Abstract
OBJECTIVES
This study evaluates the repeatability of brain perfusion using dynamic susceptibility contrast magnetic resonance imaging (DSC-MRI) with a variety of post-processing methods.
METHODS
Thirty-two patients with newly diagnosed glioblastoma were recruited. On a 3-T MRI using a dual-echo, gradient-echo spin-echo DSC-MRI protocol, the patients were scanned twice 1 to 5 days apart. Perfusion maps including cerebral blood volume (CBV) and cerebral blood flow (CBF) were generated using two contrast agent leakage correction methods, along with testing normalization to reference tissue, and application of arterial input function (AIF). Repeatability of CBV and CBF within tumor regions and healthy tissues, identified by structural images, was assessed with intra-class correlation coefficients (ICCs) and repeatability coefficients (RCs). Coefficients of variation (CVs) were reported for selected methods.
RESULTS
CBV and CBF were highly repeatable within tumor with ICC values up to 0.97. However, both CBV and CBF showed lower ICCs for healthy cortical tissues (up to 0.83), healthy gray matter (up to 0.95), and healthy white matter (WM; up to 0.93). The values of CV ranged from 6% to 10% in tumor and 3% to 11% in healthy tissues. The values of RC relative to the mean value of measurement within healthy WM ranged from 22% to 42% in tumor and 7% to 43% in healthy tissues. These percentages show how much variation in perfusion parameter, relative to that in healthy WM, we expect to observe to consider it statistically significant. We also found that normalization improved repeatability, but AIF deconvolution did not.
CONCLUSIONS
DSC-MRI is highly repeatable in high-grade glioma patients.
Introduction
Magnetic resonance imaging (MRI) has been used to quantify blood perfusion of the brain for nearly a quarter of a century [1,2]. Cerebral blood volume (CBV) and cerebral blood flow (CBF) have been recognized as biomarkers in a number of neurological diseases, in particular brain tumors [3]. In antiangiogenic therapy of brain tumors, and gliomas in particular, CBV and CBF correlate with tumor grade and response to therapy [4–7]. Changes in blood perfusion are correlated with disease-free survival, best shown with a longitudinal analysis of dynamic susceptibility contrast MRI (DSC-MRI) [8,9].
Monitoring the therapeutic response of anti-vasogenic drugs has increased the interest of DSC-MRI for brain tumors [5–9]. Some studies have shown that DSC-MRI is an imaging biomarker for cerebral perfusion, making it useful for evaluating novel anti-vasogenic agents [7,9]. More recently, dual gradient-echo (GE), spin-echo (SE) DSC-MRI pulse sequences quantify vessel caliber and distinguish low-oxygenation veins from higher oxygenation arteries within the tumor core, which may be predictive of overall survival [10].
One aspect in the evaluation of a biomarker is the reliability and accuracy of the measurements [11]. True changes in the measured perfusion parameters caused by a therapy (regardless of therapy having significant clinical outcome) may be determined by both the measurement error and physiological variations in the tissues. Recently, the Quantitative Imaging Biomarkers Alliance initiative has sought to identify the sources of variation contributing to the measurement error and to improve repeatability and reproducibility of such quantitative imaging biomarkers. This is important to translate imaging biomarkers into clinical trials and general practice. In particular, intra-scanner variations contribute to the measurement error and may be evaluated by repeatability analyses.
The repeatability of DSC-MRI is affected by both image acquisition and post-processing. Image acquisition includes signal-to-noise ratio, susceptibility artifact, temporal and spatial resolution, length of imaging, motion artifact, signal saturation [12], misregistration due to frequency shifts induced by contrast agent (CA) [13], and the degree of T1 contamination [14]. Post-processing includes the applicability of the applied kinetic model, accuracy in arterial input function (AIF) detection and deconvolution, accuracy of geometric mapping of selected anatomic regions of interest (ROIs) to the perfusion maps, and the chosen calibration and normalization techniques.
Earlier studies have evaluated the optimal DSC image acquisition and post-processing approaches [14–16,2]. Several studies have also evaluated the repeatability of DSC quantification using repeated scans [17–22]. However, most repeatability studies of brain DSC-MRI have been performed for healthy subjects. Only one repeatability study evaluated DSC in gliomas but had a small cohort of patients [17].
Here, we evaluate the repeatability of dual GE and SE DSC-based perfusion on a 3-T MRI scanner using a double baseline MRI acquisition setup in 32 patients with newly diagnosed glioblastoma. We compare CBV and CBF from two MRI exams taken within 1 to 5 days in both healthy tissue and tumor using two of the state-of-the-art leakage correction methods and an automated AIF detection algorithm. By assessing the repeatability of DSC-MRI, we quantify the variations in these measurements caused by the intra-scanner and normal biologic variations using current methodologies, with the goal of defining true changes of perfusion in human brain tumors. This will strengthen its use as an imaging biomarker. We hope this study will contribute to the understanding the sources of measurement variability in DSC-MRI.
Materials and Methods
Patients
The Institutional Review Board approved this study. Informed consent was obtained from all patients. We recruited 40 adult patients with newly diagnosed grade IV glioblastomas in a study of cediranib, a small molecule pan–vascular endothelial growth factor tyrosine kinase inhibitor, sponsored by the National Cancer Institute (R01CA129371) [23–25]. Of 40 patients, eight were eliminated because of missing a scan (n = 4) or inadequate image quality (n = 4). The remaining 32 patients (19 M, 13 F, age 23-75, mean 56) were included in the study. All patients were on a stable or decreasing dose of steroids for 5 days before each scan. The patients did not receive cediranib before or between the two scans. No tumor had significant progression between the two scans as measured by change in contrast enhancing tumor (ET) volume or fluid-attenuated inversion recovery (FLAIR) hyperintensity (FH). Most tumors were unifocal and some were midline, but these factors should not influence the results of our analysis as we compared the DSC results within patients.
Magnetic Resonance Imaging
Each subject was scanned at two time points (hereafter referred to as baselines 1 and 2) using an identical imaging protocol on a 3.0-T MRI System (TimTrio; Siemens Medical Solutions, Malvern, PA). The scans were done 1 to 5 days apart (mean ± SD = 3.7 ± 1.4) to allow the contrast to wash out between scans. AutoAlign was used to ensure that the brain was oriented the same way for every scan. DSC-MRI was acquired as follows: a 78-mm slab of tissue was imaged using a dual-echo, combined GE and SE echo planar imaging sequence, with echo times of 31 and 94 milliseconds, respectively, a repetition time of 1.48 seconds, a flip angle of 90°, and 1.2-mm in-plane (160 × 160 matrix) and 6.5-mm through-plane resolution. A total of 100 volumes were acquired and 0.1 mmol/kg Gd-DTPA (Magnivist) was injected at 5 ml/s after approximately 81 seconds of imaging. The DSC acquisition was preceded by a dynamic contrast-enhanced acquisition (0.1 mmol/kg Gd-DTPA injected at 5 ml/s), and this CA injection acted as a pre-dose to minimize errors in CBV estimates due to T1-shortening effects induced by CA extravasation in regions of blood-brain barrier breakdown or resection.
In addition, structural images including axial FLAIR with 0.43 mm × 0.43 mm × 6 mm resolution, post-contrast T1-weighted image with 0.43 mm × 0.43 mm × 6 mm resolution, and magnetization-prepared rapid GE (MPRAGE) with 1-mm isotropic voxels were acquired.
Perfusion Analysis
Before perfusion analysis, potential motion artifacts (motion between time points) in the DSC data were detected and corrected using FSL (http://www.fmrib.ox.ac.uk/fsl/). Figure 1 shows the analysis methods that were used in the experiments. All perfusion analyses as described in the Supplementary Materials were performed in a modified version of nordicICE (NordicNeuroLab AS, Bergen, Norway).
Figure 1.
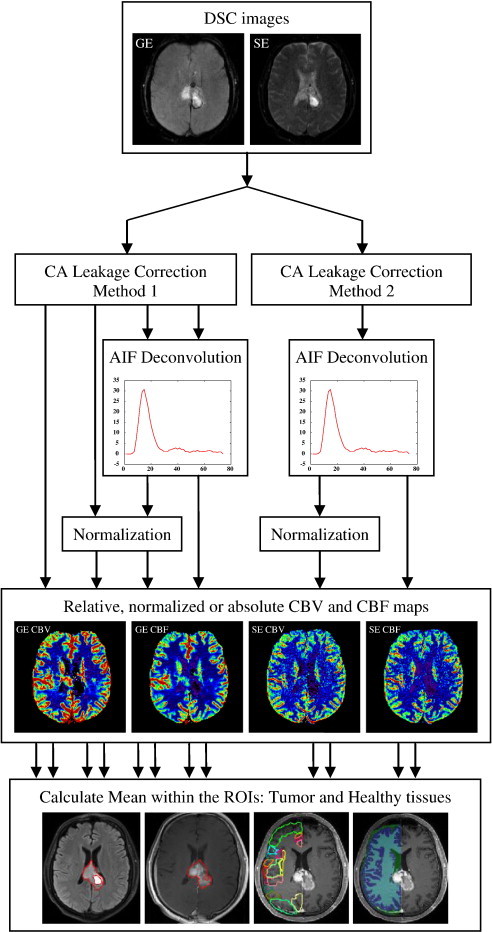
Flowchart summarizing the analysis steps for the different methods used for generation of hemodynamic images and consequent ROI analysis. The bottom row from left to right shows FH ROI, ET ROI, healthy cortical tissues, and healthy WM and GM ROIs, respectively.
Kinetic modeling
Two established kinetic models for CA extravasation correction were compared: Method 1 is based on the correction technique proposed by Weisskoff et al. [26] and later elaborated by Donahue et al. [27] and Boxerman et al. [28]. Method 2 is based on the estimation of CA extravasation from the tissue residue function [29]. AIF deconvolution is by default applied in method 2, but it is an option for method 1.
AIF determination and normalization
The AIFs were determined automatically in each patient by an established clustering method [30] that has been shown to have a good agreement with manual AIF selection and provide physiologically plausible perfusion estimates without user interaction [31].
Normalization to a reference tissue mask improves the reliability of CBV and CBF measurements [32,33]. Reference tissue masks representing unaffected gray matter (GM) and white matter (WM) were generated automatically from the DSC images as previously described [34,31]. A combined WM + GM mask was used to generate the reference tissue response curves used for CA extravasation correction according to method 1 and also to generate normalized CBV and CBF maps by dividing the respective perfusion parameters by their corresponding mean value in the WM + GM mask.
The CBV and CBF maps were calculated both with and without applying AIF deconvolution (for method 1, the residue function is corrected directly for leakage effects and CBV is then estimated from the corrected residue function rather than from the normal raw data integral ratio) and with and without normalization to the WM + GM mask for both GE and SE images (Figure 1).
Image Analysis
For each subject, tumor and healthy tissue ROIs were generated as described below for baseline 1 and mapped to baseline 2 by rigid coregistration to ensure that the same regions were evaluated in both baselines and to eliminate the inter-rater variations of ROI drawing. ET was outlined on 3-dimensional (3D) post-contrast T1-weighted axial images by an experienced neuroradiologist with 5 years of clinical experience in brain tumor imaging [35]. Likewise, FH was outlined on 3D FLAIR images. Several cortical regions were also automatically segmented by non-rigid registration of the Harvard-Oxford Atlas to the MPRAGE images using ANTS [36]. Regions contralateral to the tumor were considered “healthy” brain (see Figures 1, S1, and S2).
MPRAGE volumes were also segmented into healthy WM and GM using FSL to create a separate label volume for the healthy (or healthier) hemisphere (distinct from the WM + GM reference mask automatically generated from DSC-MRI) [37]. Sample ROIs are shown in Figure 1. Finally, all labels were mapped to the perfusion maps by rigid coregistration in SPM (http://www.fil.ion.ucl.ac.uk/spm/). The mean values of the generated perfusion maps were calculated within each ROI.
Statistical Analysis
Intra-class correlation coefficients (ICCs) and repeatability coefficients (RCs) were computed in MATLAB (The Mathworks, Natick, MA) for the tumor and healthy tissue ROIs. ICC was calculated using a two-way random effect model [38]. ICC ranges from 0 to 1 and describes how strongly two within-subject measurements resemble each other relative to the variation between subjects. RC represents the estimated range of variation between the measurements so that the difference between two measurements is expected to be between − RC and RC for 95% of patients. RC was calculated as where is equal to the standard deviation of the measurement differences. RC reflects how much change should occur to be considered significant [39]. For healthy tissues segmented by atlas-based segmentation, averages of ICCs and RCs were reported.
We also calculated the ratio of RC to the mean value of the measurement within healthy WM for selected methods. Assuming that the mean values of perfusion parameters in healthy WM are similar among patients, these values reported in percentage show how much variation, due to both measurement error and physiological variations, we may expect relative to the mean value within healthy WM. This measure is more informative than a measure such as within-subject coefficient of variation (CV, defined as σW/μ, where μ is mean value), which depends on the mean value of all measurements and thus varies with the tissue type and patient population. However, we also reported CV for select methods.
To compare the ICC of two methods, a bootstrapping approach with 5000 repetitions to generate samples was used for hypothesis testing [40]. In each repetition of the bootstrap procedure, we randomly resampled the available data with replacement such that the size of each subsample is the same as the size of the original data. Then, the ICC means of both methods were calculated from the generated bootstrapped samples and compared through a mean hypothesis testing procedure [41]. Using the false discovery rate control [42], the significance level of each hypothesis test was corrected for multiple comparisons with an overall significance level of 0.05.
Results
Very high repeatability was obtained for all perfusion metrics, with ICC values > 0.9 in tumor for both GE CBV and CBF after normalization. A comprehensive table of the ICCs and RCs is provided in the Supplementary Materials (Tables S1–S2). Overall, both methods had excellent ICCs after normalization. Method 1 had slightly higher ICCs. Using this method, the GE CBV maps had very high ICC within ET (ICC = 0.96) and FH (ICC = 0.97). The same was true for CBF (ICC = 0.97 for ET and ICC = 0.96 for FH).
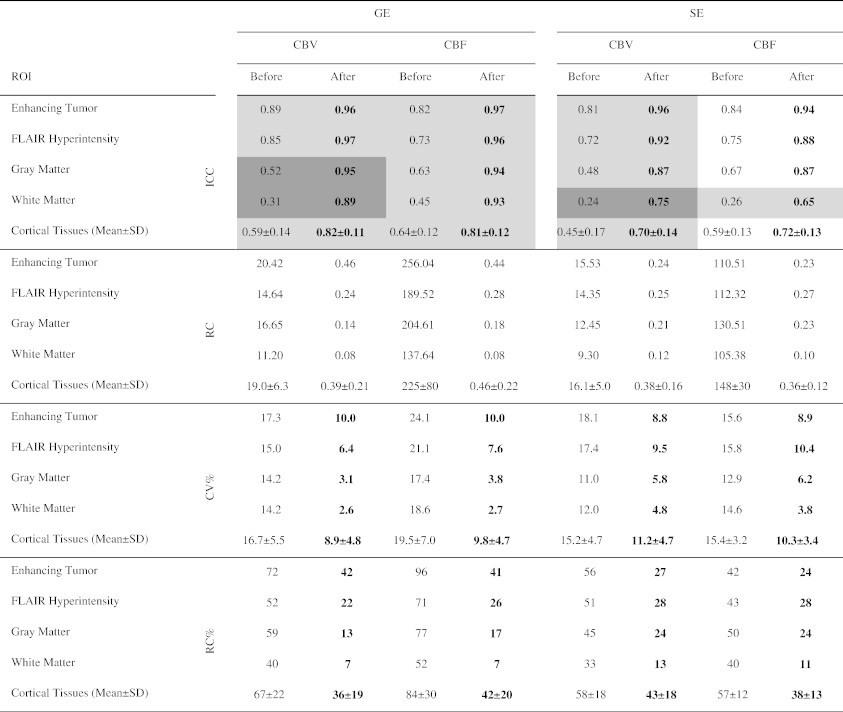
Table 1 compares the ICCs, RCs, and CVs before and after normalization using method 1 with AIF deconvolution. Normalization always improved the ICCs of both GE and SE maps (P < .05 is highlighted). False discovery rates are highlighted in darker gray (corresponding to P < .0032). After normalization, WM and GM had a lower RC compared with tumor regions, indicating that their intra-subject variability was lower compared with tumor regions. Furthermore, normalization always decreased CV. For GE CBV, GE CBF, SE CBV, and SE CBF, the CV values after normalization ranged from 8.8% to 10.0% for ET, 6.4% to 10.4% for FH, 3.1% to 6.2% for GM, and 2.6% to 4.8% for WM. However, since CV is dependent on the mean value of measurements and there is variability in tumor perfusion, we have also reported the ratios (percentages) of RCs to the mean value of measurements within healthy WM of all subjects in Table 1. As shown, normalization often reduced the variance resulting in more stable measurements. Note that the high percentages of variations for tumor regions are partly due to higher perfusion in these regions compared to the healthy WM.
Table 1.
Comparison of ICCs, RCs, CVs, and Ratios (Percentages) of RC and Mean Value of Measurement within Healthy WM before and after Normalization. The values Were Generated Using Method 1 with AIF Deconvolution. Comparisons for ICCs with P < .05 Are Highlighted in Gray. Statistically Significant Comparisons Are Highlighted in Dark Gray. Higher ICCs and Lower CV and RC Percentages Are in Boldface
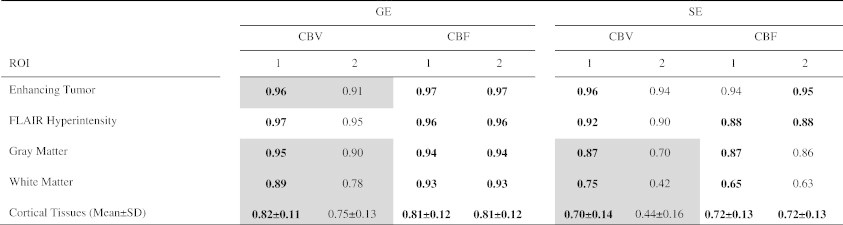
Tables 2 and 3 evaluate the effect of AIF deconvolution. Using method 1 and when normalization was not applied, the application of AIF deconvolution on average decreased the ICCs for GE sequence with some P values under .05 (Table 2). This comparison was statistically significant for SE CBF of WM. Using method 1 and when normalization was applied, the application of AIF deconvolution on average did not significantly change the ICCs (Table 3). In method 2, AIF deconvolution is applied by default. When compared with method 1 with AIF deconvolution, method 2 often trended toward lower ICCs for GE sequence (Table 4), with some P values under .05 but not statistically significant by false discovery rate control.
Table 2.
Comparison of ICCs before and after AIF Deconvolution. The ICCs Were Generated Using Method 1 without Normalization. Comparisons with P < .05 Are Highlighted in Gray. Statistically Significant Comparisons Are Highlighted in Dark Gray
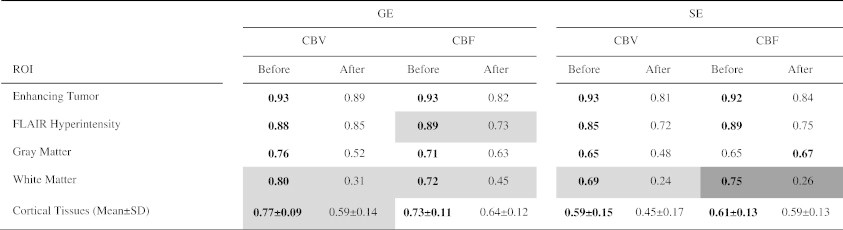
Table 3.
Comparison of ICCs before and after AIF Deconvolution. The ICCs Were Generated Using Method 1 with Normalization. The Differences between ICCs Were Not Statistically Significant
| GE |
SE |
|||||||
|---|---|---|---|---|---|---|---|---|
| CBV |
CBF |
CBV |
CBF |
|||||
| ROI | Before | After | Before | After | Before | After | Before | After |
| Enhancing Tumor | 0.96 | 0.96 | 0.96 | 0.97 | 0.96 | 0.96 | 0.95 | 0.94 |
| FLAIR Hyperintensity | 0.97 | 0.97 | 0.95 | 0.96 | 0.92 | 0.92 | 0.93 | 0.88 |
| Gray Matter | 0.95 | 0.95 | 0.93 | 0.94 | 0.87 | 0.87 | 0.87 | 0.87 |
| White Matter | 0.90 | 0.89 | 0.88 | 0.93 | 0.76 | 0.75 | 0.76 | 0.65 |
| Cortical Tissues (Mean±SD) | 0.82 ± 0.11 | 0.82 ± 0.11 | 0.83 ± 0.10 | 0.81 ± 0.12 | 0.71 ± 0.14 | 0.70 ± 0.14 | 0.74 ± 0.09 | 0.72 ± 0.13 |
Table 4.
Comparison of ICCs for Methods 1 and 2. The ICCs Were Generated with AIF Deconvolution and Normalization. Comparisons with P < .05 Are Highlighted in Gray
On average, ICC of both CBV and CBF within the FH region was comparable to the ICC of the ET after normalization. For WM and GM and healthy cortical tissues, both ICCs and RCs were generally lower than those of the tumor regions. In particular, CBV and CBF, respectively, had maximum ICCs of 0.95 and 0.94 in GM, 0.90 and 0.93 in WM, and 0.82 ± 0.11 and 0.83 ± 0.10 in cortical regions.
The Bland-Altman plots of the GE parametric maps of tumor, WM, and GM regions demonstrate that both inter-subject variability and intra-subject variability were lower in healthy WM and GM compared with tumor regions (Figures 2 and 3). Furthermore, inter-subject variability was lower within the FH compared with ET. Intra-subject variability was also slightly lower (Table 1). The mean ± SD values of GE CBV generated by method 1 with AIF deconvolution and normalization were 1.65 ± 0.83 within ET, 1.36 ± 0.48 within FH, 1.63 ± 0.22 within GM, and 1.09 ± 0.09 within WM. For SE CBV, these values were 0.99 ± 0.42, 0.95 ± 0.32, 1.31 ± 0.20, and 0.89 ± 0.09, respectively (see Table S3 for a complete list of mean ± SD values).
Figure 2.
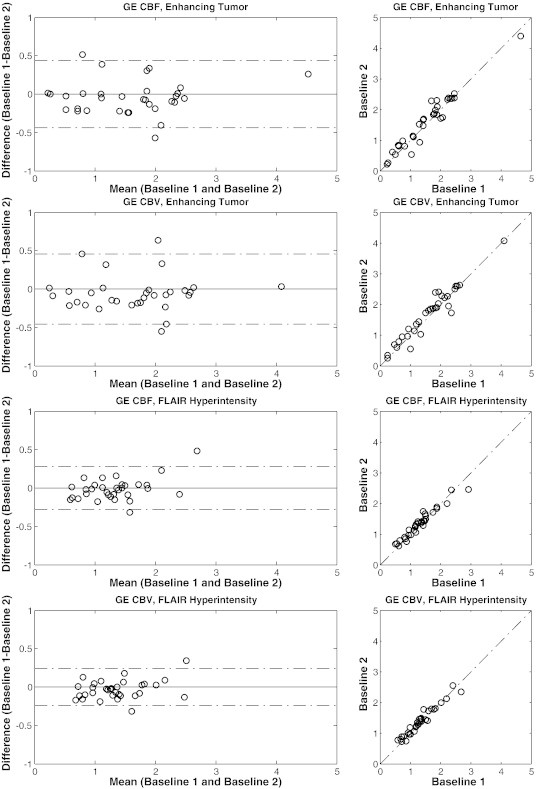
Bland-Altman scatter plots of the GE parametric maps within ET and FH ROIs generated by method 1 with AIF deconvolution and normalization. The dotted lines on the left show the 95% confidence interval.
Figure 3.
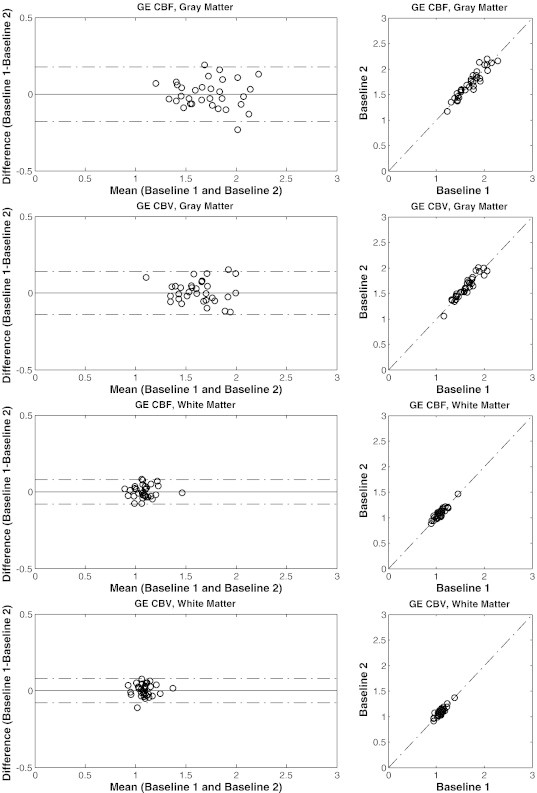
Bland-Altman scatter plots of the GE parametric maps within WM and GM ROIs generated by method 1 with AIF deconvolution and normalization. The dotted lines on the left show the 95% confidence interval.
High ICC values were also obtained for CBV and CBF derived from the SE sequence within ET and FH (ICC ≥ 0.9 after normalization) but still lower compared with the same parameters obtained from the GE data. Using method 1 with normalization for CBV, ICC = 0.96 within ET and ICC = 0.92 within FH (Table 1). For CBF, ICC = 0.94 within ET and ICC = 0.88 within FH. Similar to GE maps, the ICCs were lower within the cortical tissues in comparison with tumor regions. Normalization often improved the ICCs. The effect of AIF deconvolution for method 1 is shown for SE images in Tables 2 and 3. Using method 1 and when normalization was not applied, AIF deconvolution decreased ICC in WM (P < .05). However, when normalization was applied, AIF deconvolution did not significantly change the ICCs (Table 3). Similar to GE sequence, when comparing with method 1 with AIF deconvolution (Table 4), method 2 often trended toward lower ICCs (P < .05 for some SE CBV values, yet not under the threshold of 0.0032 set by false discovery rate control).
Discussion
Targeting blood vessels has become an attractive strategy in cancer therapeutic interventions. The dependence of tumor growth on the recruitment of new blood vessels by angiogenesis has inspired the development of antiangiogenic approaches to cancer therapy. These new agents might perform best in combination with other traditional cytotoxic drugs, indicating that a systematic process of testing and optimization is necessary—a process greatly aided by non-invasive biomarkers. DSC-MRI may indeed provide such a biomarker, notably by measuring changes in perfusion as well as microvessel caliber that accompany response [9,10].
Translation of such biomarkers into clinical trials and practice requires the identification of all sources of variation in the measurement, such as variations related to image acquisition, post-processing, and physiological variations. This is essential in determining the true changes caused by the therapy effect. The goal is to limit the measurement error related to such variations to reliably detect true changes. This study evaluated the measurement error related to within-scanner and physiological variations. This required repeated scans on the same scanner and we referred to it as repeatability and may be evaluated by the ICC and RC values [43,44]. We identified post-processing methods that have smaller contribution to the measurement error and showed that the resultant measurement error is small relative to inter-subject variation, resulting in improved repeatability. The AIF deconvolution had large contributions to the measurement error. This may be due to the variability introduced by AIF detection despite showing good agreement with manual AIF detection in previous studies [30,31]. However, normalization significantly reduced such post-processing variations. AIF deconvolution may be beneficial for the estimation of regional flow variations (delay), CA leakage correction [29], and in MRI-based vessel caliber imaging and also multimodal image comparisons where a pseudo-quantitative CBV value might be relevant [45,46,31]. In this study, the reference tissues were generated automatically. Manual ROI selection would add some user bias, as it is difficult to place the ROIs in exactly the same positions in both sessions.
The WM GE CBV was slightly higher than 1 (1.09 ± 0.09), whereas WM SE CBV was slightly lower than 1 (0.89 ± 0.09). Note that the nordicICE WM/GM segmentation algorithm removes macroscopic vessels since the method is based on segmentation from hemodynamic features derived from the voxel-wise time-intensity curves. WM segmentation from structural scans will, however, include vessels leading to an increase in relative WM CBV. As vessels are much more prominent in GE DSC than in SE DSC, they contribute to more elevated CBV values in GE compared to SE.
Note that since the ICC is related to the ratio of inter-subject variability to intra-subject variability, it measures the sensitivity of the measurement in distinguishing differences between subjects. Tumor regions had higher ICC values in comparison with healthy tissues despite having larger RC values, mainly due to larger inter-subject variance. To determine what level of change in the perfusion parameter should be observed to be confident that there is a true change in the parameter, the RC may be used [11]. The ability to determine that perfusion remains unchanged is part of the same assessment. We reported the values of RC relative to the mean value within healthy WM. The CV values were under 11%. This is very well in line with the goals of Quantitative Imaging Biomarkers Alliance that aims at reaching CV < 20%. Table 1 reveals that a change in GE CBV of ET greater than 42% relative to the mean value of CBV within WM may be considered significant with a 95% confidence (42% × 1.09/1.65 = 28% compared to the mean value within tumor). CBV RC is 22%, 13%, 7%, and 36% for GE CBV of FH, GM, WM, and healthy cortical tissues, respectively. For other perfusion parameters, relative RC values are reported in Table 1.
This study evaluated the repeatability of mean relative CBV (rCBV) and relative CBF. We chose mean value due to the variability of perfusion within the tumors. Other measurements such as maximal rCBV and relative CBF may also be used to evaluate tumor malignancy [47], but mean rCBV is the most commonly reported when measuring tumor response. We did not exclude patients who had prior surgery. Hemorrhage and post-surgery signal dropout had little impact on our measurements since such signal dropout is primarily an issue immediately after the surgery, whereas our patients were weeks/months away from the surgery. In addition, such changes affect both baselines similarly and have little impact on the repeatability analysis. Similarly, tumor hemorrhage and calcification as well as steroid have little effect on the repeatability analysis since they have similar effects on both baseline images.
To translate DSC-MRI into general practice, other sources of variations such as between-scanner variations and inter-rater variations for ROI placement need to be evaluated. This requires evaluation of a data set with each patient scanned on different scanners and is referred to as the reproducibility study. We had acquired both images on the same scanner and had eliminated the inter-rater variations by mapping the ROI of visit 1 to visit 2. This allowed us to independently evaluate the within-scanner and physiological variations. We generated the perfusion maps using readily available software. Although this makes the translation of DSC-MRI to clinical practice more feasible, reproducibility across such post-processing software tools will be evaluated in future studies.
In conclusion, GE and SE DSC maps are highly repeatable demonstrated by an optimized double baseline protocol. Normalization of the derived perfusion metrics to corresponding values in healthy tissue increases the repeatability of measurements, but AIF deconvolution may decrease it. Therefore, for most clinical scenarios, a normalized CBV or CBF without AIF deconvolution is acceptable and produce repeatable values.
Footnotes
This work was supported by NCI/NIH R21CA117079, R01CA129371, and K24CA125440 (T.B.); NIH S10RR023401, 5R01NS069696, and 5R01NS060918 (S.M.S.); Saic-Frederick Inc. grant 26XS263 (T.B.); Norwegian Research Council grant 191088/V50 and Norwegian Cancer Society Grant 3434180 (K.E.E.); Harvard Catalyst grant M01-RR-01066 (T.B.); and NIH Awards UL1 RR025758 (T.B. and B.R.) and R01NS059775 (O.W.); Harvard Catalyst | The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, NIH Award UL1 TR001102) and financial contributions from Harvard University and its affiliated academic healthcare centers. This work also involved the use of instrumentation supported by the NCRR Shared Instrumentation Grant Program (1S10RR023401 and 1S10RR023043). Disclosure/conflict of interest: K.E.E. and A.B. have intellectual property rights at NordicNeuroLab AS. A.B. is a board member at NordicNeuroLab. K.S. is a co-founder of Imaging Biometrics LLC, a company that develops medical image processing software. She and her family have equity interest in, receive income from, and own intellectual property being developed. P.Y.W. is on Advisory Boards for Genentech and Novartis. T.B., Pharmaceutical consulting: 1) Merck & Co., Inc., 2) Roche, 3) Kirin Pharmaceuticals; CME lectures/material: 1) Up to Date, Inc., 2) Robert Michael Educational Institute LLC, 3) Educational Concepts Group, 4) Research to Practice, 5) Oakstone Medical, 6) Publishing, 7) American Society of Hematology; other consulting: 1) Champions, 2) Biotechnology, 3) Advance Medical; research support: 1) Pfizer, 2) Astra Zeneca, 3) Millennium. B.R. is on the consultant-advisory board of Siemens Medical.
This article refers to supplementary materials, which are designated by Tables S1 to S3 and Figures S1 and S2 and are available online at www.transonc.com.
Appendix A. Supplementary data
Perfusion analysis, comprehensive tables of the ICCs and RCs, table of Mean±SD values of CBV and CBF maps, and figures showing the image analysis.
References
- 1.Villringer A, Rosen BR, Belliveau JW, Ackerman JL, Lauffer RB, Buxton RB, Chao YS, Wedeen VJ, Brady TJ. Dynamic imaging with lanthanide chelates in normal brain: contrast due to magnetic susceptibility effects. Magn Reson Med. 1988;6(2):164–174. doi: 10.1002/mrm.1910060205. [DOI] [PubMed] [Google Scholar]
- 2.Willats L, Calamante F. The 39 steps: evading error and deciphering the secrets for accurate dynamic susceptibility contrast MRI. NMR Biomed. 2012;26(8):913–931. doi: 10.1002/nbm.2833. [DOI] [PubMed] [Google Scholar]
- 3.Sorensen AG, Reimer P. Thieme; New York: 2000. Cerebral MR Perfusion Imaging: Principles and Current Applications. [Google Scholar]
- 4.Aronen HJ, Gazit IE, Louis DN, Buchbinder BR, Pardo FS, Weisskoff RM, Harsh GR, Cosgrove GR, Halpern EF, Hochberg FH. Cerebral blood volume maps of gliomas: comparison with tumor grade and histologic findings. Radiology. 1994;191(1):41–51. doi: 10.1148/radiology.191.1.8134596. [DOI] [PubMed] [Google Scholar]
- 5.Covarrubias DJ, Rosen BR, Lev MH. Dynamic magnetic resonance perfusion imaging of brain tumors. Oncologist. 2004;9(5):528–537. doi: 10.1634/theoncologist.9-5-528. [DOI] [PubMed] [Google Scholar]
- 6.Aronen HJ, Gazit IE, Louis DN, Buchbinder BR, Pardo FS, Weisskoff RM, Harsh GR, Cosgrove GR, Halpern EF, Hochberg FH. Glial tumor grading and outcome prediction using dynamic spin-echo MR susceptibility mapping compared with conventional contrast-enhanced MR: confounding effect of elevated rCBV of oligodendrogliomas [corrected] AJNR Am J Neuroradiol. 2004;25(2):214–221. [PMC free article] [PubMed] [Google Scholar]
- 7.Law M, Oh S, Johnson G, Babb JS, Zagzag D, Golfinos J, Kelly PJ. Perfusion magnetic resonance imaging predicts patient outcome as an adjunct to histopathology: a second reference standard in the surgical and nonsurgical treatment of low-grade gliomas. Neurosurgery. 2006;58(6):1099–1107. doi: 10.1227/01.NEU.0000215944.81730.18. [discussion-107] [DOI] [PubMed] [Google Scholar]
- 8.Gerstner ER, Sorensen AG, Jain RK, Batchelor TT. Advances in neuroimaging techniques for the evaluation of tumor growth, vascular permeability, and angiogenesis in gliomas. Curr Opin Neurol. 2008;21(6):728–735. doi: 10.1097/WCO.0b013e328318402a. [DOI] [PubMed] [Google Scholar]
- 9.Sorensen AG, Emblem KE, Polaskova P, Jennings D, Kim H, Ancukiewicz M, Wang M, Wen PY, Ivy P, Batchelor TT. Increased survival of glioblastoma patients who respond to antiangiogenic therapy with elevated blood perfusion. Cancer Res. 2012;72(2):402–407. doi: 10.1158/0008-5472.CAN-11-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emblem KE, Mouridsen K, Bjornerud A, Farrar CT, Jennings D, Borra RJ, Wen PY, Ivy P, Batchelor TT, Rosen BR. Vessel architectural imaging identifies cancer patient responders to anti-angiogenic therapy. Nat Med. 2013;19(9):1178–1183. doi: 10.1038/nm.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnhart HX, Barboriak DP. Applications of the repeatability of quantitative imaging biomarkers: a review of statistical analysis of repeat data sets. Transl Oncol. 2009;2(4):231–235. doi: 10.1593/tlo.09268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellinger R, Kremser C, Schocke MF, Kolbitsch C, Griebel J, Felber SR, Aichner FT. The impact of peak saturation of the arterial input function on quantitative evaluation of dynamic susceptibility contrast-enhanced MR studies. J Comput Assist Tomogr. 2000;24(6):942–948. doi: 10.1097/00004728-200011000-00022. [DOI] [PubMed] [Google Scholar]
- 13.Rausch M, Scheffler K, Rudin M, Radu EW. Analysis of input functions from different arterial branches with gamma variate functions and cluster analysis for quantitative blood volume measurements. Magn Reson Imaging. 2000;18(10):1235–1243. doi: 10.1016/s0730-725x(00)00219-8. [DOI] [PubMed] [Google Scholar]
- 14.Paulson ES, Schmainda KM. Comparison of dynamic susceptibility-weighted contrast-enhanced MR methods: recommendations for measuring relative cerebral blood volume in brain tumors. Radiology. 2008;249(2):601–613. doi: 10.1148/radiol.2492071659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakaie KE, Shin W, Curtin KR, McCarthy RM, Cashen TA, Carroll TJ. Method for improving the accuracy of quantitative cerebral perfusion imaging. J Magn Reson Imaging. 2005;21(5):512–519. doi: 10.1002/jmri.20305. [DOI] [PubMed] [Google Scholar]
- 16.Knutsson L, Stahlberg F, Wirestam R. Absolute quantification of perfusion using dynamic susceptibility contrast MRI: pitfalls and possibilities. MAGMA. 2010;23(1):1–21. doi: 10.1007/s10334-009-0190-2. [DOI] [PubMed] [Google Scholar]
- 17.Jackson A, Kassner A, Zhu XP, Li KL. Reproducibility of T2* blood volume and vascular tortuosity maps in cerebral gliomas. J Magn Reson Imaging. 2001;14(5):510–516. doi: 10.1002/jmri.1214. [DOI] [PubMed] [Google Scholar]
- 18.Henry ME, Kaufman MJ, Lange N, Schmidt ME, Purcell S, Cote J, Perron-Henry DM, Stoddard E, Cohen BM, Renshaw PF. Test-retest reliability of DSC MRI CBV mapping in healthy volunteers. Neuroreport. 2001;12(8):1567–1569. doi: 10.1097/00001756-200106130-00011. [DOI] [PubMed] [Google Scholar]
- 19.Shin W, Horowitz S, Ragin A, Chen Y, Walker M, Carroll TJ. Quantitative cerebral perfusion using dynamic susceptibility contrast MRI: evaluation of reproducibility and age- and gender-dependence with fully automatic image postprocessing algorithm. Magn Reson Med. 2007;58(6):1232–1241. doi: 10.1002/mrm.21420. [DOI] [PubMed] [Google Scholar]
- 20.Grandin CB, Bol A, Smith AM, Michel C, Cosnard G. Absolute CBF and CBV measurements by MRI bolus tracking before and after acetazolamide challenge: repeatabilily and comparison with PET in humans. NeuroImage. 2005;26(2):525–535. doi: 10.1016/j.neuroimage.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 21.Lankester KJ, Taylor JN, Stirling JJ. Dynamic MRI for imaging tumor microvasculature: comparison of susceptibility and relaxivity techniques in pelvic tumors. J Magn Reson Imaging. 2007;25(4):796–805. doi: 10.1002/jmri.20881. [DOI] [PubMed] [Google Scholar]
- 22.Lankester KJ, Taylor JN, Stirling JJ, Boxall J, d'Arcy JA, Collins DJ, Walker-Samuel S, Leach MO, Rustin GJ, Padhani AR. Reproducibility and correlation between quantitative and semiquantitative dynamic and intrinsic susceptibility-weighted MRI parameters in the benign and malignant human prostate. J Magn Reson Imaging. 2010;32(1):155–164. doi: 10.1002/jmri.22215. [DOI] [PubMed] [Google Scholar]
- 23.Batchelor TT, Gerstner ER, Emblem KE, Duda DG, Kalpathy-Cramer J, Snuderl M, Ancukiewicz M, Polaskova P, Pinho MC, Jennings D, et al. Improved tumor oxygenation and survival in glioblastoma patients who show increased blood perfusion after cediranib and chemoradiation. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed]
- 24.Batchelor TT, Sorensen AG, di Tomaso E, Zhang WT, Duda DG, Cohen KS, Kozak KR, Cahill DP, Chen PJ, Zhu M. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell. 2007;11(1):83–95. doi: 10.1016/j.ccr.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batchelor TT, Duda DG, di Tomaso E, Ancukiewicz M, Plotkin SR, Gerstner E, Eichler AF, Drappatz J, Hochberg FH, Benner T. Phase II study of cediranib, an oral pan-vascular endothelial growth factor receptor tyrosine kinase inhibitor, in patients with recurrent glioblastoma. J Clin Oncol. 2010;28(17):2817–2823. doi: 10.1200/JCO.2009.26.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weisskoff R, Boxerman J, Sorensen A. Proceedings of the Twelfth Annual Meeting of the Society for Magnetic Resonance Imaging. 1994. Simultaneous blood volume and permeability mapping using a single Gd-based contrast injection. [Google Scholar]
- 27.Donahue KM, Krouwer HG, Rand SD, Pathak AP, Marszalkowski CS, Censky SC, Prost RW. Utility of simultaneously acquired gradient-echo and spin-echo cerebral blood volume and morphology maps in brain tumor patients. Magn Reson Med. 2000;43(6):845–853. doi: 10.1002/1522-2594(200006)43:6<845::aid-mrm10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 28.Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol. 2006;27(4):859–867. [PMC free article] [PubMed] [Google Scholar]
- 29.Bjornerud A, Sorensen AG, Mouridsen K, Emblem KE. T1- and T2*-dominant extravasation correction in DSC-MRI: part I—theoretical considerations and implications for assessment of tumor hemodynamic properties. J Cereb Blood Flow Metab. 2011;31(10):2041–2053. doi: 10.1038/jcbfm.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mouridsen K, Christensen S, Gyldensted L, Ostergaard L. Automatic selection of arterial input function using cluster analysis. Magn Reson Med. 2006;55(3):524–531. doi: 10.1002/mrm.20759. [DOI] [PubMed] [Google Scholar]
- 31.Bjornerud A, Emblem KE. A fully automated method for quantitative cerebral hemodynamic analysis using DSC-MRI. J Cereb Blood Flow Metab. 2010;30(5):1066–1078. doi: 10.1038/jcbfm.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmainda KM, Prah M, Connelly J, Rand SD, Hoffman RG, Mueller W, Malkin MG. Dynamic-susceptibility contrast agent MRI measures of relative cerebral blood volume predict response to bevacizumab in recurrent high-grade glioma. Neuro-Oncology. 2014;16(6):880–888. doi: 10.1093/neuonc/not216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bedekar D, Jensen T, Schmainda KM. Standardization of relative cerebral blood volume (rCBV) image maps for ease of both inter- and intrapatient comparisons. Magn Reson Med. 2010;64(3):907–913. doi: 10.1002/mrm.22445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emblem KE, Bjornerud A. An automatic procedure for normalization of cerebral blood volume maps in dynamic susceptibility contrast-based glioma imaging. AJNR Am J Neuroradiol. 2009;30(10):1929–1932. doi: 10.3174/ajnr.A1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorensen AG, Patel S, Harmath C, Bridges S, Synnott J, Sievers A, Yoon YH, Lee EJ, Yang MC, Lewis RF. Comparison of diameter and perimeter methods for tumor volume calculation. J Clin Oncol. 2001;19(2):551–557. doi: 10.1200/JCO.2001.19.2.551. [DOI] [PubMed] [Google Scholar]
- 36.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 38.Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 1979;86(2):420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 39.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1(8476):307–310. [PubMed] [Google Scholar]
- 40.Efron B, Tibshirani R. Chapman & Hall; New York: 1993. An Introduction to the Bootstrap. [Google Scholar]
- 41.Montgomery DC. 7th edn. Wiley; Hoboken, NJ: 2008. Design and analysis of experiments. [Google Scholar]
- 42.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995:289–300. [Google Scholar]
- 43.Sorensen AG, Patel S, Harmath C, Bridges S, Synnott J, Sievers A, Yoon YH, Lee EJ, Yang MC, Lewis RF. Quantitative imaging biomarkers: a review of statistical methods for technical performance assessment. Stat Methods Med Res. 2014;24(1):27–67. doi: 10.1177/0962280214537344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sorensen AG, Patel S, Harmath C, Bridges S, Synnott J, Sievers A, Yoon YH, Lee EJ, Yang MC, Lewis RF. The emerging science of quantitative imaging biomarkers terminology and definitions for scientific studies and regulatory submissions. Stat Methods Med Res. 2014;24(1):9–26. doi: 10.1177/0962280214537333. [DOI] [PubMed] [Google Scholar]
- 45.Yankeelov TE, Abramson RG, Quarles CC. Quantitative multimodality imaging in cancer research and therapy. Nat Rev Clin Oncol. 2014;11(11):670–680. doi: 10.1038/nrclinonc.2014.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Troprès I, Pannetier N, Grand S, Lemasson B, Moisan A, Péoc'h M, Rémy C, Barbier EL. Imaging the microvessel caliber and density: principles and applications of microvascular MRI. Magn Reson Med. 2014;73(1):325–341. doi: 10.1002/mrm.25396. [DOI] [PubMed] [Google Scholar]
- 47.Law M, Yang S, Babb JS, Knopp EA, Golfinos JG, Zagzag D, Johnson G. Comparison of cerebral blood volume and vascular permeability from dynamic susceptibility contrast-enhanced perfusion MR imaging with glioma grade. AJNR Am J Neuroradiol. 2004;25(5):746–755. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Perfusion analysis, comprehensive tables of the ICCs and RCs, table of Mean±SD values of CBV and CBF maps, and figures showing the image analysis.


