Abstract
The use of pharmacogenomics (PGx) knowledge in treatment of individual patients is becoming a common phenomenon in the developed world. However, poorly resourced countries have thus far been constrained for three main reasons. First, the cost of whole genome sequencing is still considerably high in comparison to other (non-genomics) diagnostics in the developing world where both science and social dynamics create a dynamic and fragile healthcare ecosystem. Second, studies correlating genomic differences with drug pharmacokinetics and pharmacodynamics have not been consistent, and more importantly, often not indexed to impact on societal end-points, beyond clinical practice. Third, ethics regulatory frames over PGx testing require improvements based on nested accountability systems and in ways that address the user community needs. Thus, CYP2B6 is a crucial enzyme in the metabolism of antiretroviral drugs, efavirenz and nevirapine. More than 40 genetic variants have been reported, but only a few contribute to differences in plasma EFV and NVP concentrations. The most widely reported CYP2B6 variants affecting plasma drug levels include c.516G>T, c.983T>C, and to a lesser extent, g.15582C>T, which should be considered in future PGx tests. While the first two variants are easily characterized, the g.15582C>T detection has been performed primarily by sequencing, which is costly, labor intensive, and requires access to barely available expertise in the developing world. We report here on a simple, practical PCR-RFLP method with vast potentials for use in resource-constrained world regions to detect the g.15582C>T variation among South African and Cameroonian persons. The effects of CYP2B6 g.15582C>T on plasma EFV concentration were further evaluated among HIV/AIDS patients. We report no differences in the frequency of the g.15582T variant between the South African (0.08) and Cameroonian (0.06) groups, which are significantly lower than reported in Asians (0.39) and Caucasians (0.31). The g.15582C/T and T/T genotypes were associated with significantly reduced EFV levels (p=0.006). This article additionally presents the policy relevance of the PGX global health diagnostics and therefore, collectively makes an original interdisciplinary contribution to the field of integrative biology and personalized medicine in developing world. Such studies are, in fact, broadly important because resource-constrained regions exist not only in developing world but also in major geographical parts of the G20 nations and the developed countries.
Introduction
Identification of genetic variation that is of pharmacogenomics (PGx) relevance in African populations is an enormous endeavour and will continue to be a research priority in the next decade (Dandara et al., 2014a; 2014b). Although there is an explosion in the generation of human DNA sequences using next generation sequencing (NGS) technologies, the accessibility of these technologies in the developing world is severely limited. Simple, sensitive, and quick genotyping methods will always be required to assess whether a specific variant has an effect on a phenotype of interest, and if it does, to be able to offer genotyping for a few relevant single nucleotide polymorphisms (SNPs) at low cost to a specific patient group, for example, HIV/AIDS patients. Hence, the use of PGx for the treatment of individual patients in low and middle income countries has thus far been constrained, because genome sequencing can be costly, impractical, or has opportunity costs in regards to other more simple and effective (non-genomics) diagnostics in developing world where both science and social dynamics create a dynamic and fragile healthcare ecosystem.
A second factor of importance for PGx testing in resource-constrained settings is that, although there are recognized examples of PGx applications, studies that correlate genomic differences with drug pharmacokinetics and pharmacodynamics have not been uniformly consistent, and more importantly, often not indexed to changes in public health or societal end-points, beyond clinical practice.
A third notable factor is the ethics frames over PGx testing. They, too, need to be improved but based on nested accountability systems whereby each innovation actor and stakeholder can crosscheck transparently each other's behavior, claims of novel PGx tests and their value for society (Dove and Ozdemir, 2014; Ostrom et al. 1999), and in ways that address the local community needs, bearing in mind the inter-dependency of “local” and “global” health. Antiretroviral drugs, we suggest, are one of the prime examples where the real-life value of genomics/genetics testing for personalized medicine can be examined in resource constrained world regions. Such studies are important since resource-constrained regions exist not only in developing world but also in major geographical parts of the G20 and the developed countries (Hotez, 2013).
Genetic variation in CYP2B6 has become important because of the increasing number of different substrates it metabolizes, as well as the elusive cure for HIV/AIDS, which means that more and more people will be put on antiretroviral drugs as part of chronic medication. The two main drugs that form the backbone of antiretroviral therapy (ART), efavirenz (EFV) and nevirapine (NVP), are substrates of CYP2B6 (di Iulio et al., 2009). CYP2B6 gene polymorphisms are particularly relevant in sub-Sahara Africa, where HIV/AIDS is continuing to affect the lives of millions of patients. More than 40 SNPs have been reported in the CYP2B6 gene and its regulatory regions. The most important are the variants that affect CYP2B6 expression and activity, which have a bearing on the systemic exposure to drugs that are principally metabolized by CYP2B6 (Arab-Alameddine et al., 2009; di Iulio et al., 2009).
Several studies have reported on the contribution of the different CYP2B6 variants, culminating in the observation of qualitative and quantitative differences in the distribution of CYP2B6 variants in different geographical populations (Dandara et al., 2011; Lamba et al., 2003; Swart et al., 2013). For example, the variant c.1459C>T, which is associated with decreased CYP2B6 activity (Lang et al., 2001) and occurs in 2%–3%, 9%–14% and 1% of Africans (observed in West Africans), Caucasians, and Chinese, respectively (Crettol et al., 2005; Guan et al., 2006; Jacob et al., 2004; Klein et al., 2005; Mehlotra et al., 2006). This example clearly shows that some variants are important in some population groups and not in others.
Recent studies have narrowed down the variants that contribute most of the observed variation in CYP2B6 activity to CYP2B6 c.516G>T, c.983T>C, and the intron 3 variant, g.15582C>T (Holzinger et al., 2012; Lamba et al., 2003). The first two are exonic mutations and affect protein structure, whereas the latter is an intronic mutation and has been associated with CYP2B6 functionality. The g.15582T variant disrupts a cryptic splice acceptor and also lies within an exonic splicing enhancer (ESE), thus, enhancing the skipping of exons 4–6 in individuals who also carry the exon 4 mutation, c.516G>T (g.15631G>T) (Lamba et al., 2003).
Since CYP2B6 is involved in the metabolism of two of the most extensively used ARV drugs, EFV and NVP, it is important that inexpensive and easy-to-use genetic tests are developed. Although the genotyping for CYP2B6 c.516G>T and c.983T>C has been presented using less expensive methods, evaluation of the intron 3 variation, g.15582C>T, has been carried out using sequencing, which is more costly and more challenging to analyze and interpret. The aim of this study was to design a simple, practical PCR-RFLP method with vast potentials for use in resource-constrained world regions to detect the g.15582C>T SNP, to report on the frequency of the CYP2B6 g.15582T-allele in South Africans and Cameroonians, and finally to evaluate the effects of the CYP2B6 g.15582C>T single nucleotide polymorphism on plasma EFV levels.
Materials and Methods
The effects of CYP2B6 g.15582C>T SNP were investigated in a cohort of 321 South Africans (104 healthy individuals and 217 HIV/AIDS patients) previously described by Swart et al. (2012; 2013) and the distribution of its frequency was investigated in these plus an additional 99 healthy Cameroonian individuals (Swart et al., 2012). The study received ethics clearance and approval from the Human Research Ethics Committee, Faculty of Health Sciences, University of Cape Town (HREC REF 103/2009).
DNA isolation was performed from whole blood (400 μL) or buffy coat (200 μL) samples, depending on availability, by using either the QIAmp Blood Midi Kit (QIAGEN, Hilden, Germany) or GENelute Kit (Sigma-Aldrich, St Louis, MO, USA). Primers flanking the CYP2B6 (NCBI accession number NC_000019.10) intron 3 variation were adapted from Lang et al. (2001) and synthesized by Integrated DNA Technologies (IDT). Primer sequences are CYP2B6-F: 5′-GGTCTGCCCATCTATAAAC-3′ and CYP2B6-4AS: 5′-TCCCTCTCCGTCTCCCTG-3′. The primer set was used to amplify a 411 bp PCR fragment encompassing two restriction sites for the Mbo I restriction enzyme. The PCR reaction mixture contained 50–100 ng of DNA, 1X Green GoTaq Reaction Buffer (Promega, Madison, USA), 0.4 mM dNTPs (BioLine, London, UK), 1.5 mM MgCl2, 0.4 μM of each primer, and 0.5 U GoTaq DNA polymerase. The “T100™ thermal cycler from Bio-Rad was used for PCR amplification. The PCR amplification conditions included an initial denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 30 sec, annealing at 54.9°C for 30 sec, and extension at 72°C for 40s. This was followed by a final extension at 72°C for 10 min.
After PCR, the PCR product was digested overnight at 37°C using the restriction enzyme, Mbo I. The enzyme restriction digestion mix contained the following; 10 μL of PCR product, 1X Buffer R, and 3 U Mbo I (New England BioLabs, Inc., Ipswich, MA). PCR digests were electrophoresed on a 2.5% agarose gel for 120 min at 100V in the presence of GRGreen dye (InnoVita, Inc., Gaithersburg, MD). Digestion of the g.15582C-allele resulted in two fragments (92 bp and 319 bp), while in the presence of the g.15582T-allele, three fragments of 92 bp, 109 bp, and 210 bp were observed (Fig. 1). This PCR-RFLP method makes it easy for screening of the CYP2B6 g.15582C>T SNP and analyzing the possible association with particular phenotypes, for example, measures of response to EFV or NVP therapy.
FIG. 1.
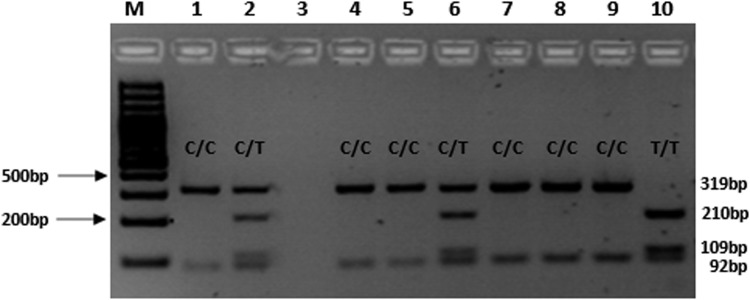
Agarose gel electrophoresis (2.5% agarose gel, 80V, 2 h) for the detection of CYP2B6 g.15582C>T SNP. M=Molecular weight marker; Lanes 1, 4, 5, 7–9=homozygous C/C genotype (319 and 92 bp); Lane 10=T/T genotype (210 bp, 109 bp, 92 bp); Lanes 2 and 6=heterozygous C/T genotype (319 bp, 210 bp, 109 bp, and 92 bp).
Allele and genotype frequencies for different populations were obtained from the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/) and compared to the frequencies observed in this study among the South Africans. Pearson's χ2-test and Fisher's exact test were used to assess if differences in frequency distribution between populations were significant (p<0.05). Plasma EFV levels were log-transformed and ANOVA was used to determine the effect of CYP2B6 g.155282C>T SNP alone and also with or without a c.516T- or c.983C-allele. The genotyping results for CYP2B6 c.516G>T and c.983T>C SNPs in this cohort and the effects on EFV levels have previously been reported on by Swart et al. (2013).
Results and Discussion
The CYP2B6 g.15582T-allele was observed among 104 healthy South Africans and 99 Cameroonians, at a frequency of 8% and 6%, respectively. The allele and genotype frequencies were further compared to those of Caucasians, Asians, and other Africans using data from HapMap and 1000 genomes project (Table 1). No significant differences were observed between the allele and genotype frequencies when comparing South Africans to Cameroonians or Nigerians (i.e., Yoruba). Despite the similarity in allele and genotype frequencies for the CYP2B6 g.15582C>T SNP between South Africans, Cameroonians, Yoruba, and African-Americans, previous studies have shown that differences in minor allele frequencies for SNPs exist between these populations (Swart et al., 2012). African populations are, however, consistently shown to be significantly different from European and Asian populations (Dandara et al., 2014b) and this has major implications for personalised medicine in Low- and Middle-Income Countries (LMIC).
Table 1.
Genotype and Allele Frequencies of the CYP2B6 g.15582C>T SNP in Different World Populations
| N | C/C | C/T | T/T | C-allele | T-allele | |
|---|---|---|---|---|---|---|
| South Africa (this study) | 104 | 89 (0.86) | 13 (0.13) | 2 (0.02) | 191 (0.92) | 17 (0.08) |
| Cameroon (this study) | 99 | 88 (0.89) | 11 (0.11) | 0 (0.00) | 187 (0.94) | 11 (0.06) |
| Yoruba (dbSNP) | 24 | 24 (1.00) | 0 (0.00) | 0 (0.00) | 48 (1.00) | 0 (0.00) |
| Hispanic* (dbSNP) | 44 | 8 (0.18) | 22 (0.50) | 14 (0.32) | 36 (0.43) | 48 (0.57) |
| European* (dbSNP) | 42 | 22 (0.52) | 14 (0.33) | 6 (0.14) | 58 (0.69) | 26 (0.31) |
| African American (dbSNP) | 30 | 24 (0.80) | 4 (0.13) | 2 (0.07) | 52 (0.87) | 8 (0.13) |
| Asian* (dbSNP) | 46 | 20 (0.44) | 16 (0.35) | 10 (0.22) | 56 (0.61) | 36 (0.39) |
Indicates overall significant differences (p=0.0001) for genotype and allele frequencies compared to frequencies among South Africans.
Linkage disequilibrium analysis was performed between CYP2B6 g.15582C>T, c.516G>T, and c.983T>C. Linkage disequilibrium parameters (D' and r2, respectively) were as follows, g.15582C>T vs. c.516G>T (0.74 and 0.04), g.15582C>T vs c.983T>C (1.00 and 0.00), and c.516G>T vs. c.983T>C (0.95 and 0.01). Holzinger et al. (2012) reported that the CYP2B6 g.15582C>T and c.983T>C SNPs were in strong LD in a Caucasian population, thus, indicating that whereas in one population, testing for one SNP may give strong clues about the other SNP, in another population, there may be a need to test for each SNP independently.
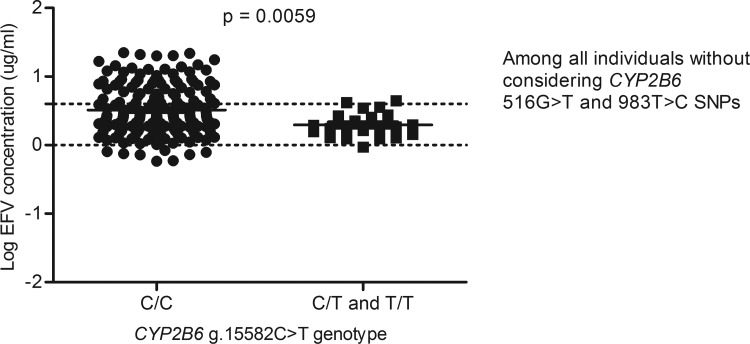
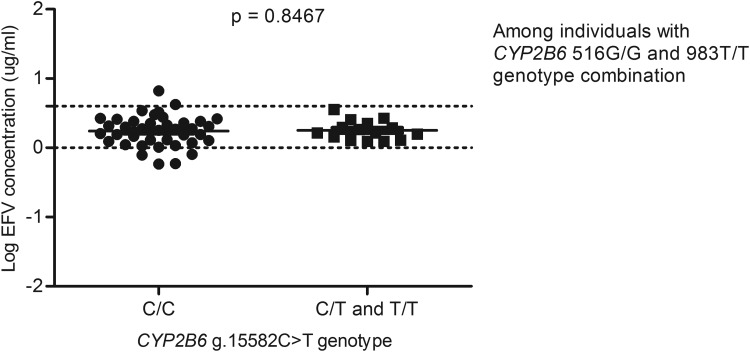
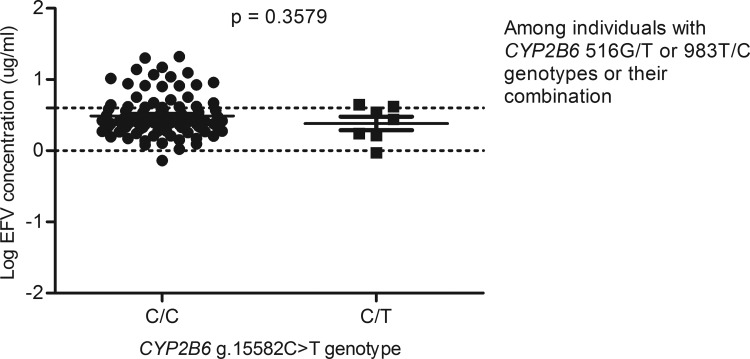
For example, in a GWAS study that aimed to identify contributors to variable EFV levels, it was observed that after taking into account the c.516G>T and the c.983T>C variations, the g.15582C>T loci was significant (Holzinger et al., 2012), whereas in this study, among the 217 South African individuals with data on EFV levels, the CYP2B6 g.15582T-allele was associated with decreased plasma EFV levels (p=0.0059) (Fig. 2), but significance was lost upon adjusting for the c.516G>T and c.983T>C SNPs (Figs. 3 and 4) and this is in agreement with a recent report by Sinxadi et al. (2015) in a separate South African HIV/AIDS cohort.
FIG. 2.
Effect of the CYP2B6 g.15582C>T variation on plasma EFV levels. This analysis was done without stratifying for either CYP2B6 c.516G>T or c.983T>C single nucleotide polymorphisms.
FIG. 3.
Effect of CYP2B6 g.15582C>T on plasma EFV levels stratified for CYP2B6 c.516G>T and 983T>C SNPs. Only individuals with homozygous 516G/G and 983T/T genotypes were included in the analysis.
FIG. 4.
Effect of CYP2B6 g.15582C>T on plasma EFV levels among individuals with CYP2B6 c.516G/T or c.983T/C genotypes or their combination.
This is in contrast to the results reported by Holzinger et al. (2012) and Lamba et al. (2003), showing an association between the CYP2B6 g.15582T-allele alone with lower CYP2B6 expression and increased plasma EFV levels even after correcting for c.516G>T and the c.983T>C variations. Lamba et al. (2003) reported LD between the CYP2B6 g.15582C>T SNP and the CYP2B6 g.-2320T>C SNP in a putative HNF4 binding site within the promoter region, and the combination of g.-2320T/15582C was associated with high levels of CYP2B6 protein in Caucasian females, while the g.-2320C/15582T combination was associated with low levels of CYP2B6 protein (Lamba et al., 2003).
The effect of the CYP2B6 g.-2320T>C SNP should be further investigated, as this SNP could potentially be involved in the high levels of CYP2B6 that gives rise to the decreased EFV levels observed in this study. The c.516T- and c.983C-alleles have consistently been associated with CYP2B6 loss-of-function and significantly high plasma EFV concentrations (Thorn et al., 2010). However, for most of the other SNPs, associations with altered plasma EFV levels have been conflicting in different populations. For example, Kwara et al. (2009) reported an association between CYP2A6 -48T>G SNP and altered plasma EFV levels, but this association was not confirmed in two other studies in different populations (Elens et al., 2010; Leger et al., 2009). These conflicting reports make translation of PGx approach to clinical healthcare setting complicated, but certainly more ethnic specific.
The availability of this quick PCR-RFLP genotyping method will enable further genotyping of the CYP2B6 g.15582C>T SNP in other African populations and also in HIV/AIDS patients receiving either EFV or NVP to establish the importance of the CYP2B6 g.15582C>T SNP conclusively. This genotyping method can contribute to addressing the need for cost-effective PGx tests in resource limited settings, while new next generation sequencing (NGS) technologies still remains pricy and not easily available in developing countries.
We report on the effect of CYP2B6 g.15582C>T, alone and in combination with c.516G>T and c.983T>C on plasma EFV levels by using an easy method for quick screening of the CYP2B6 g.15582C>T SNP. We also provide data that the frequency of CYP2B6 g.15582C>T varies between different global population groups.
Implications for policy
The use of PGx for the treatment of individual patients in low and middle income countries has thus far been limited for two main reasons. First, genotyping has proven useful, but impractical and costly within resource constrained settings. This has been the case with other notable examples such as glucose-6-phosphate dehydrogenase (G6PD) deficiency (Relling et al., 2014). Second, studies that correlate genetic differences in absorption, distribution, metabolism, and excretion (ADME), with treatment efficacy in ART therapies, have generally been inconsistent.
Drug response phenotype is a complex phenotype influenced by environmental and genetic factors. In addition to gene–environment interactions, gene–gene interactions also play an important role in drug response. The variation in microbiome composition within and between individuals and how that variability affects drug response, the new area of pharmacomicrobiomics (ElRakaiby et al., 2014), also needs to be taken into account to understand the full spectrum of PGx. The interaction between these factors and their contribution to explaining differences in ADME is not fully understood, with only a portion of the variability in plasma EFV levels explained. Failure to take into account all variables likely to affect PGx could be the reason for inconsistencies in replication of PGx association studies in various studies or populations.
Although difficult to measure, a systems biology approach supported by bioinformatics and appropriate statistical analysis is the most likely to be able to incorporate all the PGx variables (ElRakaiby et al., 2014; Pinelli et al., 2012). The varied and different approaches used in PGx have led to generation of inadequate and inconsistent evidence to warrant adoption of a PGx approach (Pavlos et al., 2012; Pavlos and Phillips, 2012). This study provides not only evidence to support a PGx approach, but a methodology that can be adapted for a more cost effective and practical application in the clinical setting.
Policy that allows translation of this work is somehow limited. The greater proportion of PGx research is carried out by industry, and is targeted towards new drug candidates. Studies concerning existing drugs such as EFV and NVP have received less funding. Studies such as this one need to be supported by further research on translation and how findings may be implemented in clinical practice. Day to day PGx practice entails the collection, storage, and use of genetic data collected from patients. Managing this data requires specialist software and database systems for digital curation, processing, and collation of data, and qualified personnel with appropriate expertise.
The addition of genetic counsellors may also be a consideration as they have proved useful in supporting families affected by inherited genetic conditions. As the use of genomic technologies within routine care increases, many more patients and their families will need support to understand the implications of particular genetic findings, and perhaps to help them make important decisions regarding treatment.
Not only do primary health care providers and public health professionals need to be “genetically literate”, but also the public, if PGx is to be used in treatment. A consistent and coordinated approach to engaging with the public is needed to promote understanding of these new technologies and what it means for healthcare. Public engagement, education, or consultation exercises can be costly, particularly if the target population is large and widely distributed, or if there are several languages in use. High levels of literacy can contribute positively to public engagement efforts. Public engagement to introduce new health interventions, may however face an uphill struggle if there are high levels of population displacement and instability, as can happen in conflict zones.
Clear and stable regulatory and governance frameworks will assist in translation. A study by Hopkins et al. (2006) looking at Europe, the US, and Japan found that there is room for further harmonization and rationalization of regulatory frameworks governing clinical research in PGx. Furthermore, controls on the application of diagnostic tests remain a neglected area. For example, Hopkins et al. (2006) recommend assessment of the social and ethical impacts of PGx testing on a case-by-case basis. Earlier, De Vries et al. (De Vries, 2004; De Vries et al., 2004) have suggested to take into account both ethics and ethics of ethics in evaluation of new knowledge emergence in society. There has been insufficient regulatory analysis done to determine whether regulatory frameworks in the global south will help or hinder the development and commercialization of the type of test demonstrated in this article.
Also underexplored is how the assessment of social and ethical implications of testing is provided for by governance frameworks. More widely, the governance of PGx testing will need to cover aspects as far reaching as marketing and direct to consumer advertising, patents, access and intellectual property, liability, and exploitation (of patients, health care professionals, and governments from exaggerating the efficacy, scope, or necessity of tests), and addressing the controversy over the use of race and ethnicity in PGx research (World Health Organization, 2007).
This article therefore collectively makes an original contribution to the emerging field of “global OMICS” using the example of PGx testing in resource-constrained settings. Such studies with the aim of addressing the needs of global health diagnostics, could conceivably be extended to the testing of noncommunicable diseases (NCDs) that are now impacting the developed and developing world alike (Agirbasli et al., 2013).
Acknowledgments
We thank the South African Medical Research Council (SA MRC), the National Research Foundation (NRF) of South Africa, and the University of Cape Town for funding.
Author Disclosure Statement
No conflicting financial interests exist.
References
- Agirbasli M, Eren F, Agirbasli D, White MJ, and Williams SM. (2013). Multi-locus candidate gene analyses of lipid levels in a pediatric Turkish cohort: Lessons learned on LPL, CETP, LIPC, ABCA1, and SHBG. OMICS 17, 636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab-Alameddine M, Di Iulio J, Buclin T, et al. (2009). Pharmacogenetics-based population pharmacokinetic analysis of efavirenz in HIV-1-infected individuals. Clin Pharmacol Therapeut 85, 485–494 [DOI] [PubMed] [Google Scholar]
- Crettol S, Deglon JJ, Besson J, et al. (2005). Methadone enantiomer plasma levels, CYP2B6, CYP2C19, and CYP2C9 genotypes, and response to treatment. Clin Pharmacol Therapeut 78, 593–604 [DOI] [PubMed] [Google Scholar]
- Dandara C, Huzair F, Borda-Rodriguez A, et al. (2014a). H3Africa and the African life sciences ecosystem: Building sustainable innovation. OMICS 18, 733–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandara C, Lombard Z, Du Plooy I, McLellan T, Norris SA, and Ramsay M. (2011). Genetic variants in CYP (-1A2, -2C9, -2C19, -3A4 and -3A5), VKORC1 and ABCB1 genes in a black South African population: A window into diversity. Pharmacogenomics 12, 1663–1670 [DOI] [PubMed] [Google Scholar]
- Dandara C, Swart M, Mpeta B, Wonkam A, and Masimirembwa C. (2014b). Cytochrome P450 pharmacogenetics in African populations: Implications for public health. Exp Opin Drug Metab Toxicol 10, 769–785 [DOI] [PubMed] [Google Scholar]
- De Vries R. (2004). How can we help? From “sociology in” to “sociology of” bioethics. J Law Med Ethics 32, 279–292 [DOI] [PubMed] [Google Scholar]
- De Vries R, DeBruin DA, and Goodgame A. (2004). Ethics review of social, behavioral, and economic research: Where should we go from here? Ethics Behav 14, 351–368 [DOI] [PubMed] [Google Scholar]
- di Iulio J, Fayet A, Arab-Alameddine M, et al. (2009). In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet Genom 19, 300–309 [DOI] [PubMed] [Google Scholar]
- Dove ES, and Ozdemir V. (2014). The epiknowledge of socially responsible innovation. EMBO Rep 15, 462–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elens L, Vandercam B, Yombi JC, Lison D, Wallemacq P, and Haufroid V. (2010). Influence of host genetic factors on efavirenz plasma and intracellular pharmacokinetics in HIV-1-infected patients. Pharmacogenomics 11, 1223–1234 [DOI] [PubMed] [Google Scholar]
- ElRakaiby M, Dutilh BE, Rizkallah MR, Boleij A, Cole JN, and Aziz RK. (2014). Pharmacomicrobiomics: The impact of human microbiome variations on systems pharmacology and personalized therapeutics. Omics 18, 402–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan S, Huang M, Chan E, Chen X, Duan W, and Zhou SF. (2006). Genetic polymorphisms of cytochrome P450 2B6 gene in Han Chinese. Eur J Pharmaceut Sci 29, 14–21 [DOI] [PubMed] [Google Scholar]
- Holzinger ER., Grady B, Ritchie MD, et al. (2012). Genome-wide association study of plasma efavirenz pharmacokinetics in AIDS Clinical Trials Group protocols implicates several CYP2B6 variants. Pharmacogenet Genom 12, 858–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins MM., Ibarreta D, Gaisser S, et al. (2006). Putting pharmacogenetics into practice. Nature Biotechnol 24, 403–410 [DOI] [PubMed] [Google Scholar]
- Hotez PJ. (2013). NTDs V.2.0: “Blue marble health”—Neglected tropical disease control and elimination in a shifting health policy landscape. PLoS Neglected Trop Dis 7, e2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob RM, Johnstone EC, Neville MJ, and Walton RT. (2004). Identification of CYP2B6 sequence variants by use of multiplex PCR with allele-specific genotyping. Clin Chem 50, 1372–1377 [DOI] [PubMed] [Google Scholar]
- Klein K, Lang T, Saussele T, et al. (2005). Genetic variability of CYP2B6 in populations of African and Asian origin: Allele frequencies, novel functional variants, and possible implications for anti-HIV therapy with efavirenz. Pharmacogenet Genom 15, 861–873 [DOI] [PubMed] [Google Scholar]
- Kwara A, Lartey M, Sagoe KW, Rzek NL, and Court MH. (2009). CYP2B6 (c.516G– >T) and CYP2A6 (*9B and/or *17) polymorphisms are independent predictors of efavirenz plasma concentrations in HIV-infected patients. Br J Clin Pharmacol 67, 427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba V, Lamba J, Yasuda K, et al. (2003). Hepatic CYP2B6 expression: Gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J Pharmacol Exp Therapeut 307, 906–922 [DOI] [PubMed] [Google Scholar]
- Lang T, Klein K, Fischer J, et al. (2001). Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics 11, 399–415 [DOI] [PubMed] [Google Scholar]
- Leger P, Dillingham R, Beauharnais CA, et al. (2009). CYP2B6 variants and plasma efavirenz concentrations during antiretroviral therapy in Port-au-Prince, Haiti. J Infect Dis 200, 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlotra RK, Ziats MN, Bockarie MJ, and Zimmerman PA. (2006). Prevalence of CYP2B6 alleles in malaria-endemic populations of West Africa and Papua New Guinea. Eur J Clin Pharmacol 62, 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom E, Burger J, Field CB, Norgaard RB, and Policansky D. (1999). Revisiting the commons: Local lessons, global challenges. Science 284, 278–282 [DOI] [PubMed] [Google Scholar]
- Pavlos R, Mallal S, and Phillips E. (2012). HLA and pharmacogenetics of drug hypersensitivity. Pharmacogenomics 13, 1285–1306 [DOI] [PubMed] [Google Scholar]
- Pavlos R, and Phillips EJ. (2012). Individualization of antiretroviral therapy. J Pharmacogenom Person Med 5, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinelli M, Scala G, Amato R, Cocozza S, and Miele G. (2012). Simulating gene-gene and gene-environment interactions in complex diseases: Gene-Environment iNteraction Simulator 2. BMC Bioinformat 13, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling MV, McDonagh EM, Chang T, et al. (2014). Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for rasburicase therapy in the context of G6PD deficiency genotype. Clin Pharmacol Therapeut 96, 169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinxadi PZ, Leger PD, McIlleron H, et al. (2015). Pharmacogenetics of plasma efavirenz exposure in HIV-infected adults and children in South Africa. Br J Clin Pharmacol. DOI: 10.1111/bcp.12590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart M, Skelton M, Ren Y, Smith P, Takuva S, and Dandara C. (2013). High predictive value of CYP2B6 SNPs for steady-state plasma efavirenz levels in South African HIV/AIDS patients. Pharmacogenet Genom 23, 415–427 [DOI] [PubMed] [Google Scholar]
- Swart M, Skelton M, Wonkam A, Kannemeyer L, Chin'ombe N, and Dandara C. (2012). CYP1A2, CYP2A6, CYP2B6, CYP3A4 and CYP3A5 polymorphisms in two Bantu-speaking populations from Cameroon and South Africa: Implications for global pharmacogenetics. Curr Pharmacogenom Person Med 10, 43–53 [Google Scholar]
- Thorn CF, Lamba JK, Lamba V, Klein TE, and Altman RB. (2010). PharmGKB summary: Very important pharmacogene information for CYP2B6. Pharmacogenet Genom 20, 520–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. (2007). The ethical, legal and social implications of pharmacogenomics in developing countries: Report of an international group of experts. pgs. 1–67 [Google Scholar]