Abstract
Green algae, Spirogyra (Chlorophyta), are found in a wide range of habitats including small stagnant water bodies, rivers, and streams. Species identification of Spirogyra based on morphological characteristics has proven to be a difficult process. An accurate identification method is required to evaluate genetic variations. This study is aimed at investigating the molecular profiling of 19 samples of Spirogyra from northern and northeastern Thailand. The morphological characteristics of each sample were recorded, viz. cell dimensions (width and length), along with the number and arrangement of chloroplast spirals/pyrenoids. With regard to a correlation of the biological and ecological parameters, conductivity was clearly significantly related to the number of pyrenoids. While DO is negatively related to the number of chloroplast spirals. Molecular studies with 10 ISSR primers were amplified to examine the DNA fingerprints. Morphological characters were determined to be significantly different by revealing 5 traits (P < 0.05) for all specimens. In addition, the DNA markers of all specimens were investigated using 10 ISSR primers. The results show that the PCR technique amplified 108 fragments. An analysis of the DNA fragments grouped all samples by ISRR-PCR, which were then separated into two groups according to their distribution.
Keywords: Spirogyra, Thailand, Morphology, Molecular profiling, ISSR-PCR
1. Introduction
Spirogyra is consumed in an uncooked form in the north and northeast of Thailand where Spirogyra is locally known as tao, thao and phakkai. Spirogyra is a filamentous type of freshwater green algae, of which the most easily recognized genus in Zygnemaceae due to its spirally coiled chloroplasts. Spirogyra spp. are filamentous, unbranched algae that have a unique mode of sexual reproduction. There are more than 400 species in the world. Spirogyra records remain limited to the generic level in floristic checklists and biodiversity inventories because of certain identification problems. The taxonomy of Spirogyra for vegetative growth consists of three characteristics: (i) type of cross walls (plane, replicate semi-replicate or colligate), (ii) cell width, and (iii) chloroplast number (Berry and Lembi, 2000; Hainz et al., 2009). The process of conjugation has to be included in species identification. Samples in sexually reproductive stages have rarely been collected. Stress from temperature, drought, and pH to the Spirogyra could induce the formation of conjugation tubes for the fertilization of male and female gametes. The morphology of this conjugation tube and zygote is also often used for identification. Little is known of its ecology and the effects it has on the morphologically distinct filamentous forms (Hainz et al., 2009). The morphology of some species in the genus Spirogyra and some related species, such as Zygnema and Cladophora, revealed them to be cell-shaped with a spiral chloroplast. Reports on the diversity of Spirogyra spp. in Thailand have been limited. Lewmanomont et al. (1995) recorded 8 Spirogyra spp. in Thailand as follows; Spirogyra crassa Kutz., Spirogyra decimina (Mull.) Kutz., Spirogyra dubia Kutz., Spirogyra fluviatilis Hilse., Spirogyra gracilis Kutz., Spirogyra neglecta Kutz., Spirogyra schmidii West & G. S. West and Spirogyra stictica (Engl. Bot). While, Thiamdao and Peerapornpisal (2011) have investigated the morphology of Spirogyra ellipsospora Transeau in northern Thailand. The vegetative cells were 118–200 × 240–600 μm. Three to five parietal chloroplast bands made 4–5 turns in each cell with numerous circular pyrenoids placed in the middle of the chloroplast band. A rough margin of chloroplast bands was observed.
In some cases, the identification of Spirogyra is mainly based on the conjugation tube process and the zygospores. However, this genus is mostly found in its vegetative stage, which complicates the studies on the ecological demand for individual species. The species identification of related Spirogyra based on the morphological characteristics can be difficult. However, in addition, Spirogyra can respond to the environmental conditions through the expression of different filament type groups (morphotypes), cell length/width and the number of chloroplast spirals which are related to the physico-chemical parameters of the water resource. At the same time, environmental stresses such as those related to temperature, drought and pH could stimulate the induction of the formation of a conjugation tube and gametes. The morphology of the conjugation tube and zygote is required for specific identification. Yoshida et al. (2003) reported that within their habitat, they are divided into two groups. One group floats in still water, and the other group lives in running water, and forms rhizoids for the purpose of anchoring to the substratum.
Therefore, molecular approaches using the PCR method have been used to resolve and support taxonomic evidence related to various organisms including algae. A number of molecular markers, such as amplified fragment length polymorphism (AFLP) (Vos et al., 1955), rDNA sequences (Wu et al., 2001), and inter-simple sequence repeats (ISSR) (Godvin et al., 1997; Wolfe and Randle, 2001) including microsatellite markers (Widmer et al., 2010), have been applied widely in the identification of the genetic diversity of many living organisms, including green algae (Shen, 2008), such as Entomophthora fungus (Lihme et al., 2009; Alaniz et al., 2009) and Gerbera spp. (Bhatia et al., 2009). The working principles of ISSR-PCR are similar to RAPD, except that the ISSR primer sequences are designed from microsatellite regions, such as (AGTG)4 or (AG)8 that are distributed widely in genomes while being considered good targets for the PCR-based fingerprinting technique. ISSR-PCR is more stable than RAPD due to the fact that the primers for ISSR-PCR are usually longer (16–20 bp) than those for RAPD (10 bp), which allows for a higher stringent condition. ISSR approach has proven that it has more reliability than RAPD. This is because the primers of ISSR repeat the sequences, which can mutate more quickly than those in the encoding region. If any differences appear in the genomes of the two species, they would be present in the polymorphic bands. ISSR markers have been applied in many research studies and it is clear that these markers have great potential and are beneficial for the study of the genetic variations among natural populations (Wolff and Morgan-Richards, 1998).
Hence, this study is aimed at determining the morphological traits, the molecular identification and genetic relationships of Spirogyra from northeastern and northern Thailand using the genotyping of the ISSR markers due to the fact that the morphological characteristics of Spirogyra may change according to the specific ecological conditions.
2. Method
2.1. Spirogyra specimens
The collecting sites were located in 19 different locations of northern and northeastern Thailand from February 2009 to May 2011 (Table 1 and Fig. 1). Fresh specimens of Spirogyra from each collection site were examined by wet mount under a light microscope and photographed with an Olympus DP 20. The length, width, vegetative cell length/width ratio, number of spirals, number of chloroplasts and shape of pyrenoids were recorded. Moreover, the zygospore of some Spirogyra specimens was investigated (shaped, size, and color). It is relatively easy to observe the morphological characteristics in order to classify the specimens according to each morphological trait. In addition, the various ecological parameters were also studied at each site (Table 2).
Table 1.
Morphological characteristic of each Spirogyra specimens in each trait.
| Details | Trait 1 | Trait 2 | Trait 3 | Trait 4 | Trait 5 |
|---|---|---|---|---|---|
| Vegetative cell width (μm) | 41–92 | 40–56 | 35–60 | 41–50 | 43–63 |
| Vegetative cell length(μm) | 115–223 | 80–185 | 90–193 | 127–164 | 93–195 |
| L/W ratio vegetative cell | 2.4–2.8 | 2.0–3.3 | 2.57–3.21 | 3.09–3.28 | 2.16–3.09 |
| Number of chloroplasts | 2–3 | 2 | 3 | 2 | 4–5 |
| Shape of zoospore | Ellipsoid | ns | ns | ns | ns |
| Zoospore width | 60–73 | ns | ns | ns | ns |
| Zoospore length | 75–95 | ns | ns | ns | ns |
| L/W ratio zoospore | 1.25–1.3 | ns | ns | ns | ns |
| Shape of pyrenoid | Discoid |
Remark: ns = not seen.
Figure 1.
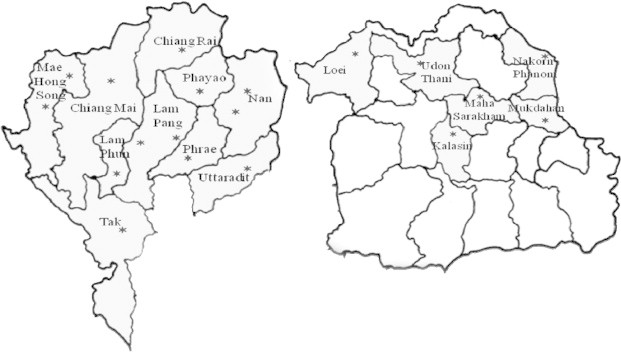
Location of 19 sampling sites (★) where the samples of Spirogyra spp. were collected.
Table 2.
Measurement and methods for investigating the ecological and biological parameters of Spirogyra spp. at field and laboratory investigations.
| Type of parameters | Parameters | Equipments and methods |
|---|---|---|
| Ecological | Conductivity | Consort™ multi-meter |
| pH | Consort™ multi-meter | |
| Total dissolved solids (TDS) | Consort™ multi-meter | |
| Salinity | Consort™ multi-meter | |
| Dissolved Oxygen (DO) | Azide modification method | |
| Biological | Width of cell | Light microscope |
| Length of cell | Light microscope | |
| Number of spiral | Light microscope | |
| Number of granule | Light microscope | |
2.2. DNA extraction
Genomic DNA of all Spirogyra specimens were extracted and purified using the modified plant tissue extraction protocol. DNA quality and quantity were determined by 1.4% gel electrophoresis and optical density was recorded using a spectrophotometer at 260 and 280 nm, respectively. All total genomic DNAs were diluted to a working concentration of 50 ng/μl and stored at −20 °C, and each 1 μl sample was then used in PCR reactions.
2.3. Inter-simple sequence repeat (ISSR) PCR protocol
Total genomic DNA of Spirogyra from each sampling site was performed by inter-simple sequence repeat (ISSR) PCR technique. Ten ISSR primers were used individually for ISSR-PCR and the reactions (Table 3) were carried out at a final volume of 25 μl, with a common PCR composition and performed in a MyCycler™ Thermocycler (Bio-RAD). PCR conditions were as follows: 1 cycle of 94 °C for 5 min, 40 cycles at 94 °C for 20 s, 51 °C for 1 min, 72 °C for 20 s and 1 cycle of final extension at 72 °C for 6 min. ISSR-PCR products were separated on 1.40% TBE agarose gel electrophoresis with 1× TBE (Tris–Boric acid–EDTA) buffer, stained with 0.5 μg/ml ethidium bromide, visualized with UV trans-illuminator and photographed with a Kodak Digital Camera Gel Logic 100. ISSR-profiles were scored and analyzed using the solutions of UPGMA in Mega 5.05 version. All the DNA patterns from the PCR products were compared for the purposes of separating the specimens into different morphological groups.
Table 3.
List of ISSR primers use to generate DNA fragment by ISSR PCR.
| Primer name | Sequence 5′ → 3′ | Length |
|---|---|---|
| UBC 807 | AGA GAG AGA GAG AGA GT | 17 |
| UBC 808 | AGA GAG AGA GAG AGA GC | 17 |
| UBC 809 | AGA GAG AGA GAG AGA GG | 17 |
| UBC 825 | ACA CAC ACA CAC ACA CT | 17 |
| UBC 826 | ACA CAC ACA CAC ACA CC | 17 |
| UBC 827 | ACA CAC ACA CAC ACA CG | 17 |
| UBC 835 | AGA GAG AGA GAG AGA GYC | 18 |
| UBC 857 | ACA CAC ACA CAC ACA CYG | 18 |
| UBC 855 | ACA CAC ACA CAC ACA CYT | 18 |
| UBC 821 | GTG TGT GTG TGT GTG TT | 17 |
2.4. Phylogenetic relationships
Phylogenetic relationships among the Spirogyra spp. samples were analyzed based on the scorable data from each primer using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) in the Mega program (version 5.0).
2.5. Statistical analysis
Data from the morphological and ecological parameters were determined by correlation coefficient and cluster analysis methods using SPSS V. 18.0 with a significantly acceptable P-value of 0.05.
3. Results
Morphological studies using a light microscope showed that, among the 19 Spirogyra specimens, 5 character groups could be defined (Fig. 2). The differences were mainly found in the number of pyrenoids and the arrangement of the chloroplast spirals, which were in condensed or scattered forms. The arrangement of the chloroplast spirals and pyrenoids of patterns 1 and 5 were highly condensed and compacted, while patterns 2, 3 and 4 were relatively scattered. Data for each morphological pattern of Spirogyra are shown in Table 1.
Figure 2.
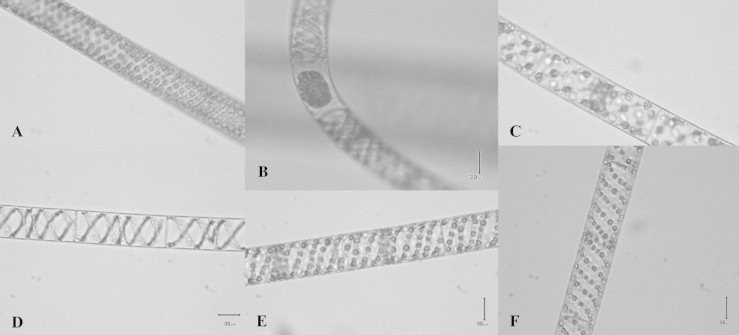
Five different morphological patterns of Spirogyra specimens collected from Thailand, (A and B) condensed and slightly compacted chloroplast spirals, (C) short cells with scattered chloroplast spirals, (D) long cells with few chloroplast spirals, (E) short cells with few chloroplast spirals, (F) long cells with condensed and compact chloroplast spirals (scale bar = 30 μm).
For molecular profiling, ten ISSR primers, viz. UBC 809, UBC 826, UBC 835, UBC 808, UBC 825, UBC 827, UBC 864, UBC 857, UBC 880, and UBC 807, were preliminarily screened for all 19 Spirogyra specimens. This generated 108 PCR fragments consisting of sizes ranging from 200 to 2500 base pairs (bp). The number of polymorphic bands that were generated varied between 3 and 16 bands, with an average of 7 bands per primer (see Figs. 3 and 4).
Figure 3.
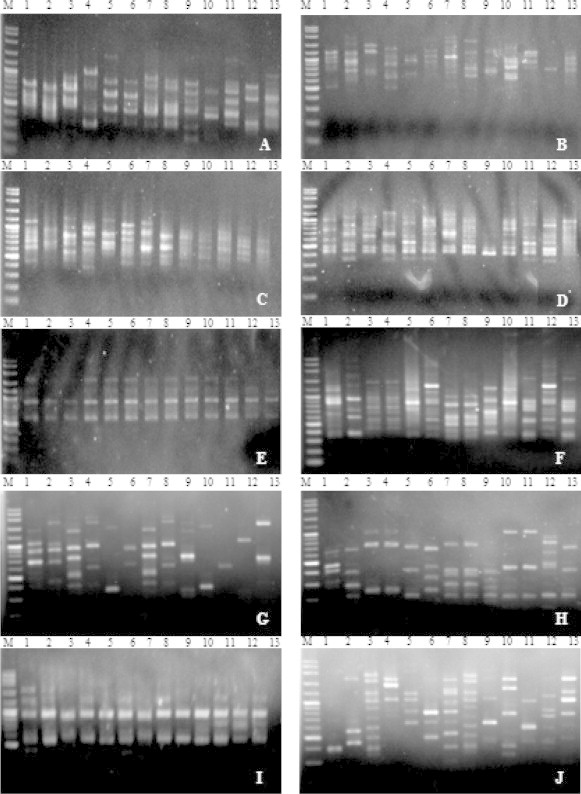
ISSR-PCR profiles of Spirogyra from northeastern Thailand generated by (A) UBC 835, (B) UBC 826, (C) UBC 809, (D) UBC 808, (E) UBC 825, (F) UBC 827, (G) UBC 864, (H) UBC 807, (I) UBC 857, and (J) UBC 880.
Figure 4.
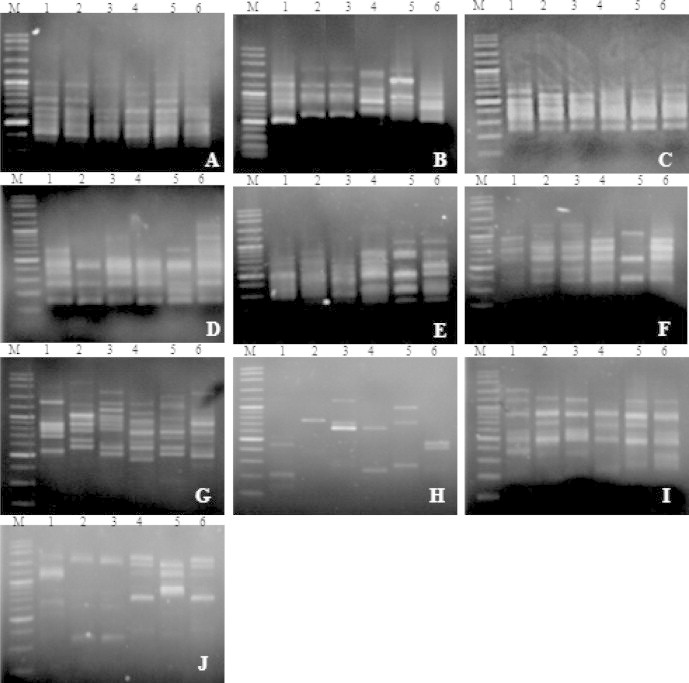
ISSR-PCR profiles of Spirogyra from northern Thailand generated by (A) UBC 835, (B) UBC 826, (C) UBC 809, (D) UBC 808, (E) UBC 825, (F) UBC 827, (G) UBC 864, (H) UBC 807, (I) UBC 857, and (J) UBC 880.
ISSR primers produced a total of 108 scorable markers. A cluster analysis of the ISSR markers separated the Spirogyra specimens into two distinct clusters; Cluster 1: northern, and Cluster 2: northeastern (Fig. 5).
Figure 5.
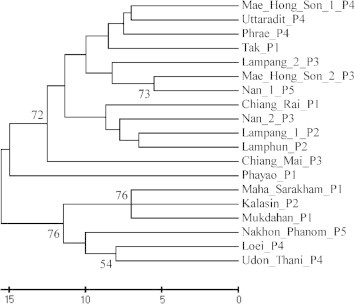
Cladogram of Spirogyra using ISSR-PCR.
pH values, conductivity, TDS, salinity, and DO ranged from 4.09 to 9.04, 113 to 752 μs, 63 to 671 ppm, 0.1 to 0.8, and 5.5 to 11.2 mg/l, respectively. The biological parameters, which included cell width, cell length, number of chloroplast spirals, and number of pyrenoids, ranged from 40 to 90 μm, 85 to 263 μm, 5.5 to 16 spiral, and 50 to 240 pyrenoids, respectively. The results of a study on the correlation coefficient analysis showed that the conductivity was significantly related to the number of pyrenoids with relative values (r) = 0.571 (P < 0.01). Negative DO was related to the number of chloroplast spirals with relative values (r) = −0.443 (P < 0.01) (Fig. 6).
Figure 6.
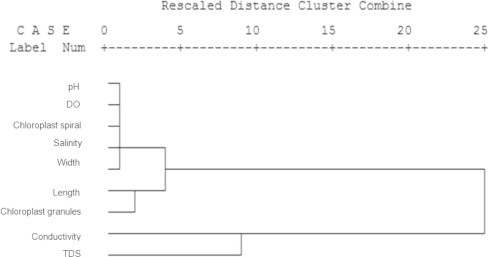
Dendrogram of hierarchical cluster analysis showing the correlation of each parameter.
4. Discussion
There are classical morphological and molecular methods that were used in the identification of the Spirogyra specimens. Distribution of Spirogyra in Thailand is typically limited to cosmopolitan areas and it is abundant during the hot, dry season and before the rainy season. Recent research has reported on the morphology of S. ellipsospora from northern Thailand under light and transmission electron microscopes, but only a few species have been identified due to the lack of scientific references (Thiamdao and Peerapornpisal, 2011). Moreover, other parts of Thailand must be included in further studies because of the differences in the geographical distribution, which may induce variations at the genetic level, as well as to identify the effects of different preferred habitats and surrounding climatic features that seem to be reliable causative variation factors (Shen, 2008).
Therefore, phenotypic traits may lead to incidences of misidentification and the results would then be less accurate than with molecular identification. The known distribution of S. ellipsospora author (pattern 1) is found throughout all parts of Thailand. The morphological characteristics of this species correspond to Kim et al. (2004). This morphological pattern consists of 2–3 chloroplasts per cell, helices consisting of 6–16 turns, and ellipsoid zygospores.
Pattern 2 represented the most common type of Spirogyra found in Thailand: Spirogyra neglecta. Pattern 3 consisted of 9 collecting sites from the north. Patterns 4 and 5 were not widely distributed. More than 20 species of Spirogyra are known to be from Pakistan and California (Zarina et al., 2007), while 82 species came from the Netherlands (Simons et al., 1990). Thiamdao and Peerapornpisal (2011) reported on S. ellipsospora from the north and northeast of Thailand. The knowledge of Spirogyra populations in Thailand has been limited and has been less documented in terms of distribution, diversity and habitats.
Compared with other reports, more than 20 species of Spirogyra have been reported to be from Pakistan and California (Zarina et al., 2007), while 82 species were recorded from the Netherlands (Simons et al., 1990). Moreover, Thiamdao and Peerapornpisal (2011) reported on the identification of S. ellipsospora in the North and Northeast of Thailand. This mention of the results indicates that the information and knowledge on the Spirogyra population of Thailand still have been less documented in terms of geographic distribution, diversity and ecological habitat.
The morphological patterns constructed in this study covered most of the genus variability described in the literature. Each morphological pattern is comprised of a well-defined cell width, cell length, number of chloroplast spirals, and number of pyrenoids, thus, lending itself to water quality assessors.
Very little is known regarding the ecological and physical parameters with regard to the distribution of Spirogyra (McCourt et al., 1989). Ecological parameters affect Spirogyra growth, and diversity (Goldman and Horne, 1983). Water temperature was not a decisive variable for the morphological characteristics of Spirogyra. It could be concluded that an increased level of nutrient supply is beneficial for the development and growth of Spirogyra filaments. Morphological patterns with short cells occurred at sites with low nutrient availability.
Polyploidy of Spirogyra, demonstrated by Allen (1985), has been recognized as a serious problem for the species concept. Identification of closely related species of Spirogyra based only on the morphological characteristics can be confusing or result in incidences of misidentification.
After ISSR amplification was performed with 10 primers for the analysis of the genetic relationships of all 36 Spirogyra populations, most primers were found to give an adequate number of amplified DNA fragments, which were enough to reconstruct a genetic relationship tree. A dendrogram was developed for 19 Spirogyra populations and indicated 2 main clusters by an analysis of the ISSR patterns from all ten primers. Each clade was separated by location. ISSR-PCR was then used to study the diversity of the Spirogyra populations.
Metais et al. (2000) considered using ISSR-PCR for the analysis of other organisms. Songdong (2008) screened ISSR primers of the green alga, Chlorella vulgaris genomic DNA, where 18 primers were found to give reproducibly amplified products. Our ten ISSR primers (UBC 809, UBC 826, UBC 835, UBC 808, UBC 825, UBC 827, UBC 864, UBC 857, UBC 880, and UBC 807) were used to investigate the genetic diversity of the Spirogyra specimens. All ISSR primers can be used as molecular markers of the Spirogyra species. ISSR primers generated highly reproducible fragments and were further used to study the genetic relationships between the Spirogyra populations from each region of Thailand.
Filippis et al. (1996) commented upon the importance of doing a reproducibility test. He advised that the genetic markers usually have limitations, mainly because the reproducibility from the samples is difficult. From our optimization experiment, the results showed that all distinctively major ISSR fragments were still reproduced.
ISSR-PCR plays an increasingly important role in the analysis of the genetic diversity of living organisms, such as beans (Phaseolus vulgaris) (Galvan et al., 2003), green algae (Shen, 2008), chickpeas (Cicer arietinum) (Bhagyawant and Srivastava, 2008), Entomophthora fungus (Lihme et al., 2009; Alaniz et al., 2009) and gerbera plants (Bhatia et al., 2009), as well as to detect fungal and algal symbionts of lichen (Widmer et al., 2010). These results indicate that any one of the ISSR primers was sufficient for the purposes of analyzing and organizing a cluster of the Spirogyra specimens.
Conflict of interest
None declared.
Acknowledgements
We would like to thank the Applied Technology in Biodiversity Research Unit, Institute for Science and Technology Research and the Economic Plant Genomes Research and Service Center, Faculty of Science, Chiang Mai University for allowing access to their valuable facilities. Finally, we would like to thank Dr. J.F. Maxwell for editing our manuscript.
Footnotes
Peer review under responsibility of King Saud University.
References
- Alaniz S., Armengol J., León M., A-Jimé N.G.J., Abad-Campos P. Analysis of genetic and virulence diversity of Cylindrocarpon liriodendri and C. macrodidymum associated with black foot disease of grapevine. Mycol. Res. 2009;113:16–23. doi: 10.1016/j.mycres.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Allen, M.A., 1985. The Biology of a Species Complex in Spirogyra (Ph.D. thesis). University Bloomington, Indiana, USA.
- Berry H.A., Lembi C.A. Effect of temperature and irradiance on the seasonal variation of a Spirogyra (Chlorophyta) population in a Midwestern Lake (U.S.A.) J. Phycol. 2000;36:841–851. [Google Scholar]
- Bhagyawant S.S., Srivastava N. Genetic fingerprinting of chickpea (Cicer arietinum L.) germplasm using ISSR markers and their relationships. Afr. J. Biotechnol. 2008;7:4428–4431. [Google Scholar]
- Bhatia R., Singh K., Jhang T., Sharma T.R. Assessment of clonal fidelity of micropropagated gerbera plants by ISSR markers. Sci. Hortic. 2009;119:208–211. [Google Scholar]
- Filippis L.D., Hoffmann E., Hampp R. Identification of somatic hybrids of tobacco generated by electrofusion and culture of protoplasts using RAPD-PCR. Plant Sci. 1996;121:39–46. [Google Scholar]
- Galván M.Z., Bornet B., Balatti P.A., Branchard M. Inter simple sequence repeat (ISSR) markers as a tool for the assessment of both genetic diversity and gene pool origin in common bean (Phaseolus vulgaris L.) Euphytica. 2003;132:297–301. [Google Scholar]
- Godvin I.D., Aitken E.A., Smith L.W. Application of inter simple sequence repeat (ISSR) markers to plant genetics. Electrophoresis. 1997;18:1524–1528. doi: 10.1002/elps.1150180906. [DOI] [PubMed] [Google Scholar]
- Goldman C.R., Horne H.J. McGraw-Hill Book Company; New York: 1983. Limnology. [Google Scholar]
- Hainz R., Wöber C., Schagerl M. The relationship between Spirogyra (Zygnematophyceae, Streptophyta) filament type groups and environmental conditions in Central Europe. Aquat. Bot. 2009;91:173–180. [Google Scholar]
- Kim J.H., Kim Y.H., Lee I.K. Morphotaxonomy of the Genus Spirogyra (Zygnemataceae, Chlorophyta) in Korea. Algae. 2004;19:91–105. [Google Scholar]
- Lewmanomont K., Wongrat L., Supanwanid C. Office of Environmental Policy and Planning; 1995. Algae in Thailand. p. 334. [Google Scholar]
- Lihme M., Jensen A.B., Rosendahl S. Local scale population genetic structure of Entomophthora muscae epidemics. Fungal Ecol. 2009;2:81–86. [Google Scholar]
- McCourt R.M., Hoshaw R.W., Wang J.C. Distribution, morphological diversity and evidence for polyploidy in North American Zygnemataceae (Chlorophyta) J. Phycol. 1989;22:307–315. [Google Scholar]
- Métais I., Aubry C., Hamon B., Jalouzot R., Peltier D. Description and analysis of genetic diversity between commercial bean lines (Phaseolus vulgaris L.) Theory Appl. Genet. 2000;101:1207–1214. [Google Scholar]
- Shen S. Genetic diversity analysis with ISSR PCR on green algae Chlorella vulgaris and Chlorella pyrenoidosa. Chin. J. Oceanol. Limnol. 2008;26:380–384. [Google Scholar]
- Simons J., Van Beem A.P., de Vries P.J.R. Induction of conjugation and spore formation in species of Spirogyra, Chlorophyceae, Zygnematales. Acta Bot. Neerl. 1990;33:323–334. [Google Scholar]
- Songdong S. Genetic diversity analysis with ISSR PCR on green algae Chlorella vulgaris and Chlorella pyrenoidosa. Chin. J. Oceanol. Limnol. 2008;26:380–384. [Google Scholar]
- Thiamdao S., Peerapornpisal Y. Morphological observation of Spirogyra ellipsosppora transeau, an edible freshwater macroalgae. J. Microsc. Soc. Thai. 2011;4:94–97. [Google Scholar]
- Vos P., Hogers R., Bleeker M., Reijans M., Vandelee T., Hornes M., Frijter A., Pot J., Peleman J., Kuiper M., Zabeau M. AFLP – a new technique for DNA fingerprinting. Nucleic Acids Res. 1955;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer I., DalGrande F., Cornejo C., Scheidegger C. Highly variable microsatellite markers for the fungal and algal symbionts of the lichen Lobaria pulmonaria and challenges in developing biont-specific molecular markers for fungal associations. Fungal Biol. 2010;114:538–544. doi: 10.1016/j.funbio.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Wolfe A.D., Randle C.P. Relationships within and among species of the holoparasitic genus Hyobanche (Orobanchaceae) inferred from ISSR banding patterns and nucleotide sequences. Syst. Bot. 2001;26:120–130. [Google Scholar]
- Wolff K., Morgan-Richards M. PCR markers distinguish Plantago major subspecies. Theor. Appl. Genet. 1998;96:282–286. [Google Scholar]
- Yoshida K., Inove N., Sonobe S., Shimmen T. Involvement of microtubules in rhizoid differentiation of Spirogyra species. Protoplasma. 2003;221:227–235. doi: 10.1007/s00709-002-0078-8. [DOI] [PubMed] [Google Scholar]
- Zarina A., Hazan M.U., Shameel M. Diversity of the genus Spirogyra (Zygnemophyceae Shameel) in the North-Eatern areas of Pakistan. Proc. Pakistan Acad. Sci. 2007;44:225–248. [Google Scholar]


