Abstract
MicroRNAs (miRNAs) are a family of single-stranded RNA molecules about 22 nt in length, which can regulate protein-coding gene expression in various organisms by post-transcriptional repression of messenger. In this research, the potential miRNAs and their target genes were analyzed and predicted by computational methods from the EST and GSS databases of eleven fish species, 43 potential miRNAs were identified, they belong to 38 miRNA families, some miRNAs are highly conserved in animal kingdom, the predicted target genes are involved in development, signal transduction, response to environmental stress and pathogen invasion. Taken together, our data suggest that there are a plentiful of miRNAs in these eleven fish species, these miRNAs may play some important roles by regulating their target genes, and the data provide important information for further functional studies.
Keywords: MicroRNA, Computational prediction, Fish, Target, Function
1. Introduction
MicroRNAs (miRNAs) are a class of endogenous, evolutionary conserved, single strand non-coding RNAs with approximately 22 nucleotides (nts), which involved in the regulation of gene expression by translational repression and mRNA destabilization (Ambros, 2004; Ambros and Chen, 2007; Kloosterman and Plasterk, 2006). Mature miRNAs are generated from the stem portion of single stranded stem-loop precursors (pre-miRNAs), which is processed by ribonuclease III-like enzyme from primary miRNA (pri-miRNA) transcript. Pre-miRNAs are exported into the cytoplasm where cleavage of the loop by the RNase Dicer generates a duplex of two about 22 nt long mature miRNA (miRNA and miRNA-star) duplex. And then mature miRNAs are incorporated into the RNA-induced silencing complex (RISC) and guide RISC to complementary miRNA targets. Finally, the RISC inhibits translation elongation or triggers the degradation of target mRNAs (Bartel, 2005; Kim et al., 2009; Liu et al., 2008; Mallanna and Rizzino, 2010). Due to miRNAs playing various regulatory roles in gene regulation, several studies have indicated that they take part in a wide variety of biological processes including organ development, cell proliferation and death, apoptosis and fat metabolism, cell differentiation, signal transduction, fat metabolism and adaptive immune responses as well as diseases (Bartel, 2004; Belver et al., 2010; Ladomery et al., 2011; Rogers and Chen, 2013; Sun and Lai, 2013).
Most of the known miRNAs are highly evolutionarily conserved from species to species, ranging from insects to humans in animal kingdom (Daido et al., 2014; Maher et al., 2006; Niwa and Slack, 2007; Takane et al., 2010; Tanzer and Stadler, 2004). Conservation among species became one of the most important properties of miRNAs. So, this feature will facilitate us to perform the computational search for miRNAs based on the highly conserved sequence in the mature miRNAs and long hairpin structures in miRNA precursors (Mishra and Lobiyal, 2011; Ren et al., 2012; Saetrom et al., 2006). There are several significant advantages of identifying miRNAs, because it is accurate, fast, and inexpensive compared to the experimental method. For this reason, computational approaches provide an ideal way for identifying miRNAs in animals by using expressed sequence tags (EST) and genome survey sequence (GSS) databases, especially in organisms in which genome sequences are not available. Using this method, a large number of miRNAs have been successfully identified in some plant and animal species (Akter et al., 2014; Barozai, 2012b; Dong et al., 2012; Luo and Zhang, 2009; Paul and Chakraborty, 2013; van der Burgt et al., 2009; Yousef et al., 2009).
To date, over 28,645 miRNA genes have been deposited in the public database, miRBase (Release 21, 2014, http://www.mirbase.org); however, only 1637 miRNAs are in the database, they are just a small portion of the miRNAs described. Till now, little is known about experimental or computational identification of miRNAs in the eleven fish species. In this research, we carried out computational prediction to identify miRNAs in these eleven fish species. The study will make a substantial supplement to the known miRNA in fish species and it also provides a foundation for further research on miRNAs.
2. Materials and methods
2.1. Availability of databases
To search for potentially conserved miRNAs in the eleven fish species miRNAs, a total of 6.893 previously known animal miRNAs were retrieved from miRBase and defined as a reference set of miRNA sequences. To avoid the redundant or overlapping miRNAs, the repeated sequences of miRNAs within the above animal species were removed and the remaining sequences were used as query sequences for BLAST search. The ESTs and GSSs sequences from the 11 studied species were downloaded from the GenBank nucleotide databases of National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/). There are 187 GSSs from Mylopharyngodon piceus (mpi); 3.968 GSSs and 20.122 ESTs from Ctenopharyngodon idellus (cid); 2.272 GSSs from Hypophthalmichthys molitrix (hmo); 1.367 GSSs from Aristichthys nobilis (ano); 5.006 GSSs and 4.200 from Pseudosciaena crocea (pcr); 98.880 GSSs and 10.128 ESTs from Cynoglossus semilaevis (cse); 425 GSSs from Channa argus (car); 1.266 GSSs and 5.361 ESTs from Siniperca chuatsi (sch); 248 GSSs and 3.385 ESTs from Acipenser sinensis (asi); 676 GSSs and 937 ESTs from Monopterus albus (mal); 850 GSSs from Pelteobagrus fulvidraco (pfu), respectively.
2.2. Computational identification of the conversed miRNAs
The alignment tool BLAST version 2.2.27 was used to identify the potentially conserved miRNAs and was downloaded from the NCBI website. BLASTN parameters were set as follows: an expect value cut-off of 10; the window size 7; a low-complexity sequence filter; number of descriptions and alignments was 1000. All BLAST results were saved and used for further analysis. Procedure of search for potential miRNAs in the 11 fish species is shown in Fig. 1. The following five criteria were raised to identify the potential miRNAs: (1) mature miRNAs were allowed to have only 0–4 nucleotide mismatches in sequence with all previously known animal mature miRNAs; (2) the potential pre-miRNA could be folded into a typical stem-loop hairpin secondary structure, such that one arm of the hairpin contains the ∼22 nt mature miRNA sequence; (3) there are no loops in the miRNA/miRNA star duplex; (4) the predicted secondary structure of the miRNA pre cursor should have lower minimal free energy (MFE) and minimal free energy index (MFEI) than other types of RNA; (5) the predicted pre-miRNAs should have an A + U content of 30–80% by SVM (support vector machine) (Ding et al., 2010; Wu et al., 2011; Xu et al., 2008). If one sequence met these criteria, we considered it as a miRNA. Finally, some possible false sequences of pre-miRNAs should be deleted by manual inspection.
Figure 1.
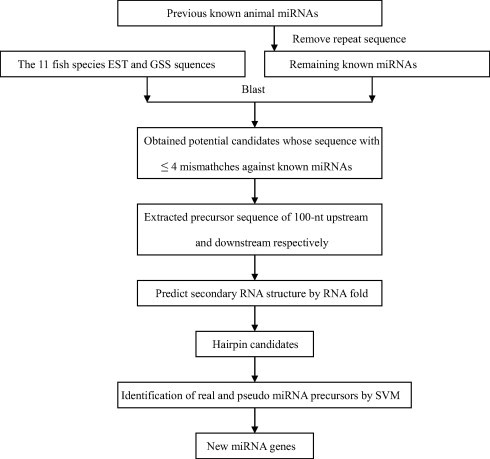
Procedure for prediction of the potential miRNAs from 11 fish species.
2.3. Phylogenetic analysis of the identified miRNAs
Because most of animal mature miRNAs and their precursor sequences are derived from the same gene families, they are strongly conserved and have high sequence identity, even between distantly related species. The mature and precursor sequences of the identified 11 fish species miRNAs were aligned and phylogenetically analyzed with the MEGA5.0 software (Tamura et al., 2011). Evolutionary distances were calculated by the neighbor-joining (NJ) method following 1000 bootstrapped replicates.
2.4. Target prediction for identified miRNAs
The mRNA database of the 11 fish species downloaded from NCBI database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unigene) and their 3′-UTR sequences which ⩾20 nt in length were extracted and used for target prediction. Potential targets of the predicted miRNAs were identified using RNAhybrid program (Rehmsmeier et al., 2004). The parameters employed are described as follows: (1) P-value cutoff of 0.05, target duplex free energy △G ⩽ −24 kcal/mol; (2) no mismatches in the seed region (5′ region of mature miRNA, from second to eighth nt position); (3) only one G:U pairing in the seed region; (4) the miRNA sequences and potential mRNAs targets were no more than four gaps at positions 9–21 from miRNA 5′ end. Subsequently, miRNA-target duplexes were checked manually.
3. Results and discussion
3.1. Identification of putative miRNAs from 11 fish species
In the present study, a strategy based on homology searching and secondary structure evaluation was employed to screen for potential miRNAs in 11 fish species. After the redundant sequences of the same genes were removed, and then the protein-coding sequences were also removed, a total of 43 potential miRNAs were identified. The 43 identified potential miRNAs represent 38 miRNA families in these 11 fish species. Among the 43 predicted miRNAs, 16 miRNAs were identified from the ESTs and 26 miRNAs from the GSSs. Among these, four miRNAs were identified in mpi, five miRNAs were identified in cid, two miRNAs were identified in hmo, three miRNAs were identified in ano, four miRNAs were identified in pfu, five miRNAs were identified in mal, one miRNA were identified in sch, two miRNAs were identified in car, five miRNAs were identified in cse, eight miRNAs were identified in pcr, and the rest four miRNAs were identified in asi, respectively (Table 1).
Table 1.
43 newly identified miRNAs in 11 fish species.
| miRNAs name | Source miRNA homologous | Gene source | Predicted mature sequence (5′–3′) | Loc | Strand | LP (nt) | A + U (%) | MFE | MFEI |
|---|---|---|---|---|---|---|---|---|---|
| mpi-miR-3245 | bmo-miR-3245 | DQ026435(GSS) | UAGUCACUUGGGAGAGGCUAAUC | 3′ | Minus | 130 | 58.46 | −33.80 | 0.63 |
| mpi-miR-4054 | cin-mir-4054 | AY704462(GSS) | UAUCAUUGAUGUCCUAUGGC | 5′ | Minus | 64 | 65.62 | −12.80 | 0.58 |
| mpi-miR-6835–3p | hsa-miR-6835–3p | GQ406278(GSS) | GUUGAACCUUUUCUGUCUCCCAU | 3′ | Minus | 117 | 65.81 | −29.80 | 0.73 |
| mpi-miR-222 | hsa-miR-222–5p | GU217957(GSS) | UUCAGUAGCCAGUGUACUCUAC | 3′ | Plus | 132 | 52.27 | −39.80 | 0.65 |
| cid-miR-2437 | bta-miR-2437 | GT223130(EST) | UGUGGUUUUUUGUUUUCGUAU | 5′ | Minus | 113 | 61.94 | −25.70 | 0.62 |
| cid-miR-5192 | hsa-miR-5192 | GT224283(EST) | GGAGAGUGGAUUCCAGAUAUC | 5′ | Minus | 93 | 54.83 | −26.90 | 0.64 |
| cid-miR-3198 | hsa-miR-3198 | GT223053(EST) | UUGGAUUCCUGGGGAAUGGAGA | 5′ | Plus | 82 | 43.90 | −31.40 | 0.61 |
| cid-miR-223 | bta-miR-223 | GR942893(EST) | UGUCAGUUUGUCAAAUACCCCA | 5′ | Plus | 77 | 46.75 | −25.80 | 0.63 |
| cid-miR-1814b | bta-miR-1814b | GR946702(EST) | GGUUUGUUUAGUUUUGUUUG | 3′ | Plus | 107 | 72.89 | −23.70 | 0.82 |
| hmo-miR-2192 | dre-miR-2192 | JX499811(GSS) | AAAGUGAAAGGUGACUGAGGC | 3′ | Minus | 79 | 55.69 | −28.40 | 0.67 |
| hmo-miR-2293 | bta-miR-2293 | DQ136011(GSS) | UGACUUUUGUUGUUUUGUAU | 5′ | Plus | 143 | 69.93 | −34.10 | 0.79 |
| ano-miR-2800 | bmo-miR-2800 | HM012521(GSS) | AGAAUAUUGUGUCUUGCAAGCCA | 5′ | Minus | 134 | 64.17 | −31.90 | 0.68 |
| ano-miR-2293 | bta-miR-2293 | DQ136011(GSS) | GACUUUUGUUGUUUUGUAUG | 5′ | Plus | 143 | 60.13 | −36.10 | 0.63 |
| ano-miR-1603 | bta-miR-1603 | KC191355(GSS) | GGUGUUUGUUUUGUGUUUUU | 5′ | Plus | 96 | 66.66 | −20.00 | 0.63 |
| pfu-miR-29 | cin-miR-29 | DY450843(EST) | ACCCUCUCCUUUUGGUUUGC | 3′ | Minus | 95 | 53.68 | −26.80 | 0.78 |
| pfu-miR-2304 | bta-miR-2304 | EU439604(GSS) | AUGUGUGUGGUUGUGUGUGU | 3′ | Minus | 171 | 45.61 | −57.60 | 0.62 |
| pfu-miR-297 | hsa-miR-297 | FJ851155(GSS) | GUGUGUGUGUGCAUGUGCAUG | 5′ | Plus | 188 | 45.21 | −77.90 | 0.77 |
| pfu-miR-669 | bta-miR-669 | FJ851155(GSS) | UGUGCGUGUGUGCAUGUGCGUG | 5′ | Plus | 147 | 46.25 | −57.20 | 0.73 |
| mal-miR-4040–3p | cin-miR-4040–3p | GW584894(EST) | CAACCAGAUCAGAAAGACCU | 3′ | Plus | 73 | 50.68 | −21.00 | 0.58 |
| mal-miR-4709 | hsa-miR-4709 | AY363652(GSS) | AUGAAGAGGAGGUGCUCAUGUCA | 5′ | Minus | 103 | 46.60 | −37.60 | 0.69 |
| mal-miR-297 | hsa-miR-297 | DQ987572(GSS) | AUGUAUGUGUGCAUGUGAAGG | 5′ | Minus | 142 | 48.59 | −47.20 | 0.65 |
| mal-miR-42 | cel-miR-42 | NC003192(GSS) | AGUGGUGUUUGCUUUUUCUGCGGCU | 3′ | Minus | 166 | 52.40 | −49.70 | 0.64 |
| mal-miR-4194–3p | cin-miR-4194–3p | DQ987581(GSS) | AUAUAUAUAUGUGUGUGG | 3′ | Minus | 72 | 59.72 | −16.70 | 0.58 |
| sch-miR-2437 | bta-miR-2437 | EU659698(GSS) | UCUCUUUUUUUGUUUUCCUUU | 5′ | Plus | 104 | 56.73 | −28.80 | 0.64 |
| car-miR-4433b-3p | hsa-miR-4433b-3p | KC823604(GSS) | UAGGAGUGGGGGGUGGGCGGU | 3′ | Minus | 117 | 47.00 | −39.60 | 0.65 |
| car-miR-125b | dre-miR-125b | HQ404190(GSS) | UCCCUGAGACCCUAACUUGUGA | 5′ | Minus | 82 | 46.34 | −39.60 | 0.91 |
| cse-miR-2191 | dre-miR-2191 | EU907211(GSS) | UCACACCUACAAUCCCCCCCC | 3′ | Plus | 127 | 48.03 | −43.60 | 0.67 |
| cse-miR-2316 | bta-miR-2316 | EF683116(GSS) | ACGUGGGCCUGGACUGCGGCGAG | 5′ | Plus | 141 | 37.17 | −54.90 | 0.63 |
| cse-miR-203b-3p | dre-miR-203b-3p | GQ426771(GSS) | GUGAAAUGUUCAGGACCACUGA | 3′ | Plus | 97 | 53.60 | −38.40 | 0.86 |
| cse-miR-190a-3p | hsa-miR-190a-3p | JQ003879(GSS) | AUUUAUAUCAAACAUAUUCAU | 3′ | Plus | 127 | 76.37 | −23.40 | 0.80 |
| cse-miR-2444 | bta-miR-2444 | JQ003879(GSS) | UUUGUGUUGUUUUUUGUUUU | 5′ | Minus | 154 | 75.32 | −30.30 | 0.79 |
| pcr-miR-431-3p | hsa-miR-431-3p | GO651700(EST) | CAGGUCGUCUUGCAGGGGAUCA | 3′ | Minus | 110 | 43.63 | −38.10 | 0.62 |
| pcr-miR-6837 | hsa-miR-6837 | GO652159(EST) | UGCUCACUGUGACUCUGCUGGAA | 5′ | Minus | 89 | 43.80 | −37.60 | 0.75 |
| pcr-miR-147 | bta-miR-147 | CX348533(EST) | GUGUGCGGAAAUGCUUCUGCUC | 3′ | Plus | 87 | 50.57 | −34.50 | 0.81 |
| pcr-miR-34 | cel-miR-34 | CX348881(EST) | UGCUAGUGUGGUUAGCUGGUGA | 3′ | Plus | 69 | 40.57 | −33.20 | 0.76 |
| pcr-miR-4695-5p | hsa-miR-4695–5p | GO652832(EST) | GAGGAUGAGGAGGAGGUGGAGG | 5′ | Minus | 81 | 44.44 | −36.90 | 0.83 |
| pcr-miR-2444 | bta-miR-2444 | CX348588(EST) | UUUGUUUUGUUUUUUGUUUU | 3′ | Minus | 73 | 61.64 | −21.90 | 0.79 |
| pcr-miR-297 | hsa-miR-297 | CX348877(EST) | GUGUGUGUGUGCAUGUGCAUU | 3′ | Minus | 85 | 48.23 | −30.70 | 0.71 |
| pcr-miR-2415 | bta-miR-2415 | ASJX01000025(GSS) | CCAGGCCUGCUGGACCGAAGC | 5′ | Plus | 94 | 30.53 | −45.20 | 0.69 |
| asi-miR-965-5p | bmo-miR-965–5p | EV824426(EST) | AGGGAGAAGCUAUAGCGAAAAUGU | 5′ | Plus | 125 | 56.80 | −42.30 | 0.79 |
| asi-miR-2304 | bta-miR-2304 | ES698401(EST) | GUGUGUGUGGUUGUGUGUGU | 5′ | Plus | 65 | 47.69 | −26.40 | 0.78 |
| asi-miR-374a | hsa-miR-374a | KC984851(GSS) | CUUAUCAGAUUGUAUGCAGUGU | 5′ | Plus | 77 | 57.14 | −22.30 | 0.68 |
| asi-miR-86 | cel-miR-86 | JN099311(GSS) | GUGGGCUCAGAUUCGCCGGUUG | 5′ | Minus | 98 | 35.71 | −47.10 | 0.75 |
Abbreviations: NM = number of mismatches; LP, Length of precursor; Loc = location; MFE, minimal folding free energy (kcal/mol); MFEI, minimal folding free energy index. The shaded letters indicate nucleotide mismatches.
All of the precursors for those mature miRNAs fold into the typical secondary structure of miRNAs and they are postulated to be important validation parameters for the miRNA genes predicted (Fig. 1S). The length of the precursors vary from 64 nt to 188 nt with an average of 108 nt. Mature miRNA sequences have been reported to be evenly located on the two arms of the stem-loop hairpin structures of potential pre-miRNAs (Gorodkin et al., 2006). These 43 identified fish species miRNAs also have a similar situation, of which 24 (55.81%) were found to be located on the 5′-arms of the stem-loop hairpin structures, while the other 19 (44.19%) were located on the 3′-arms (Table 1 and Fig. 1S). The A + U contents of these predicted fish species pre-miRNA sequences ranged from 30.53% to 76.37%, with an average of 52.90%, which closely matched the results of previous studies (Ambros et al., 2003; Keshavan et al., 2010; Neutelings et al., 2012).
MFE values are important for evaluating the stability of RNA secondary structures. In general, the lower the MFE, the more stable the secondary structure of an RNA sequence. The MFE values of the identified 11 fish species miRNA precursors varied broadly from −77.90 kcal/mol to −12.80 kcal/mol, with an average of −35.07 kcal/mol. The MFEI of each potential miRNA precursor was calculated for the precise discrimination of the miRNA from other types of small RNAs. Since other RNAs such as mRNA, rRNA, tRNA may also form similar hairpin structures, we used the minimal fold energy index (MFEI) to distinguish other RNAs or RNA fragments. In the present prediction, the newly identified pre-miRNAs from 11 fish species have MFEI values ranging from 0.58 to 0.91, with an average of about 0.71 (Table 1). These values were significantly higher compared to those reported for tRNAs (0.64), rRNAs (0.59), and mRNAs (0.62–0.66), indicating that newly predicted potential fish species miRNAs are probably true miRNAs than any other type of RNA molecules.
3.2. Phylogenetic analysis of the identified miRNAs
Mature miRNA sequences, along with their corresponding precursor sequences, are highly conserved among distantly related animal species (Chen et al., 2012; Lee et al., 2007). This phenomenon provides opportunities for the investigation of evolutionary relationships of miRNAs belonging to the same families in different animal species. In this study, a comparison of the precursor sequences of the predicted two miRNAs families (miR-147 and miR-203) with other members in the same family showed that most members could be found to have a high degree of sequence similarity with others (Fig. 2). These two miRNA precursor families were further considered for phylogenetic analyses, respectively. The results revealed that pcr-miR-147 and hhi-miR-147 were clustered into 1 group indicating that these two families are possibly highly conserved in marine fishes, and which have evolutionary relatedness (Fig. 3A). Similarly, the phylogenetic trees for the miR-203 family revealed that predicted miR-203b-3p grouped with the closely related species miR-203b and miR-203b (Fig. 3B).
Figure 2.
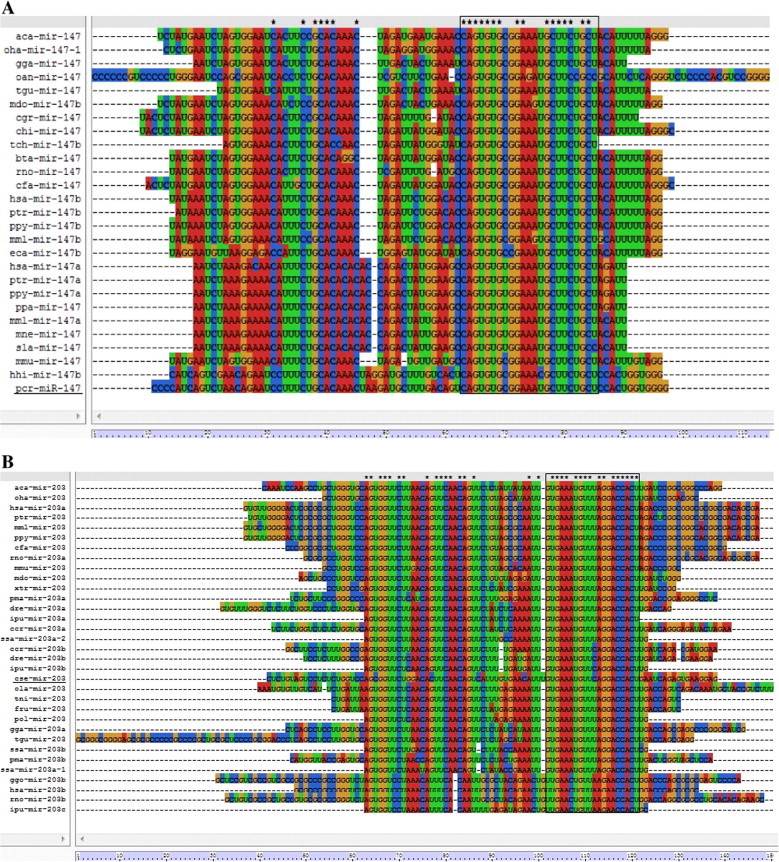
Sequence alignment of pre-miRNAs in each miRNA family. Alignments of known animal miRNAs and their newly annotated homologs are presented as follows: (A) miR147; (B) miR203. The names of the miRNAs identified in this study are underlined. Asterisks indicate conserved region in mature sequences.
Figure 3.
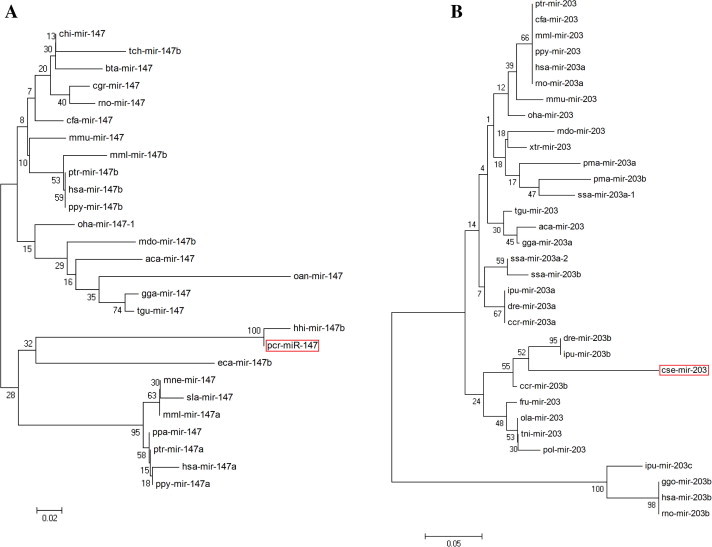
Phylogenetic tree for the newly identified miRNA showing homology. Identified fish miRNA is shown in red box. (A) miR-147; (B) miR-203.
In addition, in these newly identified miRNAs, miR-2444 was found in two fish species, cse and pcr; miR-2293 was found in hmo and ano; miR-297 was found in pfu, mal and pcr; miR-2304 was found in pfu and asi, respectively; which are presumably considered to be evolutionarily conserved regulators of gene expression. Our current findings indicate that the miRNAs from these lower vertebrates lineages were complex, and more data are urgently required to better understand their evolution.
3.3. Prediction of potential targets of identified miRNA
Target identification is essential for understanding the biological functions of miRNAs. Using a combination of BLAST and RNA-hybrid online software, a total of 42 putative target genes were identified in eleven fish species, and these targets belong to a variety of gene families that partake in various biological and physiological functions (Table 2). Studies’ estimate has stated that miRNAs have approximately 100 target sites within the protein-coding genes (Brennecke et al., 2005). Additionally, miRNAs are thought to target more than 30% of protein-coding genes in humans and this number is expected to rise as more miRNAs are discovered (Lewis et al., 2005). So, some miRNAs, more than one potential target gene were predicted in our research. Among 43 identified miRNAs, nine failed to predict their target genes, which are mpi-miR-6835-3p, cid-miR-5192, pfu-miR-29, pfu-miR-2304, pfu-miR-297, mal-miR-4709, car-miR-4433b-3p, pcr-miR-297 and asi-miR-2304. The situation may result from these factors: (a) the lack of genomic information in related fish species and their targets cannot be predicted; (b) the target gene prediction program was struck and probably some miRNA targets were missed.
Table 2.
List of potential targets of our identified miRNAs in 11 fish species.
| miRNA | Targeted protein | Target function | Genes ID |
|---|---|---|---|
| mpi-miR-3245 | Mitochondrial antiviral signaling protein | Signal transduction | 521311590 |
| mpi-miR-4054 | Zinc finger and BTB domain containing 22 protein | Transcription factor | 319429530 |
| Glycosyltransferase | Metabolism | 319429441 | |
| mpi-miR-222 | Beta-actin protein | Development | 31323261 |
| cid-miR-2437 | Metallothionein | Metabolism | 459463736 |
| cid-miR-3198 | Trypsinogen | Development | 241911727 |
| cid-miR-223 | Nonspecific cytotoxic cell receptor protein | Transcription factor | 327344086 |
| Toll-like receptor 21 | Signal transduction | 506956260 | |
| cid-miR-1814b | Cytosolic malate dehydrogenase | Metabolism | 186908741 |
| hmo-miR-2192 | Glucose phosphate isomerase | Metabolism | 337255732 |
| Copper/zinc superoxide dismutase | Metabolism | 300087118 | |
| hmo-miR-2293 | Lipoprotein lipase | Metabolism | 253317430 |
| Putative interleukin-8 like protein | Immunoregulation | 205278402 | |
| ano-miR-2800 | Glutathione reductase-like protein | Metabolism | 239950053 |
| ano-miR-2293 | Parvalbumin | Metabolism | 204324084 |
| ano-miR-1603 | Transmembrane protein 120B | Signal transduction | 226358576 |
| pfu-miR-669 | Ribosomal protein L15 | Development | 254908960 |
| mal-miR-4040–3p | Glutamate dehydrogenase | Metabolism | 371491860 |
| mal-miR-297 | Insulin-like growth factor 1 receptor | Transcription factor | 663440153 |
| mal-miR-42 | Na+/K+-ATPase | Signal transduction | 540352503 |
| mal-miR-4194–3p | MHC class II antigen | Immunoregulation | 51256194 |
| sch-miR-2437 | Nucleocapsid protein | Environmental stress response | 4443086 |
| RNA-dependent RNA polymerase | Development | 4443091 | |
| car-miR-125b | NADH dehydrogenase | Metabolism | 10251172 |
| cse-miR-2191 | Interleukin enhancer binding factor 2 | Transcription factor | 103394462 |
| cse-miR-2316 | Transfer RNA glutamic acid | Metabolism | 103352779 |
| cse-miR-203b-3p | Interferon regulatory factor 1 | Immunoregulation | 103394766 |
| cse-miR-190a-3p | (Asp-Glu-Ala-Asp) box polypeptide | Metabolism | 103389588 |
| IKAROS family zinc finger 1 | Transcription factor | 103387497 | |
| cse-miR-2444 | Growth hormone receptor | Transcription factor | 103397680 |
| pcr-miR-431–3p | G-lysozyme | Environmental stress response | 150034872 |
| Immunoglobulin IgL light chain precursor protein | Immunoregulation | 113197015 | |
| pcr-miR-6837 | NADH dehydrogenase | Metabolism | 7095387 |
| pcr-miR-147 | ATP synthase | Development | 709538 |
| pcr-miR-34 | Tumor necrosis factor alpha protein | Environmental stress response | 121044680 |
| pcr-miR-4695–5p | Growth hormone | Signal transduction | 11231167 |
| pcr-miR-2444 | Proteasome activator | Transcription factor | 95105543 |
| Interferon-inducible protein 56 | Immunoregulation | 164422176 | |
| pcr-miR-2415 | Growth differentiation factor-8 | Development | 74099690 |
| asi-miR-965–5p | Cytochrome | Metabolism | 7804435 |
| asi-miR-374a | Nanos1 | Transcription factor | 401709452 |
| asi-miR-86 | Neuroendocrine protein (7B2) | Signal transduction | 315506996 |
These predicted targets are found to be involved in immune-related, signaling, transcription factors, metabolism, transportation, growth and development, responses to diseases and environmental stresses and others proteins (Table 2). For example, mpi-miR-4054 targets the zinc finger and BTB domain containing 22 protein transcription factors, which may play a role in gene regulation of fish growth and development. Pcr-miR395 targets the ATP synthase, which may involve in oxidative phosphorylation, oxidation–reduction/redox reactions in fish organism. Several miRNAs can target genes involved in signal transduction, especially hormone signaling pathways. The growth hormone protein which are thought to regulate transcription in response to auxin, contain potential pcr-miR-4695-5p binding sites. In addition, some targets of miRNAs are involved in metabolism, development, responses to diseases and environmental stress, such as cid-miR-2437 targets metallothionein, sch-miR-2437 targets nucleocapsid protein, pfu-miR-669 targets ribosomal protein L15, mal-miR-4194–3p targets MHC class II antigen, respectively. Similar findings were reported by many groups in different animal species (Barozai, 2012a; Carrington and Ambros, 2003; Gong et al., 2010; Jagadeeswaran et al., 2010). Future experimental validation will determine how many of these predicted targets are genuinely targeted by miRNAs in these eleven fish species.
4. Conclusions
In this report, a bioinformatics pipeline was applied to discover the existence of miRNAs in eleven fish species from EST and GSS sequences, all miRNAs are not reported before. By using the sequences of the known animal miRNAs, we identified 43 new miRNAs with high confidence belonging to 38 miRNA families. A total of 42 potential targets are also identified. These findings of miRNA will be helpful to understand the gene regulation concept in these fish species. Moreover, it shows an easy approach for the prediction and analysis of miRNAs to those species whose genomes are not available.
Acknowledgements
This research was supported by the Natural Science Foundation of China (31302013) and Doctoral Science Foundation (09001578) and Natural Science Innovation and Development Foundation (2013ZCX014) of Henan University of Science and Technology.
Footnotes
Peer review under responsibility of King Saud University.
Appendix A. Supplementary data
Supplementary Figure S1.
References
- Akter A., Islam M.M., Mondal S.I., Mahmud Z., Jewel N.A., Ferdous S., Amin M.R., Rahman M.M. Computational identification of miRNA and targets from expressed sequence tags of coffee (Coffea arabica) Saudi J. Biol. Sci. 2014;21:3–12. doi: 10.1016/j.sjbs.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ambros V., Bartel B., Bartel D.P., Burge C.B., Carrington J.C., Chen X., Dreyfuss G., Eddy S.R., Griffiths-Jones S., Marshall M., Matzke M., Ruvkun G., Tuschl T. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V., Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134:1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- Barozai M.Y. Identification and characterization of the microRNAs and their targets in Salmo salar. Gene. 2012;499:163–168. doi: 10.1016/j.gene.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Barozai M.Y. The novel 172 sheep (Ovis aries) microRNAs and their targets. Mol. Biol. Rep. 2012;39:6259–6266. doi: 10.1007/s11033-012-1446-x. [DOI] [PubMed] [Google Scholar]
- Bartel B. MicroRNAs directing siRNA biogenesis. Nat. Struct. Mol. Biol. 2005;12:569–571. doi: 10.1038/nsmb0705-569. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Belver L., de Yebenes V.G., Ramiro A.R. MicroRNAs prevent the generation of autoreactive antibodies. Immunity. 2010;33:713–722. doi: 10.1016/j.immuni.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington J.C., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chen J., Chen W., Li Y. Conservation of gene order in human microRNA-neighboring regions. Genome. 2012;55:701–704. doi: 10.1139/g2012-055. [DOI] [PubMed] [Google Scholar]
- Daido Y., Hamanishi S., Kusakabe T.G. Transcriptional co-regulation of evolutionarily conserved microRNA/cone opsin gene pairs: implications for photoreceptor subtype specification. Dev. Biol. 2014;392:117–129. doi: 10.1016/j.ydbio.2014.04.021. [DOI] [PubMed] [Google Scholar]
- Ding J., Zhou S., Guan J. MiRenSVM: towards better prediction of microRNA precursors using an ensemble SVM classifier with multi-loop features. BMC Bioinf. 2010;11(Suppl. 11):S11. doi: 10.1186/1471-2105-11-S11-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q.H., Han J., Yu H.P., Wang C., Zhao M.Z., Liu H., Ge A.J., Fang J.G. Computational identification of MicroRNAs in strawberry expressed sequence tags and validation of their precise sequences by miR-RACE. J. Hered. 2012;103:268–277. doi: 10.1093/jhered/esr127. [DOI] [PubMed] [Google Scholar]
- Gong P., Xie F., Zhang B., Perkins E.J. In silico identification of conserved microRNAs and their target transcripts from expressed sequence tags of three earthworm species. Comput. Biol. Chem. 2010;34:313–319. doi: 10.1016/j.compbiolchem.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Gorodkin J., Havgaard J.H., Enstero M., Sawera M., Jensen P., Ohman M., Fredholm M. MicroRNA sequence motifs reveal asymmetry between the stem arms. Comput. Biol. Chem. 2006;30:249–254. doi: 10.1016/j.compbiolchem.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran G., Zheng Y., Sumathipala N., Jiang H., Arrese E.L., Soulages J.L., Zhang W., Sunkar R. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics. 2010;11:52. doi: 10.1186/1471-2164-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavan R., Virata M., Keshavan A., Zeller R.W. Computational identification of Ciona intestinalis microRNAs. Zool. Sci. 2010;27:162–170. doi: 10.2108/zsj.27.162. [DOI] [PubMed] [Google Scholar]
- Kim S., Hwang do W., Lee D.S. A study of microRNAs in silico and in vivo: bioimaging of microRNA biogenesis and regulation. FEBS J. 2009;276:2165–2174. doi: 10.1111/j.1742-4658.2009.06935.x. [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Plasterk R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Ladomery M.R., Maddocks D.G., Wilson I.D. MicroRNAs: their discovery, biogenesis, function and potential use as biomarkers in non-invasive prenatal diagnostics. Int. J. Mol. Epidemiol. Genet. 2011;2:253–260. [PMC free article] [PubMed] [Google Scholar]
- Lee C.T., Risom T., Strauss W.M. Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol. 2007;26:209–218. doi: 10.1089/dna.2006.0545. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Liu X., Fortin K., Mourelatos Z. MicroRNAs: biogenesis and molecular functions. Brain Pathol. 2008;18:113–121. doi: 10.1111/j.1750-3639.2007.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Zhang S. Computational prediction of amphioxus microRNA genes and their targets. Gene. 2009;428:41–46. doi: 10.1016/j.gene.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Maher C., Stein L., Ware D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006;16:510–519. doi: 10.1101/gr.4680506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallanna S.K., Rizzino A. Emerging roles of microRNAs in the control of embryonic stem cells and the generation of induced pluripotent stem cells. Dev. Biol. 2010;344:16–25. doi: 10.1016/j.ydbio.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A.K., Lobiyal D.K. MiRNA prediction using computational approach. Adv. Exp. Med. Biol. 2011;696:75–82. doi: 10.1007/978-1-4419-7046-6_8. [DOI] [PubMed] [Google Scholar]
- Neutelings G., Fenart S., Lucau-Danila A., Hawkins S. Identification and characterization of miRNAs and their potential targets in flax. J. Plant Physiol. 2012;169:1754–1766. doi: 10.1016/j.jplph.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Niwa R., Slack F.J. The evolution of animal microRNA function. Curr. Opin. Genet. Dev. 2007;17:145–150. doi: 10.1016/j.gde.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Paul P., Chakraborty S. Computational prediction of submergence responsive microRNA and their binding position within the genome of Oryza sativa. Bioinformation. 2013;9:858–863. doi: 10.6026/97320630009858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Chen L., Zhang Y., Kang X., Zhang Z., Wang Y. Identification of novel and conserved Populus tomentosa microRNA as components of a response to water stress. Funct. Integr. Genomics. 2012;12:327–339. doi: 10.1007/s10142-012-0271-6. [DOI] [PubMed] [Google Scholar]
- Rogers K., Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25:2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetrom P., Snove O., Nedland M., Grunfeld T.B., Lin Y., Bass M.B., Canon J.R. Conserved microRNA characteristics in mammals. Oligonucleotides. 2006;16:115–144. doi: 10.1089/oli.2006.16.115. [DOI] [PubMed] [Google Scholar]
- Sun K., Lai E.C. Adult-specific functions of animal microRNAs. Nat. Rev. Genet. 2013;14:535–548. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takane K., Fujishima K., Watanabe Y., Sato A., Saito N., Tomita M., Kanai A. Computational prediction and experimental validation of evolutionarily conserved microRNA target genes in bilaterian animals. BMC Genomics. 2010;11:101. doi: 10.1186/1471-2164-11-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer A., Stadler P.F. Molecular evolution of a microRNA cluster. J. Mol. Biol. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- van der Burgt A., Fiers M.W., Nap J.P., van Ham R.C. In silico miRNA prediction in metazoan genomes: balancing between sensitivity and specificity. BMC Genomics. 2009;10:204. doi: 10.1186/1471-2164-10-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wei B., Liu H., Li T., Rayner S. MiRPara: a SVM-based software tool for prediction of most probable microRNA coding regions in genome scale sequences. BMC Bioinf. 2011;12:107. doi: 10.1186/1471-2105-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.H., Li F., Sun Q.F. Identification of microRNA precursors with support vector machine and string kernel. Genomics Proteomics Bioinf. 2008;6:121–128. doi: 10.1016/S1672-0229(08)60027-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef M., Showe L., Showe M. A study of microRNAs in silico and in vivo: bioinformatics approaches to microRNA discovery and target identification. FEBS J. 2009;276:2150–2156. doi: 10.1111/j.1742-4658.2009.06933.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1.


