Accuracy (%) | 91.8 | 89.8 | 89.8 | 69.4 |
| Precision | 0.918 | 0.879 | 0.9 | 0.629 | |
| Recall | 0.918 | 0.898 | 0.898 | 0.694 | |
| F-measure | 0.913 | 0.888 | 0.895 | 0.644 | |
| Thale Cress (196) | Accuracy (%) | 91.3 | 89.3 | 89.3 | 71.4 |
| Precision | 0.913 | 0.89 | 0.897 | 0.675 | |
| Recall | 0.913 | 0.893 | 0.893 | 0.714 | |
| F-measure | 0.913 | 0.891 | 0.894 | 0.687 | |
| Wheat (72) | Accuracy (%) | 98.6 | 93.1 | 95.8 | 91.7 |
| Precision | 0.986 | 0.934 | 0.961 | 0.924 | |
| Recall | 0.986 | 0.931 | 0.958 | 0.917 | |
| F-measure | 0.986 | 0.932 | 0.959 | 0.901 |
We have conducted experiments to compare the performance of NM with J48, MLP and LibSVM classification models on the five datasets mentioned above. The J48, MLP and LibSVM classification models have been implemented in Weka for Windows which is a popular open source software consisting of state-of-the-art algorithms for pre-processing, feature selection, clustering, classification, regression and association of data (Hall et al. 2009).
J48 is an implementation of the decision tree algorithm C4.5, which is found in Weka (Hall et al. 2009). C4.5 builds decision trees from a set of training data using the concept of information entropy. At each node of the tree, C4.5 chooses the attribute of the data that most effectively splits its set of samples into subsets enriched in one class or the other. The splitting criterion is the normalized information gain (difference in entropy) (Quinlan 1993). The attribute with the highest normalized information gain is chosen to make the decision. Multilayer perceptron is a feed forward artificial neural network. It consists of 3 or more layers of nodes (including an input layer, output layer and one or more hidden layers) and each node is associated with a nonlinear activation function. The supervised learning technique back propagation is used for training the network. We have used the MLP neural network with learning rate = 0.3, and momentum = 0.2. Support vector machine is a supervised learning classifier. It constructs a hyper plane in a high-dimensional space which is used for classification. A good separation is achieved by the hyper plane that has the largest distance to the nearest training data point of any class. We have classified the data using C-SVC type of LibSVM with radial basis function exp(−gamma*|u-v|^2) as kernel (Chan and Lin 2011).
The performances of NM, J48, LibSVM and MLP have been compared on the five datasets and the classification measures viz. accuracy, precision; recall and F-measure have been presented (Table 2). The NM algorithm has achieved higher accuracy in all the five datasets followed by MLP, J48 and LibSVM.
The Wilcoxon matched-pairs rank sum test has been used to compare the accuracies of NM with that of the other three algorithms over the five datasets (Table 3). On the basis of the p-values it can be can be observed that NM has performed better than the other algorithms in 3 datasets namely that of wheat, castor bean and thale cress. In the case of rice and maize datasets, it is better than all the others except for MLP. In these two datasets, accuracies of NM are not significantly different from those of MLP.
Table 3.
Comparison of the performance of NM with other methodologies using Wilcoxon matched-pairs rank sum test
| Dataset | Method | Rank sum (+, −) | p-value |
|---|---|---|---|
| Castor bean | J48 | 36.0, 0.0 | 0.008 |
| MLP | 52.5, 2.5 | 0.005 | |
| LibSVM | 55, 0.0 | 0.002 | |
| Maize | J48 | 45.0, 0.0 | 0.004 |
| MLP | 13.0, 23.0 | 0.32 | |
| LibSVM | 55.0, 0.0 | 0.002 | |
| Rice | J48 | 43.5, 1.5 | 0.006 |
| MLP | 27.0, 18.0 | 0.367 | |
| LibSVM | 55.0, 0.0 | 0.002 | |
| Thale Cress | J48 | 55.0, 0.0 | 0.002 |
| MLP | 45.0, 0.0 | 0.004 | |
| LibSVM | 55.0, 0.0 | 0.002 | |
| Wheat | J48 | 45.0, 0.0 | 0.004 |
| MLP | 21.0, 0.0 | 0.015 | |
| LibSVM | 24.0, 4.0 | 0.04 |
A Case study for assessing the performance of NM on the Arabidopsis dataset
We have attempted to explain the methodology in assessing the performance of the classification model using NM, which is based on the nearest neighbour approach, for the classification of seed storage proteins of the Arabidopsis project (Swarbreck et al. 2007). This project is an assembly project of the plant Arabidopsis thaliana which is widely used as a model for other plants for the study of a variety of fundamental biological processes (Swarbreck et al. 2007). The various steps involved in this method are explained, which are in line with the NM algorithm (Fig 1).
-
Downloading the sequences of Arabidopsis:
The fasta sequences of seed storage proteins (globulin, glutelin, prolamin and albumin) of the Arabidopsis were downloaded from National Centre for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/) by searching the protein database of NCBI using the search terms globulin, glutelin, prolamin and albumin. All the sequences were collected in a text file (Table 4). The identical sequences were identified and only one copy of the sequence was retained.
-
Calculating the amino acid compositions and length of the sequences:
The length of a sequence is defined as the number of amino acids in the sequences. For instance length of the sequence ‘AAMPQ’ is 5, that of the sequence ‘MGSGMIRTLVILAIAL’ is 16 and so on. The amino acid compositions of the sequences were calculated by dividing the total number of occurrences of an amino acid by the length of the sequence (Table 5). For example the composition of the amino acid M in the sequence MGSGMIRTLVILAIAL is 2/16 = 0.125.
-
Normalization of the attributes:
All the attributes are normalized according to the formula in step 4 of our algorithm (Fig 1.). The normalized values lie between 0 and 1. Table 6 shows the normalized values of the Arabidopsis dataset.
-
Attribute selection:
The most influential features were determined using correlation based feature selection algorithm. This was obtained using the supervised attribute filter cfsSubsetEval of Weka data mining software (Hall et al. 2009). For the Arabidopsis dataset, the length of the sequence and amino acid compositions of A, C, E, F, G, H, I, M, Q, R, S, T, V, W were found to be the influential attributes.
-
Generating training and testing sets:
A 10-fold cross validation method was used for developing the classification model. The data file was randomly divided into 10 approximately equal parts. 10 sets of training and testing datasets were obtained by choosing 9 parts as training dataset and 1 part as testing dataset. The Arabidopsis dataset, consisting of 196 records, was divided into 10 parts. 6 parts consisted of 20 records each and 4 parts consisted of 19 records each. An example of a partition of the Arabidopsis dataset into 10 parts is given in Table 6.
-
Calculating similarity index of members of testing set
For a given set of training and testing datasets, the distance of each member of the testing set is calculated. For example, if the tuple x belongs to the testing set and y belongs to the training set where,
x = (0.044755245, 0.787107946, 0.703349282, 0.24506579, 0.217105264, 0.597039472, 0, 0.425837319, 0.299825348, 0.187643021, 0.219298246, 0.183356895, 0.186409686, 0.372180452, 0)
y = (0.074825175, 0.327055353, 0.453966416, 0.593152868, 0.210191083, 0.157643312, 0.472399153, 0.876085695, 0.28968964, 0.181667129, 0.159235669, 0.355198768, 0.295288102, 0.315286625, 0.1507431)
The distance between x and y is computed using the definition
d(x,y) = [(0.44755245−0.074825175)^2 + (0.787107946−0.327055353)^2 + … + (0.221052633−0.32101911)^2]1/2 = √1.086000853 = 1.042113647
Distance of x was computed from all members of the training set. Minimum of all these distances was noted and the class of the instance of the training set corresponding to the minimum distance was designated as the predicted class of x. The classes of all the members of the testing set were determined in this manner.
-
Calculating the performance metrics
The metrics TP, FP, TN and FN were computed using the formulae described in NM algorithm (Fig 1). Taking the 1st part of Table 7 as testing set and the remaining parts as training set, the values for TP, FP, TN and FN were obtained for the albumin class as 14, 1, 5 and 0 respectively, where,- TP
- Number of albumin sequences of the testing set classified as albumin as per the similarity index with respect to the training set
- FP
- Number of non-albumin sequences of the testing set classified as albumin
- TN
- Number of non-albumin sequences of the testing set classified as non-albumin
- FN
- Number of albumin sequences of the testing set classified as non-albumin
The metrics TP, FP, TN and FN were computed for the remaining 10 cross-folds and the average of the values for TP, FP, TN and FN over all the folds were obtained as 13.17, 0.87, 4.73 and 0.83 respectively. Using the formulae (I – IV) for accuracy, precision, recall and F-measure described in the Performance Evaluation section of Materials and Methods, we obtained the values 91.33%, 0.93, 0.94 and 0.94 for accuracy, precision, recall and F-measure respectively.
The accuracy, precision, recall and f-values were calculated for the remaining seed storage classes in a similar way. The weighted averages of accuracy, precision, recall and F-measure values were obtained as 91.3%, 0.913, 0.913 and 0.913 respectively.
The accuracy, precision, recall and F-measures obtained by NM were higher compared to J48, MLP and LibSVM in the Arabidopsis (Table 1). Hence we can conclude that NM algorithm, based on the length and the amino acid compositions A, C, E, F, G, H, I, M, Q, R, S, T, V, W of the seed storage protein sequences, can be used for the classification of seed storage proteins in Arbidopsis.
Classification accuracy of 91.3% achieved by NM on the Arabidopsis dataset indicates that 179 sequences of the 196 seed storage proteins have been correctly classified by the NM algorithm. Precision of 0.913 has been achieved by NM which means that approximately 91% of the instances predicted as a certain type of seed storage protein are actually of that type. Precision and recall are equal which indicates that FP = FN. Since precision = recall, F-measure (which is the harmonic mean of precision and recall) is equal to precision and recall.
Table 4.
Text file containing fasta sequences of Arabidopsis seed storage proteins
| >gi|332660845|gb|AEE86245.1| bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein [Arabidopsis thaliana] |
| MGSGMIRTLVILAIALFMIGSDNVHVAKAQVCGANLSGLMNECQRYVSNAGPNSQPPSRSCCALIRPIDVPCACRYVSRDVTNYIDMDKVVYVARSCGKKIPSGYKCGSYTIPAA |
| >gi|332660844|gb|AEE86244.1| bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein [Arabidopsis thaliana] |
| MGSGMIRTLVILAIALFMIGSDNVHVAKAQVCGANLSGLMNECQRYVSNAGPNSQPPSRSCCALIRPIDVPCACRYVSRDVTNYIDMDKVVYVARSCGKKIPSGYKCGSKYLSCFSYYSFVIVKHIIIWI |
| >gi|332659909|gb|AEE85309.1| seed storage albumin 4 [Arabidopsis thaliana] |
| MANKLFLVCAALALCFILTNASVYRTVVEFDEDDASNPIGPIQKCQKEFQQDQHLRACQRWMRKQMWQGRGGGPSLDDEFDMEDDIENPQRRQLLQKCCSELRQEEPVCVCPTLRQAAKAVRFQGQQHQPEQVRKIYQAAKYLPNICKIQQVGVCPFQIPSIPSYY |
| >gi|332659908|gb|AEE85308.1| seed storage albumin 3 [Arabidopsis thaliana] |
| MANKLFLVCATLALCFLLTNASIYRTVVEFEEDDASNPVGPRQRCQKEFQQSQHLRACQRWMSKQMRQGRGGGPSLDDEFDFEGPQQGYQLLQQCCNELRQEEPVCVCPTLKQAARAVSLQGQHGPFQSRKIYQSAKYLPNICKIQQVGECPFQTTIPFFPPYY |
| >gi|332659907|gb|AEE85307.1| seed storage albumin 2 [Arabidopsis thaliana] |
| MANKLFLVCATFALCFLLTNASIYRTVVEFDEDDASNPMGPRQKCQKEFQQSQHLRACQKLMRMQMRQGRGGGPSLDDEFDLEDDIENPQGPQQGHQILQQCCSELRQEEPVCVCPTLRQAARAVSLQGQHGPFQSRKIYKTAKYLPNICKIQQVGECPFQTTIPFFPPY |
| >gi|332659906|gb|AEE85306.1| seed storage albumin 1 [Arabidopsis thaliana] |
| MANKLFLVCAALALCFLLTNASIYRTVVEFEEDDATNPIGPKMRKCRKEFQKEQHLRACQQLMLQQARQGRSDEFDFEDDMENPQGQQQEQQLFQQCCNELRQEEPDCVCPTLKQAAKAVRLQGQHQPMQVRKIYQTAKHLPNVCDIPQVDVCPFNIPSFPSFY |
Table 5.
Table containing features (length and amino acid compositions) of Arabidopsis seed storage protein sequences
| Sequence number | LEN | A | C | D | E | T | V | W | Y | CLASS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 114 | 0.14 | 0.079 | 0.044 | 0.026 | 0.044 | 0.053 | 0 | 0.018 | A |
| 2 | 96 | 0.125 | 0.083 | 0.021 | 0.042 | 0.052 | 0.115 | 0 | 0.01 | A |
| 3 | 119 | 0.076 | 0.059 | 0.008 | 0.042 | 0.025 | 0.059 | 0 | 0.008 | P |
| 4 | 95 | 0.168 | 0.084 | 0.032 | 0.032 | 0.042 | 0.084 | 0 | 0 | A |
| 5 | 82 | 0.11 | 0.061 | 0.049 | 0.037 | 0.049 | 0.098 | 0.012 | 0.012 | P |
| 192 | 151 | 0.079 | 0.046 | 0.073 | 0.066 | 0.066 | 0.06 | 0.007 | 0.033 | P |
| 193 | 151 | 0.053 | 0.04 | 0.073 | 0.066 | 0.066 | 0.053 | 0.007 | 0.033 | P |
| 194 | 157 | 0.064 | 0.051 | 0.076 | 0.064 | 0.064 | 0.045 | 0.006 | 0.025 | P |
| 195 | 172 | 0.064 | 0.035 | 0.047 | 0.093 | 0.029 | 0.058 | 0.006 | 0.041 | P |
| 196 | 149 | 0.054 | 0.047 | 0.054 | 0.067 | 0.034 | 0.067 | 0.007 | 0.04 | P |
Table 6.
Table containing normalized values of the features of Arabidopsis seed storage protein sequences
| Sequence number | LEN | A | C | D | E | T | V | W | Y | CLASS |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.04 | 0.79 | 0.7 | 0.57 | 0.25 | 0.19 | 0.37 | 0 | 0.22 | A |
| 2 | 0.03 | 0.69 | 0.74 | 0.27 | 0.39 | 0.23 | 0.81 | 0 | 0.13 | A |
| 3 | 0.05 | 0.4 | 0.52 | 0.11 | 0.39 | 0.08 | 0.42 | 0 | 0.11 | P |
| 4 | 0.03 | 0.96 | 0.75 | 0.41 | 0.29 | 0.18 | 0.6 | 0 | 0 | A |
| 5 | 0.02 | 0.6 | 0.54 | 0.64 | 0.34 | 0.21 | 0.69 | 0.29 | 0.15 | P |
| 192 | 0.08 | 0.34 | 0.43 | 0.79 | 0.51 | 0.15 | 0.51 | 0.14 | 0.23 | A |
| 193 | 0.07 | 0.42 | 0.41 | 0.95 | 0.62 | 0.31 | 0.42 | 0.16 | 0.42 | P |
| 194 | 0.07 | 0.26 | 0.35 | 0.95 | 0.62 | 0.31 | 0.37 | 0.16 | 0.42 | P |
| 195 | 0.07 | 0.33 | 0.45 | 1 | 0.59 | 0.3 | 0.32 | 0.15 | 0.32 | P |
| 196 | 0.09 | 0.33 | 0.31 | 0.61 | 0.87 | 0.11 | 0.41 | 0.14 | 0.51 | P |
Table 7.
A partition of the Arabidopsis dataset into 10 approximately equal parts
| Part No. | No. of records | Serial numbers of the records chosen |
|---|---|---|
| 1 | 20 | 8, 13, 18, 31, 33, 53, 61, 74, 96, 98, 108, 116, 118, 145, 158, 159, 162, 178,182, 189 |
| 2 | 20 | 7, 17, 20, 25, 36, 42, 56, 60, 77, 90, 103, 111, 114, 134, 135, 148, 149, 161, 174, 188 |
| 3 | 20 | 19, 32, 37, 57, 66, 70, 76, 78, 80, 84, 91, 119, 129, 130 137, 142, 150, 167, 171, 195 |
| 4 | 20 | 5, 11, 16, 49, 51, 59, 63, 68, 88, 95, 100, 122, 132, 138, 156, 164, 170, 173, 183, 184 |
| 5 | 20 | 6, 9, 21, 35, 62, 67, 75, 82, 94, 97, 107, 109, 115, 131, 140, 143, 146, 152, 179, 181 |
| 6 | 20 | 3, 12, 15, 30, 34, 41, 52, 55, 69, 81, 83, 92, 102, 104, 124, 144, 154, 163, 168, 175 |
| 7 | 19 | 24, 26, 39, 43, 44, 46, 54, 85, 89, 121, 126, 128, 133, 153, 155, 180, 186, 191, 194 |
| 8 | 19 | 10, 14, 23, 38, 45, 48, 71, 73, 105, 106, 112, 120, 123, 127, 141, 151, 166, 169, 196 |
| 9 | 19 | 1, 22, 28, 40, 50, 58, 65, 72, 79, 86, 93, 101, 110, 117, 125, 172, 177, 190, 192 |
| 10 | 19 | 2, 4, 27, 29, 47, 64, 87, 99, 113, 136, 139, 147, 157, 160, 165, 176, 185, 187, 193 |
Discussion and conclusion
In the present study few methodologies have been used to classify seed storage proteins of albumin, glutelin, globulin and prolamin using specific sequences available in public databases and the best classification method has been short listed. In this context, we have reviewed literature pertaining to improvement of a particular protein/nutrient in seed storage proteins with respect to transgenic plants. An attempt has been made to present a brief note on type of research being conducted elsewhere. Saalbach et al. (1988) attempted to improve the methionine content of seed proteins through transgenic approach. The promising strategy was to introduce genes encoding sulphur-rich and lysine-rich proteins under the control of an efficient promoter into legumes and cereals, respectively. Improvement of Methionine content of grain legumes could be achieved in the best transgenic lines especially soybean to the extent of 100 % of WHO standard for nutritionally balanced protiens (Müntz et al. 1998). S-rich 2S albumin gene from sunflower has been introduced (Rafiqul et al. 1996; Sharma et al. 1998) into the forage crop clover (Trifolium subterranean) resulted into expression of protein up to maximum of 0.3 % of the total extractable protein in mature leaves. Similarly in cereal breeding programmes, several thousand collections from wild germplasm were screened for high lysine and highproly lines in maize and barley. The basic objective of the programme was to reduce the prolamin content of the grains with concomitant increase in lysine content of the grains; however, the negative effect of increased lysine content was reduced starch content of the grain (Munck and Shewry 1992). However, this effect was alleviated by the transgenic approach by introducing beta phaseolin (6 % lysine) gene of common bean (Phaseolus vulgaris) in rice glutelin promoter and lysine was expressed to the level of 4 % of total endosperm protein in transgenic rice seeds (Zheng et al. 1995). In a study involving the production of transgenic rice for more lysine content than wild varieties by knocking down the genes resulted in a reduction of 13 kD prolamin levels (Kawakatsu et al. 2010, b). A study was conducted recently in which expression of genes pertaining to glucoronidase (gus A) reporter gene was enhanced by 3.12, 2.45 and 2.14 fold in stable transgenic rice lines (Li et al. 2012). Using RNA interference technology, silencing of genes responsible for seed storage proteins in soybean resulted in rebalanced protein composition preserving seed protein content without major collateral changes in metabolome or transcriptome (Schmidt et al. 2011).
In this study, we have developed a classification model based on nearest neighbor approach which can classify the seed storage protein sequences in wheat, castor bean, thale cress, rice and maize with greater accuracy.
The input features play a very important role in the development of a classification model. In various studies on protein classification, the amino acid compositions or di-peptide compositions or physico-chemical properties of the protein or different combinations of these three feature sets have been used as the input features. In our study we have taken length of the sequence and 20 amino acid compositions as the input features. Feature selection is an important step in data mining. It removes the redundant features, thus decreasing the time required for data file preparation and model development and improves the performance of the model. We have selected the important features utilizing the correlation based feature selection method (Hall 1999). The numbers of features have been reduced to less than 50 % in the case of the 4 datasets viz. that of castor bean, maize, rice and wheat. The features Len and R have been selected by CFS in all the five cases while A, C, E, G, P and Q have been selected in at least 3 datasets. As seed storage proteins are a reserve of nitrogen for the seedling, they are generally rich in asparagines (N), glutamine (Q) and arginine (R) or proline (P) (Higgins 1984). It can be observed that at least two of the proteins P, Q and R have been selected as influential features and this indicates that their composition has a role in determining the type of the seed storage protein.
All the measures viz. accuracy, precision, recall and F-measure are higher in the case of NM in comparison to the other algorithms for all the five datasets. By Wilcoxon signed-rank test we can observe that NM has performed better than the other algorithms in 3 datasets namely that of wheat, castor bean and thale cress. In the case of rice and maize datasets, it is better than all the others except for MLP. In these two datasets, accuracies of NM are not significantly different from those of MLP. This indicates that NM has achieved superior classification accuracy over the other algorithms
In a previous study of this kind, Marla et al. (2010) have shown that MLP neural network can be used for classification of seed storage proteins in rice. In comparison to the above study, we have studied the performance of various classification models in rice, wheat, maize, thale cress and castor bean. Hence this study is an extension of the previous work carried out by Marla et al. (2010). Moreover, Marla et al. (2010) has not compared the MLP neural network classification model with any other state-of the art methods. While we have compared the performance of NM with a few state-of-the-art methods like J48, MLP and LibSVM. In our study we have shown that nearest neighbor based classification has achieved the highest accuracy compared to J48, MLP and LibSVM in all the datasets employed in this study. Hence this algorithm can be used for classification of seed storage proteins in various plants.
This study is unique of its kind and has displayed that nearest neighbor approach is suitable for classification of seed storage proteins. The algorithm developed in present study will pave ways to classify seed storage proteins by in silico methods from the available sequences. The methodology proposed in these studies will help the breeders to screen large number of grain samples obtained either by plant breeding experiments or transgenic crops for assessing the quality of different seed storage proteins. It can also be extrapolated to transgenic plants reared for altering the proportions of seed storage proteins.
A further extension of this study would deal with addressing missing values in bioinformatics datasets, which arise due to various factors and pose a problem in the development of classification models.
Acknowledgments
The first author is thankful to Director, Indian Institute for Horticultural Research (IIHR) for providing facilities for carrying out the study. The research of the second author is supported by a grant from the Foundation for Scientific Research and Technological Innovation (FSRTI). The authors are thankful to Dr. M. Naresh Kumar, Head, Information Management Systems, National Remote Sensing Centre (ISRO), India, for his suggestions in improving this work. The authors would also like to thank the anonymous reviewers for many helpful suggestions which led to the improvement of this paper.
Contributor Information
V. Radhika, Email: radhika.vanama@gmail.com
V. Sree Hari Rao, Email: vshrao@gmail.com.
References
- Anderson OD, Huo N, Gu YQ. The gene space in wheat: the complete γ-gliadin gene family from the wheat cultivar Chinese Spring. Funct Integr Genomics. 2013;13(2):261–273. doi: 10.1007/s10142-013-0321-8. [DOI] [PubMed] [Google Scholar]
- Chang CC, Lin CJ. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol. 2011;2(27):1–27. doi: 10.1145/1961189.1961199. [DOI] [Google Scholar]
- Frank W. Individual comparisons by ranking methods. Biom Bull. 1945;1(6):80–83. doi: 10.2307/3001968. [DOI] [Google Scholar]
- Hall M (1999) Correlation-based Feature Selection for Machine Learning. http://www.cs.waikato.ac.nz/∽mhall/thesis.pdf
- Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: An update. SIGKDD Explor. 2009;11(1):10–18. doi: 10.1145/1656274.1656278. [DOI] [Google Scholar]
- Hilton H, Gaut BS. Speciation and domestication in maize and its wild relatives: evidence from the globulin-1 gene. Genetics. 1998;150(2):863–872. doi: 10.1093/genetics/150.2.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Hirose S, Yasuda H, Takaiwa F. Reducing rice seed storage protein accumulation leads to changes in nutrient quality and storage organelle formation. Plant Physiol. 2010;154:1842–1854. doi: 10.1104/pp.110.164343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Hirose S, Yasuda H, Takaiwa F (2010) Reducing Rice Seed Storage Protein Accumulation Leads to Changes in Nutrient Quality and Storage Organelle Formation, [DOI] [PMC free article] [PubMed]
- Li WJ, Dai LL, Chai ZJ, Yin ZJ, Qu LQ. Evaluation of seed storage protein gene 30-untranslated regions in enhancing gene expression in transgenic rice seed. Transgenic Res. 2012;21:545–553. doi: 10.1007/s11248-011-9552-4. [DOI] [PubMed] [Google Scholar]
- Mandal S, Mandal RK. Seed storage proteins and approaches for improvement of their nutritional quality by genetic engineering. Curr Sci. 2000;79(5):576–589. [Google Scholar]
- Marla S, Bharatiya D, Bala M, Singh V, Kumar A. Classification of rice seed storage proteins using neural networks. J Plant Biochem Biotechnol. 2010;19(1):123–126. doi: 10.1007/BF03323450. [DOI] [Google Scholar]
- Munck L, Shewry PR. The case of high-lysine barley breeding. In: Shewry PR, editor. Barley: genetics, biochemistry, molecular biology and biotechnology. Wallingford: CAB International; 1992. pp. 573–601. [Google Scholar]
- Müntz K, Christov V, Saalbach G, Saalbach I, Waddell D, Pickardt T, Schieder O, Wüstenhagen T. Genetic engineering for high methionine grain legumes. Food Nahrung. 1998;42(03–04):125–127. doi: 10.1002/(SICI)1521-3803(199808)42:03/04<125::AID-FOOD125>3.3.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Osborne TB. The vegetable proteins second edition. Longmans, green and Co. London Plant Physiol. 1924;154:1842–1854. [Google Scholar]
- Quinlan JR. C4.5: programs for machine learning. San Francisco: Morgan Kaufmann Publishers Inc; 1993. [Google Scholar]
- Rafiqul M, Khan I, Ceriotti ATL, Aryan A, Rafiqul M, Khan I, Ceriotti ATL, Aryan A, Mc Nabb W, Moore A, Craig S, Spencer D, Higgins TJV. Accumulation of a sulphur-rich seed albumin from sunflower in the leaves of transgenic subterranean clover (Trifolium subterraneum L.) Transgenic Res. 1996;5:179–185. doi: 10.1007/BF01969707. [DOI] [PubMed] [Google Scholar]
- Resource Coordinators NCBI. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2013;41(D1):D8–D20. doi: 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Annotation Project et al. The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res. 2007;36(Database issue):D1028–D1033. doi: 10.1093/nar/gkm978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivarola M, Jeffrey T, Foster JT, et al. Castor bean organelle genome sequencing and worldwide genetic diversity analysis. PLoS One. 2011;6(7):e21743. doi: 10.1371/journal.pone.0021743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saalbach G, Jung E, Saalbach I, Muntz K. Construction of storage protein genes with increased number of methionine codons and their use in transformation experiments. Biochem Physiol Pflanz. 1988;183:211–218. doi: 10.1016/S0015-3796(88)80103-3. [DOI] [Google Scholar]
- Schmidt MA, Barbazuk WB, Sandford M, May G, Song Z, Zhou W, Nikolau BJ, Herman EM. Silencing of soybean seed storage proteins results in a rebalanced protein composition preserving seed protein content without major collateral changes in the Metabolome and Transcriptome. Plant Physiol. 2011;156:330–345. doi: 10.1104/pp.111.173807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, et al. The B73 maize genome: complexity, diversity and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Sharma SB, Hancock KR, Ealing PM, White DWR. Expression of a sulfur-rich maize seed storage protein, δ-zein, in white clover (shape Trifolium repens) to improve forage quality. Mol Breed. 1998;4:435–448. doi: 10.1023/A:1009656002068. [DOI] [Google Scholar]
- Shewry PR, Halford NG. Cereal seed storage proteins: structures, properties and role in grain utilization. J Exp Bot. 2002;53(370):947–958. doi: 10.1093/jexbot/53.370.947. [DOI] [PubMed] [Google Scholar]
- Spencer D, Boulter D. The physiological role of storage proteins in seeds. Phil Trans R Soc B. 1984;304(1120):275–285. doi: 10.1098/rstb.1984.0023. [DOI] [Google Scholar]
- Swarbreck D, Wilks C, Lamesch P, et al. The Arabidopsis information resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2007;36(Database issue):D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, Sawkins MC, Long AD, et al. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.) Proc Natl Acad Sci U S A. 2001;98(16):9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisstock JC, Lesk AM. Prediction of protein function from protein sequence and structure. Q Rev Biophys. 2003;36(3):307–340. doi: 10.1017/S0033583503003901. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Sumi K, Tanaka K, Murai N. The bean seed storage protein [beta]-phaseolin is synthesized, processed, and accumulated in the vacuolar type-II protein bodies of transgenic rice endosperm. Plant Physiol. 1995;109:777–786. doi: 10.1104/pp.109.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]