Abstract
A total of 205 wheat cultivars from the Yellow and Huai valley of China were used to identify allelic variations of vernalization and photoperiod response genes, as well as the copy number variations (CNVs) of Ppd-B1 and Vrn-A1 genes. A novel Vrn-D1 allele with 174-bp insertion in the promoter region of the recessive allele vrn-D1 was discovered in three Chinese wheat cultivars and designated as Vrn-D1c. Quantitative real-time polymerase chain reaction showed that cultivars with the Vrn-D1c allele exhibited significantly higher expression of the Vrn-D1 gene than that in cultivars with the recessive allele vrn-D1, indicating that the 174-bp insertion of Vrn-D1c contributed to the increase in Vrn-D1 gene expression and caused early heading and flowering. The five new cis-elements (Box II-like, 3-AF1 binding site, TC-rich repeats, Box-W1 and CAT-box) in the 174-bp insertion possibly promoted the basal activity level of Vrn-D1 gene. Two new polymorphism combinations of photoperiod genes were identified and designated as Ppd-D1_Hapl-IX and Ppd-D1_Hapl-X. Association of the CNV of Ppd-B1 gene with the heading and flowering days showed that the cultivars with Ppd-B1_Hapl-VI demonstrated the earliest heading and flowering times, and those with Ppd-B1_Hapl-IV presented the latest heading and flowering times in three cropping seasons. Distribution of the vernalization and photoperiod response genes indicated that all recessive alleles at the four vernalization response loci, Ppd-B1_Hapl-I at Ppd-B1 locus, and Ppd-D1_Hapl-I at the Ppd-D1 locus were predominant in Chinese winter wheat cultivars. This study can provide useful information for wheat breeding programs to screen wheat cultivars with relatively superior adaptability and maturity.
Keywords: bread wheat, vernalization genes, photoperiod genes, copy number variations, allelic variation
Introduction
Heading and flowering times of bread wheat, mainly modulated by vernalization and photoperiod genes, are important factors that determine the adaptation of wheat plants to different environmental conditions and influence the growth and productivity of wheat (Law and Worland, 1997). Vernalization is the acquisition of a plant's ability to flower in the spring by exposure to the prolonged cold of winter, or by an artificial equivalent. Photoperiod insensitivity is widespread in the world's wheat varieties and predominates in regions where spring wheat is grown as a crop over the winter period and where autumn-sown winter wheat needs to mature in the following year before the onset of high summer temperatures.
To date, three fundamental vernalization response genes (Vrn-1, Vrn-2, and Vrn-3) have been discovered in polyploid wheat and barley. Vrn-1 genes, encompassing Vrn-A1, Vrn-B1, and Vrn-D1 genes on the long arms of chromosome 5 (Law et al., 1975; Galiba et al., 1995; Dubcovsky et al., 1998; Barrett et al., 2002; Iwaki et al., 2002), are upregulated by vernalization treatment, and their overexpression can accelerate flowering and maturity of wheat (Yan et al., 2003). The Vrn-2 gene, a dominant repressor of flowering, is downregulated by vernalization treatment. Loss of functional mutation of Vrn-2 resulted in the spring growth of bread wheat to head and flower under non-vernalization treatment. The Vrn-3 gene is an ortholog of the Arabidopsis FT (flowering time) gene and is upregulated by vernalization treatment (Yan et al., 2006). A molecular model explaining the Vrn-1/Vrn-2/Vrn-3 epistatic interaction in winter wheat was proposed by Yan et al. (2003, 2004a,b) and Chen and Dubcovsky (2012). Moreover, different vernalization alleles at Vrn-1 and Vrn-3 loci were discovered. The Vrn-A1a allele exhibits an insertion of a foldback repetitive element, as well as a duplicated region in the promoter, causing a strong effect on vernalization response and resulting in the complete elimination of the vernalization requirement. The Vrn-A1b, Vrn-A1c, Vrn-A1d, and Vrn-A1e have been subsequently described in polyploid wheat (Yan et al., 2004a; Fu et al., 2005). The different Vrn-B1 alleles (Vrn-B1a, Vrn-B1b, and Vrn-B1c) and Vrn-D1 alleles (Vrn-D1a, Vrn-D1b, and Vrn-D1s) are mostly results of insertion and/or deletion in the intron1 region (Yan et al., 2004a; Fu et al., 2005; Santra et al., 2009; Milec et al., 2012; Shcherban et al., 2012; Zhang et al., 2012; Muterko et al., 2014). Plants homozygous for the Vrn-D1b allele headed 32 days later than plants homozygous for the Vrn-D1a allele, and that theVrn-D1b gene is associated with facultative growth habit (Zhang et al., 2012). The Vrn-B3a allele is a 5300-bp insertion in the promoter region, and Vrn-B3b and Vrn-B3c alleles were recently discovered in bread wheat (Yan et al., 2006; Chen et al., 2013a). Chen et al. (2013a) indicated that Vrn-B3b significantly reduced the expression level of the gene and caused later heading and flowering compared with the vrn-B3 gene. Additionally, copy number variation (CNV) slightly influenced the vernalization gene expression and showed little effect on the phenotype. The predominant result is a C/T double peak in sequence trace files in exon 4 on Vrn-A1, and plants with an increased CNV showed an increased requirement for vernalization, thereby requiring longer cold treatment to potentiate flowering (Diaz et al., 2012).
Photoperiod response is another important factor influencing the start and length of the flowering period. The important photoperiod response genes are Ppd-A1, Ppd-B1, and Ppd-D1, which are located in 2A, 2B, and 2D of the Ppd-1 loci, respectively (Worland et al., 1998; Shitsukawa et al., 2007). Genetic studies showed that the most effective photoperiod insensitivity gene is the Ppd-D1 gene, followed by Ppd-B1 and Ppd-A1. Beales et al. (2007) detected a 2089-bp deletion in the promoter region of the Ppd-D1a gene, which is the photoperiod-insensitive allele. The second polymorphism was a mariner-like transposable element (TE) present in intron 1; the third was a 5-bp deletion in exon 7; the fourth was a SNP (A/G) in exon 7; the fifth was a 16-bp deletion including the last two bases of the CCT domain in exon 8. Guo et al. (2010) described these alleles to six polymorphisms, involving one photoperiod-insensitive haplotype and five photoperiod-sensitive haplotypes in Ppd-D1. Chen et al. (2013a) also identified two new polymorphism combinations in Ppd-D1 of common wheat. Diaz et al. (2012) reported that the Ppd-B1 gene contained three CNVs, namely, truncated “Chinese Spring” Ppd-B1 allele, intact “Chinese Spring” Ppd-B1 allele, and intact “Sonora64” Ppd-B1 allele, and proved that the alleles with higher copy number of Ppd-B1 confer an early flowering day neutral phenotype.
China is the largest wheat consumer and producer worldwide. Winter wheat occupies more than 85% of the total area and production of Chinese wheat. China has 10 major agro-ecological zones that are further divided into 26 subzones (Zhuang, 2003). Among the agro-ecological zones, the Yellow and Huai wheat production region is the most important and largest wheat production zone, with 60–70% of both total harvested area and total wheat production. In a previous study (Chen et al., 2013a), we characterized the vernalization and photoperiod response genes in currently popular cultivars and landraces from the Yellow and Huai wheat region. In the present research, we further identified the molecular characterization of the vernalization and photoperiod response genes in backbone parents in the wheat breeding program of this wheat region and found a new Vrn-D1 allele, a Vrn-D1 null allele, two new polymorphism combinations of photoperiod genes, and several CNVs. Our results provide useful information for wheat breeding programs to screen relatively superior wheat germplasms in view of their adaptability to diverse agronomic environments.
Materials and methods
Plant materials
A total of 205 winter wheat cultivars and advanced lines were planted in 2011–2012, 2012–2013, and 2013–2014 cropping seasons, respectively, at the Zhengzhou Scientific Research and Education Center of Henan Agricultural University (N34.9; E113.6) under local management practices. All surveyed cultivars were vernalized through winter with an average temperature of 1.3°C (December, January, and February) in 2012, 2013, and 2014. The wheat germplasms used were important landrace, historical, and introduced cultivars in China, especially in the Yellow and Huai wheat region. Different from the materials we previously used (Chen et al., 2013a), these germplasms were mainly used as backbone parents and had played important roles in wheat breeding programs in China. The field experiment was conducted using a completely randomized design. Each plot contained four 200 cm-long rows with 23 cm between neighboring rows and 10 cm between neighboring plants. All surveyed cultivars grew very well with the supporting net without lodging. The heading and flowering times of each cultivar were investigated in April 2012, April 2013, and April 2014, and their heading and flowering days were calculated from the sowing day to the heading and flowering days.
The wheat plants of four Chinese cultivars (Yanzhan 4110, Jinmai 50, Lumai 19, and Yunong 876) were grown in a greenhouse under 16 h light at 25°C–28°C (day) and 8 h dark at 20°C–22°C (night) to investigate the heading and flowering days under the condition of non-vernalization treatment.
Polymerase chain reaction (PCR) parameters and DNA sequencing
The genomic DNA of each cultivar surveyed was individually extracted from three pulverized kernels as described by Chen et al. (2011). The PCR reactions and programs were performed according to Chen et al. (2013a) (detailed annealing temperatures in Supplemental Table 1). PCR products were separated on a 1.5–2.5% agarose gel stained with ethidium bromide and visualized with UV light or on a 6% polyacrylamide gel and resolved by silver staining.
After purification using Quick DNA Extraction Kit (Takara, http://www.takara.com.cn/), targeted PCR products were ligated into pGEM-T Easy vector and transformed into competent cells of an Escherichia coli DH-5α strain. Plasmids with targeted fragments, detected by colony PCR, were extracted by Plasmid Rapid Isolation Kit (Biodev-tech Company, http://biodev.technew.cn/). Five subclones for each PCR product were sequenced from both strands by SinoGenoMax Co., Ltd. (http://www.sinogenomax.com/). Analysis and multiple alignments of sequences were performed by DNAMAN Version 6.0, and graphic data were analyzed to check the reliability of the sequencing results by using Chromas Version 1.4.5 and FinchTV version 1.4.0.
Real-time quantitative reverse transcription PCR
Total RNAs of the four Chinese cultivars (Yanzhan 4110, Jinmai 50, Lumai 19, and Yunong 876) with different Vrn-D1 alleles were extracted from 2-month-old seedlings for real-time quantitative reverse transcription PCR (qRT-PCR) as described by Chen et al. (2013b). The primer set Vrn-P17F/R (Supplemental Table 1) was designed by Software Primer Premier 5.0 for qRT-PCR amplification. Amplification with β-actin primers was used as internal control to normalize all the data. Relative quantification method (2−ΔΔCT) was used to evaluate the quantitative variation among the three replicates.
Results
Discovery of a novel dominant Vrn-D1 allele in chinese winter wheat
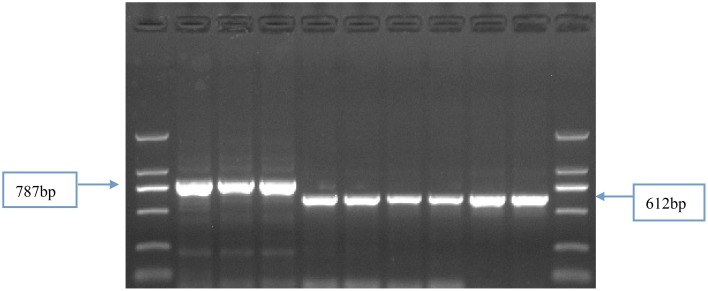
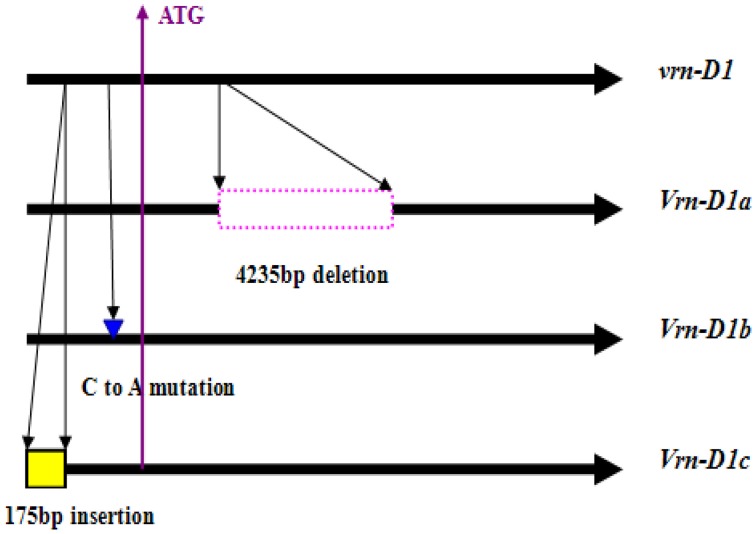
Identification of the 205 Chinese winter wheat cultivars by four primer sets (Vrn-P8F/R, Vrn-P9F/R, Vrn-P10F/R, and Vrn-P11F/R; Supplemental Table 1) indicated that 119 cultivars (58.0%) contained the recessive allele vrn-D1, and 59 (28.8%) and 24 (11.7%) cultivars contained the dominant alleles Vrn-D1a and Vrn-D1b, respectively. However, the remaining three cultivars showed an approximately 800-bp fragment when amplified with the primer set Vrn-P10F/R. The presence of this fragment indicated a nearly 200-bp insertion in these three cultivars compared with the recessive vrn-D1 allele (Figure 1). The sequencing results indicated that a 174-bp fragment was inserted into the 5′-UTR at -601-bp (relative to ATG) of the vrn-D1 gene (Table 1 and Figure 2). This new Vrn-D1 allele with 174-bp insertion was designated as Vrn-D1c allele (submitted to NCBI No.: KP721800) in accordance with the nomenclature of vernalization response genes by Fu et al. (2005), Yan et al. (2004a, 2006) and Chen et al. (2013a).
Figure 1.
Identification of vernalization response alleles by Vrn-P10F/R markers in the Chinese winter wheat cultivars. Primer set Vrn-10F/R for identification of Vrn-D1c from Vrn-D1 promoter with 787-bp fragment. From L to R: DNA ladder DL2000. 876 for three lanes, Shuangji 2, Shuangji 4, Jingshuang 16, Lumai 14, Ji 87 guan 739. DL2000.
Table 1.
Molecular characterization of vernalization response alleles in polyploid wheat.
| Locus | Allele | NCBI No. | Molecular characterization | References |
|---|---|---|---|---|
| Vrn-A1 | vrn-A1 | AY747600 | – | Fu et al., 2005 |
| Vrn-A1a | AY616458, AY616459 | 231-bp and 140-bp insertions at −439 and −348 bp, respectively | Yan et al., 2004a | |
| Vrn-A1b | AY616461 | 20-bp deletion at −157 bp | Yan et al., 2004a | |
| Vrn-A1c | AY747599 | 5504-bp deletion at +1349 bp | Fu et al., 2005 | |
| Vrn-A1d | AY616462 | 32-bp deletion at −214 bp | Yan et al., 2004a | |
| Vrn-A1e | AY616463 | 54-bp deletion at −220 bp | Yan et al., 2004a | |
| Vrn-B1 | vrn-B1 | AY747604 | – | Fu et al., 2005 |
| Vrn-B1a | AY747603 | 6850-bp deletion at +836 bp | Fu et al., 2005 | |
| Vrn-B1b | FJ766015 | 6850-bp deletion at +836 bp and 37-bp deletion at +7992 bp | Santra et al., 2009 | |
| Vrn-B1c | HQ593668, HQ130482 | 817-bp deletion and 0.4-kb duplication at +798 bp | Milec et al., 2012; Shcherban et al., 2012 | |
| Vrn-B3 | vrn-B3 | DQ890162 | – | Yan et al., 2006 |
| Vrn-B3a | DQ890165 | 5300-bp insertion at −592 bp | Yan et al., 2006 | |
| Vrn-B3b | JN627519 | 890-bp insertion at −429 bp | Chen et al., 2013a | |
| Vrn-B3c | JQ082311 | 5300-bp insertion at −592 bp but 20-bp and 4-bp deletions at −3543 and −3591 bp | Chen et al., 2013a | |
| Vrn-D1 | vrn-D1 | AY747606 | – | Fu et al., 2005 |
| Vrn-D1a | AY747597 | 4235-bp deletion at +810 bp | Fu et al., 2005 | |
| Vrn-D1b | C mutation to A at −161 bp | Zhang et al., 2012 | ||
| Vrn-D1s | KF800714 | 844-bp deletion at +1044 bp | Muterko et al., 2014 | |
| Vrn-D1c | KP721800 | 174-bp deletion at −601 bp | In this paper |
Figure 2.
Schematic of Vrn-D1 alleles identified in the Chinese bread wheat cultivars.
Expression level of the Vrn-D1 gene associated with DH and DF in winter wheat
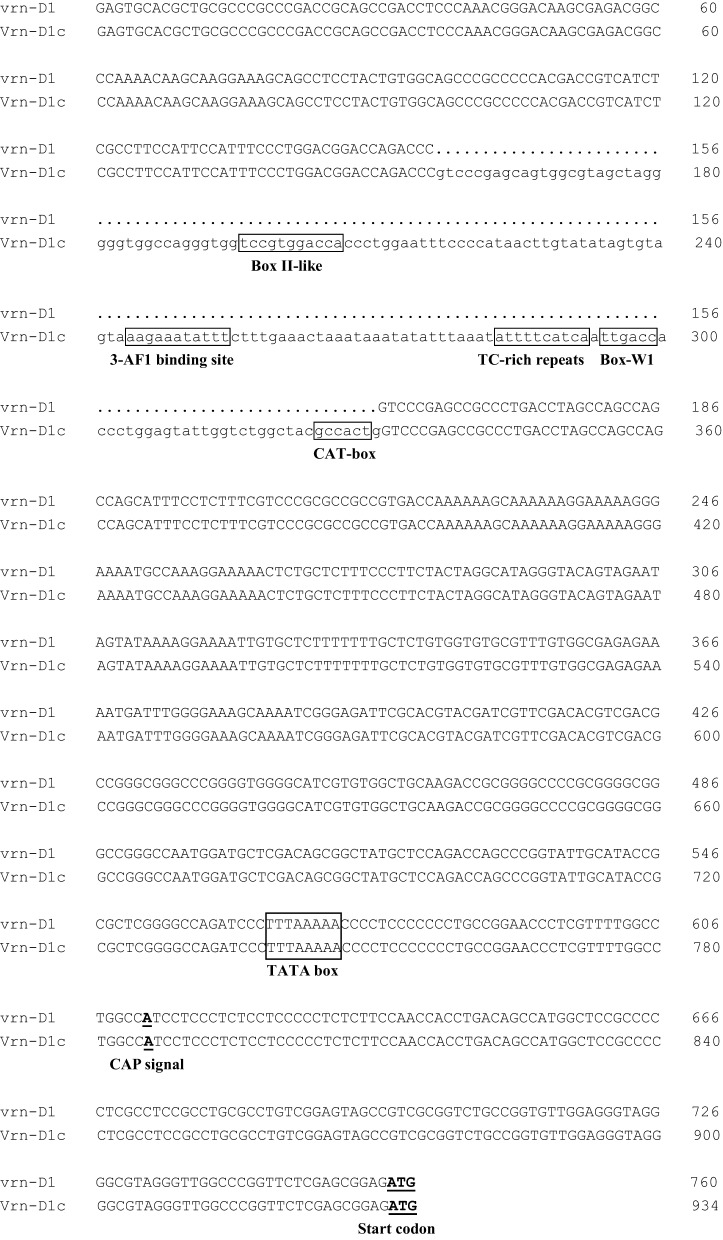
Four cultivars, namely, Lumai19 (with the recessive allele vrn-D1), Yanzhan 4110 (with Vrn-D1a allele), Jinmai 50 (with Vrn-D1b allele), and Yunong 876 (with Vrn-D1c allele), were selected to analyze the expression levels of different Vrn-D1 alleles by qRT-PCR (Figure 3) owing to their same Vrn-A1, Vrn-B1, Vrn-B3, and Ppd-D1 alleles. The qRT-PCR results indicated that Lumai 19 exhibited the lowest expression level of Vrn-D1 gene. Jinmai 50 demonstrated a significantly higher expression than that of Lumai 19, suggesting that a single point mutation significantly increased the Vrn-D1 gene expression (C-to-A mutation in Jinmai 50 allele in Figure 2). Vrn-D1 gene in Yunong 876 showed significantly higher expression levels than those of Lumai19 and Jinmai 50, suggesting that the 174-bp insertion possibly contributed to the increased expression. Among the four cultivars, Yanzhan 4110 showed the highest expression level of the Vrn-D1 gene. The prediction of cis elements in promoters of Vrn-D1 genes by software PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) indicated that five new cis-elements (Box II-like, 3-AF1 binding site, TC-rich repeats, Box-W1 and CAT-box) were generated in the 174-bp insertion of Vrn-D1c gene promoter (Figure 4). Presence of these five cis elements possibly resulted in the significantly increased basal activity level of Vrn-D1c when compared with the recessive vrn-D1 allele.
Figure 3.
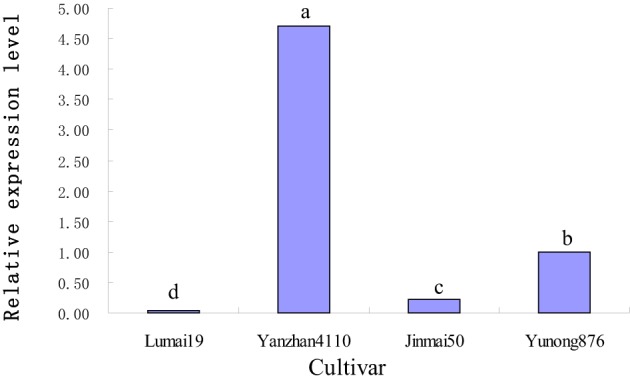
Comparison of relative expression levels of four cultivars with different Vrn-D1 alleles by real-time PCR.
Figure 4.
Sequence alignment in Vrn-D1 promoter region between the recessive vrn-D1 allele and the dominant Vrn-D1c allele. The ATG start codon and the putative CAP signal for transcription initiation are indicated in bold and underlined. Five predicted cis-elements (Box II-like, 3-AF1 binding site, TC-rich repeats, Box-W1, and CAT-box) and the TATA box are indicated in a rectangle.
Phenotype investigation of these four cultivars under vernalization treatment showed that the heading and flowering days of the cultivar Lumai 19 with recessive allele vrn-D1 were 195 and 200 days, respectively. Jinmai 50 (194 DH, 199 DF) with the Vrn-D1b allele headed and flowered 1 days ahead of Lumai 19. Yanzhan 4110 (192 DH, 196 DF) with the Vrn-D1a allele headed 3 days ahead and flowered 4 days ahead of Lumai 19. Yunong 876 (193 DH, 200 DF) with the Vrn-D1c allele headed 2 days ahead of Lumai 19 but flowered on the same day. These results suggested that the 174-bp insertion of the Vrn-D1c allele possibly prolonged the heading to flowering days of bread wheat under vernalization treatment.
Moreover, phenotype investigation of the four cultivars under non-vernalization treatment showed that the heading and flowering days of the cultivar Lumai 19 were 120 and 122 days, respectively. Jinmai 50 (111 DH, 114 DF) headed 9 days ahead and flowered 8 days ahead of Jinmai 50. Yanzhan 4110 (61 DH, 72 DF) headed 59 days ahead and flowered 50 days ahead of Lumai 19. Yunong 876 (76 DH, 80DF) with the Vrn-D1c allele headed 44 days ahead and flowered 46 days ahead of Lumai 19. These results suggested that the expression of the Vrn-D1 genes promotes early heading and flowering under non-vernalization treatment.
Distribution of vernalization response alleles in chinese winter wheat cultivars
Identification with four allele-specific primer sets (Vrn-P1F/R, Vrn-P2F/R, Vrn-P3F/R, and Vrn-P4F/R in Supplemental Table 1) showed that 198 out of 205 (96.6%) winter wheat cultivars contained the recessive vrn-A1 allele, and five and two wheat cultivars contained Vrn-A1a and Vrn-A1b alleles in the Vrn-A1 locus (Table 2), respectively. Identification with three primer sets (Vrn-P5F/R, Vrn-P6F/R, and Vrn-P7F/R) indicated that 172 out of 205 (83.90%) winter wheat cultivars contained the vrn-B1, and 24 and 9 cultivars contained Vrn-B1a and Vrn-B1b alleles in the Vrn-B1 locus (Table 2), respectively. Identification with four primer sets (Vrn-P8F/R, Vrn-P9F/R, Vrn-P10F/R, and Vrn-P11F/R) showed that 118 out of 205 (57.56%) winter cultivars contained the vrn-D1, and 60 (29.27%) and 23 (11.22%) out of the remaining 87 cultivars contained Vrn-D1a and Vrn-D1b alleles in the Vrn-D1 locus (Table 2), respectively. Three cultivars exhibited the Vrn-D1c allele as mentioned above. Surprisingly, cultivar Pincun 16 did not yield any PCR amplification result when amplified with primer sets Vrn-P8F/R, Vrn-P9F/R, Vrn-P10F/R, and Vrn-P11F/R, indicating the lack of Vrn-D1 gene. Identification results with the four primer sets (Vrn-P12F/R, Vrn-P13F/R, Vrn-P14F/R, and Vrn-P15F/R) showed that Yanda 1817, which has been widely used as a parent in Chinese wheat breeding programs, contained a rare allele Vrn-B3b (recently reported by Chen et al., 2013a), and three cultivars contained the dominant allele Vrn-B3a. All the remaining cultivars (98.05%) contained the recessive allele vrn-B3 in the Vrn-B3 locus. These results suggested that the recessive allele vrn-A1, vrn-B1, vrn-D1, and vrn-B3 were predominant in the Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 loci. But the dominant Vrn-D1a and Vrn-D1b alleles were also prevalent genotypes (Table 2). These results were consistent with the previous findings of Zhang et al. (2008) and Chen et al. (2013a).
Table 2.
The distribution of vernalization and Ppd-D1 genes in the Chinese bread wheat cultivars surveyed.
| Locus | Allele | Number of cultivars with an allele | Frenquency (%) | Typical cultivars |
|---|---|---|---|---|
| Vrn-A1 | vrn-A1 | 198 | 96.6 | Abbondanza; Orofen; Nonglin 10 |
| Vrn-A1a | 5 | 2.44 | SW604; SW601; SW618 | |
| Vrn-A1b | 2 | 0.98 | 605; Triumph | |
| Vrn-B1 | vrn-B1 | 172 | 83.9 | Nonglin 10; Shuiyuan 86; Lovin 10 |
| Vrn-B1a | 24 | 11.7 | Lankao 4; Orofen; SW604 | |
| Vrn-B1b | 9 | 4.39 | Beijing 841; Xinong 164; Yangmai 4 | |
| Vrn-D1 | vrn-D1 | 120 | 58.5 | Jingshuang 16; Beijing 6; Zhongnong 28 |
| Vrn-D1a | 59 | 28.8 | Yanzhan 1; XJ1; Mianyang 8640 | |
| Vrn-D1b | 23 | 11.2 | Jinmai 50; Zhengmai 9201; Fengyou 8 | |
| Vrn-D1c | 3 | 1.46 | Shannong 413863; Xinmai 18*; Yunong 876 | |
| Vrn-B3 | vrn-B3 | 201 | 98.1 | Jimai 13; Beijing 841; Nongda 311 |
| Vrn-B3a | 3 | 1.46 | Xiaobaimai; Pingyang 181; Lovin 10 | |
| Vrn-B3b | 1 | 0.49 | Yanda 1817 | |
| Ppd-D1a | 184 | 89.8 | Jinmai 50, Lumai 19, Yanzhan 4110 | |
| Ppd-D1 | Ppd-D1b | 21 | 10.2 | Nongda 311, Beijing 6, Jinan 4 |
| 2089-bp deletion in exon 8 | 193 | 94.1 | Jinmai 50, Lumai 19, Yanzhan 4110 | |
| TE insertion in intron 1 | 17 | 8.29 | Lumai 14, Nongda 311, Yanda 1817 | |
| 5-bp insertion in exon 7 | 8 | 3.90 | Beijing 6, Jinan 4, Pinchun 16 |
Up to 19 allelic combinations of vernalization response genes were discovered in terms of the Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 loci in the wheat cultivars (Table 3). Distribution of different allelic combinations of vernalization genes suggested that the recessive allelic combination vrn-A1/vrn-B1/vrn-D1/vrn-B3 was predominant, and three allelic combinations (vrn-A1/vrn-B1/Vrn-D1a/vrn-B3, vrn-A1/vrn-B1/Vrn-D1b/vrn-B3, and vrn-A1/Vrn-B1a/vrn-D1/vrn-B3) were also prevalent in Chinese winter wheat cultivars. Association of four above-mentioned allelic combinations of vernalization genes with heading and flowering times showed that cultivars with vrn-A1/vrn-B1/vrn-D1/vrn-B3 possessed the latest heading and flowering times across 3 years, and cultivars with vrn-A1/vrn-B1/Vrn-D1a/vrn-B3 possessed relatively earlier heading and flowering times than cultivars with vrn-A1/vrn-B1/Vrn-D1b/vrn-B3 (Table 3).
Table 3.
The distribution of allelic combinations of vernalization genes in the Chinese bread wheat cultivars surveyed.
| Allelic combination | Sample No. | Frequency (%) | 2011–2012 | 2012–2013 | 2013–2014 | |||
|---|---|---|---|---|---|---|---|---|
| HD | FD | HD | FD | HD | FD | |||
| vrn-A1/vrn-B1/vrn-D1/vrn-B3 | 90 | 43.90 | 196.8 | 201.7 | 192.5 | 200.1 | 184.3 | 190.3 |
| vrn-A1/vrn-B1/Vrn-D1a/vrn-B3 | 51 | 24.88 | 193.2 | 198.5 | 189.6 | 195.9 | 180.9 | 186.8 |
| vrn-A1/vrn-B1/Vrn-D1b/vrn-B3 | 19 | 9.27 | 195.0 | 200.2 | 191.1 | 198.2 | 182.3 | 187.9 |
| vrn-A1/Vrn-B1a/vrn-D1/vrn-B3 | 14 | 6.83 | 193.9 | 199.4 | 189.5 | 196.9 | 182.1 | 188.6 |
| vrn-A1/vrn-B1/Vrn-D1c/vrn-B3 | 3 | 1.46 | 194.0 | 200.0 | 190.0 | 198.7 | 182.0 | 189.3 |
| vrn-A1/Vrn-B1b/vrn-D1/vrn-B3 | 6 | 2.93 | 192.3 | 198.7 | 189.8 | 198.0 | 180.2 | 186.8 |
| vrn-A1/Vrn-B1a/Vrn-D1a/vrn-B3 | 6 | 2.93 | 193.5 | 200.3 | 190.7 | 195.5 | 181.0 | 187.3 |
| Vrn-A1a/Vrn-B1a/vrn-D1/vrn-B3 | 3 | 1.46 | 195.4 | 199.6 | 191.4 | 198.6 | 182.6 | 188.0 |
| Vrn-A1a/vrn-B1/vrn-D1/vrn-B3 | 2 | 0.98 | 195.5 | 200.0 | 192.0 | 199.5 | 183.0 | 188.5 |
| vrn-A1/Vrn-B1b/Vrn-D1a/vrn-B3 | 2 | 0.98 | 189.5 | 194.5 | 186.5 | 193.0 | 178.0 | 183.0 |
| vrn-A1/vrn-B1/vrn-D1/Vrn-B3a | 1 | 0.49 | 206 | 210 | 205 | 211 | 194 | 206 |
| vrn-A1/vrn-B1/vrn-D1/Vrn-B3b | 1 | 0.49 | 207 | 210 | 205 | 213 | 194 | 206 |
| Vrn-A1b/vrn-B1/vrn-D1/vrn-B3 | 1 | 0.49 | 207 | 210 | 208 | 212 | 194 | 207 |
| Vrn-A1b/vrn-B1/Vrn-D1b/vrn-B3 | 1 | 0.49 | 195 | 200 | 191 | 199 | 181 | 185 |
| vrn-A1/Vrn-B1a/Vrn-D1b/vrn-B3 | 1 | 0.49 | 190 | 198 | 188 | 192 | 186 | 193 |
| vrn-A1/Vrn-B1b/Vrn-D1b/vrn-B3 | 1 | 0.49 | 190 | 197 | 189 | 196 | 182 | 187 |
| vrn-A1/vrn-B1/Vrn-D1a/Vrn-B3a | 1 | 0.49 | 193 | 198 | 190 | 198 | 179 | 187 |
| vrn-A1/vrn-B1/Vrn-D1b/Vrn-B3a | 1 | 0.49 | 203 | 206 | 200 | 204 | 184 | 190 |
| vrn-A1/vrn-B1/vrn-D1_Null/vrn-B3 | 1 | 0.49 | 194 | 198 | 189 | 197 | 183 | 189 |
Distribution of Ppd-D1 gene in chinese winter wheat cultivars
A series of molecular markers (Ppd-P1–Ppd-P7; Supplemental Table 1) was used to identify the polymorphisms of the Ppd-D1 gene as described by Guo et al. (2010). Three polymorphisms were found in bread wheat cultivars, namely, a 2089-bp deletion in exon 8, an TE insertion in intron 1, and a 5-bp deletion in exon 7. Two polymorphisms were absent, namely, a 16-bp insertion in exon 8 and a 24-bp plus a 15-bp insertion in the 2-kb upstream region. Distribution of polymorphisms of the Ppd-D1 gene (Table 2) indicated that Ppd-D1a with the percentage of 89.8% and a 2089-bp deletion in exon 8 with the percentage of 94.1% were the most two popular genotypes in cultivars from the Yellow and Huai wheat region (detailed in Supplemental Data Sheet 1).
Furthermore, six combinations (Table 4) of the three Ppd-D1 polymorphisms mentioned above were examined. Of the 205 cultivars surveyed, 184, 2, and 9 contained Ppd-D1_Hapl-I (presence of 5-bp insertion in exon 7 and absence of others), Ppd-D1_Hapl-II (presence of 5-bp insertion in exon 7 and 2089-bp in exon 8, and absence of others), and Ppd-D1_Hapl-III (presence of transposable element in intron 1, 5-bp insertion in exon 7, 2089-bp in exon 8, and absence of others), respectively. Two cultivars contained Ppd-D1_Hapl-VIII (absence of all the five polymorphisms) previously named by Chen et al. (2013a). Moreover, two new combinations of Ppd-D1 polymorphisms were identified in this study and designated as Hapl_IX (presence of 2089-bp in exon 8 and TE in intron 1 and absence of others) in seven cultivars and Ppd-D1_Hapl-X (presence of TE in intron 1, and absence of others) in cultivar Beijing 6 in accordance with the nomenclature of Guo et al. (2010) and Chen et al. (2013a). Four known Ppd-D1 haplotypes (Ppd-D1_Hapl-IV–Ppd-D1_Hapl-VII) were absent in these cultivars. The results suggested that Ppd-D1_Hapl-I was the most prevalent (90.24%) among the six combinations of Ppd-D1 polymorphisms (Table 5).
Table 4.
Ppd-D1 haplotypes identified in the Chinese bread wheat cultivars surveyed.
| Haplotype | Sample no. | 24 bp + 15 bp | 2089 bp | TE | 5 bp | 16 bp |
|---|---|---|---|---|---|---|
| Ppd−D1_Hapl−I | 184 | − | − | − | + | − |
| Ppd−D1_Hapl−II | 2 | − | + | − | + | − |
| Ppd−D1_Hapl−III | 9 | − | + | + | + | − |
| Ppd−D1_Hapl−VIII | 2 | − | − | − | − | − |
| Ppd−D1_Hapl−V | 0 | − | + | − | + | + |
| Ppd−D1_Hapl−VI | 0 | + | + | − | + | + |
| Ppd−D1_Hapl−VII | 0 | − | − | + | + | − |
| Ppd−D1_Hapl−IX | 7 | − | + | + | − | − |
| Ppd−D1_Hapl−X | 1 | − | − | + | − | − |
Insertions and deletions are indicated by + and −, respectively.
Table 5.
Ppd-B1 haplotypes identified in the Chinese bread wheat cultivars surveyed.
| Haplotype | Sample no. | 425 bp | 994 bp | 223 bp | 2011–2012 | 2012–2013 | 2013–2014 | |||
|---|---|---|---|---|---|---|---|---|---|---|
| HD | FD | HD | FD | HD | FD | |||||
| Ppd-B1_Hapl-I | 89 | No | No | No | 195.9 | 201.0 | 192.3 | 199.3 | 183.8 | 189.8 |
| Ppd-B1_Hapl-II | 52 | No | No | Yes | 194.6 | 199.9 | 190.8 | 198.2 | 182.4 | 188.6 |
| Ppd-B1_Hapl-III | 13 | Yes | No | No | 194.7 | 199.8 | 190.8 | 197.5 | 182.5 | 188.5 |
| Ppd-B1_Hapl-IV | 2 | Yes | No | Yes | 199.0 | 203.5 | 193.5 | 202.0 | 185.0 | 190.5 |
| Ppd-B1_Hapl-V | 40 | Yes | Yes | No | 194.3 | 199.8 | 190.4 | 197.4 | 181.8 | 187.9 |
| Ppd-B1_Hapl-VI | 4 | Yes | Yes | Yes | 191.7 | 197.7 | 187.0 | 192.3 | 179.5 | 184.0 |
| Ppd-B1_Hapl-VII | 4 | No | Yes | No | 198.5 | 203.3 | 193.3 | 201.3 | 183.8 | 190.0 |
| Ppd-B1_Hapl-VIII | 1 | No | Yes | Yes | 194.0 | 200.0 | 192.0 | 199.0 | 182.0 | 188.0 |
Copy number variations in Ppd-B1 and Vrn-A1 loci
The CNV is an important component of genomic diversity that has been proven to play a significant role in wheat adaptation (Diaz et al., 2012). In this study, three primer sets were used to identify the CNV of Ppd-B1 gene in Chinese winter wheat cultivars following the method of Diaz et al. (2012); the Ppd-P8F/R (an expected 425-bp band for the gene/transposon junction in the truncated Ppd-B1 copy of cultivar Chinese Spring), Ppd-P9F/R (an expected 994-bp band for the junction between intact Ppd-B1 gene copies in Chinese Spring), and Ppd-P10F/R (an expected 223-bp band for the junction between intact Ppd-B1 gene copies in cultivars Sonora64/Timstein/C591) were all identified (Supplemental Table 1). Identification results showed that the 425-, 994-, and 223-bp bands were successfully amplified in 59, 49, and 59 cultivars, respectively. Up to 89 cultivars without any bands when amplified with the abovementioned primer sets were identified. Subsequently, eight combinations were generated and designated as Ppd-B1_Hapl-I–Ppd-B1_Hapl-VIII (detailed in Table 5). Among these combinations, Ppd-B1_Hapl-I is the most popular genotype (43.4%). Ppd-B1_Hapl-II and Ppd-B1_Hapl-V are also prevalent genotypes (25.4 and 19.5%, respectively). Percentages of the remaining genotypes were <7% in the Chinese wheat cultivars surveyed (Ppd-B1_Hapl-III with 6.3%, Ppd-B1_Hapl-IV with 0.98%, Ppd-B1_Hapl-VI with 1.96%, Ppd-B1_Hapl-VII with 1.96%, and Ppd-B1_Hapl-VIII with 0.49%). Furthermore, four Ppd-B1 genotypes with sample numbers of >10 (Ppd-B1_Hapl-I, Ppd-B1_Hapl-II, Ppd-B1_Hapl-III, and Ppd-B1_Hapl-V) were associated with heading time and flowering days. The results indicated that the cultivars with Ppd-B1_Hapl-VI (simultaneous presence of the 425-, 994-, and 223-bp bands) showed the shortest heading (179 days) and flowering days (184 days), whereas the cultivars with Ppd-B1_Hapl-I (simultaneous absence of the 425-, 994-, and 223-bp bands) demonstrated the longest heading time (184 days) and flowering time (190 days) under the condition of vernalization treatment (Supplemental Table 2).
Sequencing fragments amplified with the primer set Vrn_P16F/R (Supplemental Table 1) were used to identify the CNV of the Vrn-A1 gene in this study. As described by Eagles et al. (2011), cultivars with single nucleotide C at +10,429 bp (relative to ATG) in exon 4 of Vrn-A1 gene contained one copy of Vrn-A1 gene and cultivars with double nucleotides C plus T at this locus contained two copies of Vrn-A1 gene. Sequencing results showed that 47 wheat cultivars (22.9%) contained one copy of Vrn-A1 gene and the remaining 158 cultivars (77.1%) contained two copies of Vrn-A1 gene (Supplemental Table 2). These results suggested that the CNV of Vrn-A1 gene was very prevalent in winter wheat cultivars from the Yellow and Huai valley of China.
Discussion
The Yellow and Huai valley is the most important wheat region in China, accounting for 60–70% of the total harvested area and total wheat production of China. Backbone parents play an important role in increasing wheat yield as precious germplasms in wheat breeding programs. These precious germplasms are mainly composed of currently popular cultivars, historical cultivars, and landraces, as well as introduced cultivars from other countries or regions (e.g., International Maize and Wheat Improvement Center, USA, Europe, Australia, Canada, etc.). Chinese landrace Yanda 1817 is one of the most prevalent backbone parents, and the cross combination of Yanda 1817 and Triumph was regarded as the basis of the wheat breeding program of the Northern wheat region of China owing to their early maturity, good resistance, high yield, and strong adaptability (Zhuang, 2003; Han et al., 2009). Since the 1960s, 53 cultivars have been derived from Yanda 1817, including Nongda 183, Huabei 187, and Shijiazhuang 407. In the current study, we found that Yanda 1817 (194 DH, 206 DF) contained the rare allele Vrn-B3b, which possibly contributed to the relatively strong adaptability of this cultivar. Therefore, Vrn-B3b genotype may be fully considered as one of the relatively superior genotypes for breeding wheat cultivars with wide adaptability in bread wheat cultivars from the Yellow and Huai valley. Some introduced cultivars also played very important roles as core parents and contained many derivatives in Chinese wheat breeding programs (e.g., Lovin 10 from Romania, Nanda 2419, Funo and Abbondanza from Italy, Orofen from Chile, etc.). Molecular characterization of vernalization and photoperiod response genes for these core parents can provide useful information to further utilize the wheat germplasms in view of their adaptability and maturity in Chinese wheat breeding programs.
Previous studies indicated that increased expression of the Vrn-D1 gene contributes to early flowering and maturity (Yan et al., 2004a; Chen and Dubcovsky, 2012). The dominant Vrn-D1a allele was found to result in early heading in a large set of Australian wheat genotypes (Eagles et al., 2010; Cane et al., 2013). Fu et al. (2005) suggested that the promoter and intron 1 regulatory sequences both affected the vernalization response, and mutations in the regulatory sequences reduced the expression of this gene. Zhang et al. (2012) indicated that the Vrn-D1b allele, with a single-nucleotide mutation at the promoter region, accelerated the heading and flowering times. However, the plants with Vrn-D1b homozygous allele headed 32 days later than the plants with Vrn-D1a homozygous allele without vernalization. In the present study, we found that cultivars with the dominant Vrn-D1 allele headed earlier by an average of 3 days than cultivars with the recessive vrn-D1 allele, which is consistent with the reports of Zhang et al. (2012) and Wang et al. (2015). Moreover, cultivars with Vrn-D1a allele headed and flowered ≈ 2 days earlier than cultivars with Vrn-D1b with vernalization. Cultivars with the novel allele Vrn-D1c showed significantly earlier heading and flowering times than cultivars with the recessive allele vrn-D1 under non-vernalization treatment. This phenotype was ascribed to a 174-bp insertion contributing to higher expression level of Vrn-D1 gene. However, these cultivars showed prolonged heading to flowering times under vernalization treatment. These results can provide useful information to further understand the molecular and genetic bases of vernalization in bread wheat. These results possibly suggested that Vrn-D1c genotype is relatively preferable among known Vrn-D1 alleles in view of early maturity in bread wheat cultivars from the Yellow and Huai valley.
In the Yellow and Huai wheat region, low temperature is one of the important reasons to reduce wheat production. Cultivars with frost resistance were usually preferable for wheat breeders due to low temperature exposure of wheat plants for more than 3 months in this wheat region. Predominance of the recessive allelic combination vrn-A1/vrn-B1/vrn-D1/vrn-B3 in bread wheat cultivars from the Yellow and Huai wheat region are possibly resulted from association of vernalization genes with frost resistance (Galiba et al., 1995; Zhu et al., 2014). Wheat plants with the recessive allelic combination vrn-A1/vrn-B1/vrn-D1/vrn-B3 possibly possessed relatively stronger frost resistance than others (unpublished data), which possibly caused strong selection for this genotype in wheat breeding program in the Yellow and Huai wheat region.
To date, CNV has been recognized as a common type of polymorphism in the genomes of humans, animals, and plants (Żmieñko et al., 2014). CNVs of vernalization and photoperiod response genes significantly influence wheat flowering and maturity (Diaz et al., 2012). Cultivars with Ppd-B1_Hapl-VI demonstrated the earliest heading and flowering times in three cropping seasons among cultivars with eight different Ppd-B1 polymorphisms. These results suggested that these cultivars (Yanshi 93(13)-1-1-0-1-1, R25, Shaan 89150, and Yunong 205), as well as cultivars with Vrn-D1c (Yunong 876, Shannong 418363, and Xinmai 18*) were useful wheat germplasms to develop relatively superior cultivars with early flowering and maturity. Diaz et al. (2012) showed that an increased copy number of Vrn-A1 was strongly correlated with later flowering. In the current study, cultivars with two copies of Vrn-A1 headed and flowered 1.4 days earlier than cultivars with only one copy of Vrn-A1. Diaz et al. (2012) also proved that a higher copy number of the Ppd-B1 gene is responsible for photoperiod insensitivity. In the current study, wheat with the “Sonora64” allele showed flowering times slightly later than wheat with the “Chinese Spring” alleles. Moreover, the cultivars with both “Chinese Spring” allele and “Sonora64” allele headed the earliest among other cultivars. Cane et al. (2013) found the zero copy genotype, which is a new type of CNV. We also indentified this genotype with a high ratio (43.41%). Overall, we identified eight combinations of these three alleles. However, several new haplotypes remain unpublished. Therefore, further systematic research on CNV should be conducted.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was funded by the 973 projects (2014CB138105 and 2014CB160303), National Natural Science Foundation (31370031), and Program for New Century Excellent Talents in University (NCET-13-0776) of China.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00470
PCR Primers for detecting vernalization response and photoperiod genes in the Chinese bread wheat cultivars.
Molecular characterization of vernalization and photoperiod response genes in all wheat cultivars surveyed from the Yellow and Huai valley of China.
References
- Barrett B., Bayram M., Kidwell K. (2002). Identifying AFLP and microsatellite markers for vernalization response gene Vrn-B1 in hexaploid wheat (Triticum aestivum L.) using reciprocal mapping populations. Plant Breed. 121, 400–406. 10.1046/j.1439-0523.2002.732319.x [DOI] [Google Scholar]
- Beales J., Turner A., Griffiths S., Snape J. W., Laurie D. A. (2007). A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L). Theor. Appl. Genet. 115, 721–733. 10.1007/s00122-007-0603-4 [DOI] [PubMed] [Google Scholar]
- Cane K., Eagles H. A., Laurie D. A., Trevaskis B., Vallance N., Eastwood R. F., et al. (2013). Ppd-B1 and Ppd-D1 and their effects in southern Australian. Crop Pasture. Sci. 64, 100–114. 10.1071/CP13086 [DOI] [Google Scholar]
- Chen A., Dubcovsky J. (2012). Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS. Genet. 8:e1003134. 10.1371/journal.pgen.1003134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Gao M. X., Zhang J. H., Zuo A. H., Shang X. L., Cui D. Q. (2013a). Molecular characterization of vernalization response and photoperiod response genes in bread wheat from the Yellow and Huai valley of China. BMC Plant Biol. 13:199. 10.1186/1471-2229-13-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Xu H. X., Zhang F. Y., Xia X. C., He Z. H., Wang D. W., et al. (2011). Physical mapping of puroindoline b-2 genes and molecular characterization of a novel variant in durum wheat (Triticum turgidum L.). Mol. Breed. 28, 153–161. 10.1007/s11032-010-9469-2 [DOI] [Google Scholar]
- Chen F., Zhang F. Y., Li H. H., Morris C. F., Cao Y. Y., Shang X. L., et al. (2013b). Allelic variation and distribution independence of Puroindoline b-B2 variants and their association with grain texture in wheat. Mol. Breed. 32, 399–409. 10.1007/s11032-013-9879-z [DOI] [Google Scholar]
- Diaz A., Zikhali M., Turner A. S., Isaac P., Laurie D. A. (2012). Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associatedwith altered flowering time in wheat (Triticum aestivum). PLoS ONE 7:e33234. 10.1371/journal.pone.0033234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J., Lijavetzky D., Appendino L., Tranquilli G. (1998). Comparative RFLP mapping of Triticum monococcum genes controllingvernalization requirement. Theor. Appl. Genet. 97, 968–975. 10.1007/s001220050978 [DOI] [Google Scholar]
- Eagles H. A., Cane K., Kuchel H., Hollamby G. J., Vallance N., Eastwood R. F., et al. (2010). Photoperiod and vernalization gene effects in southern Australian wheat. Crop Pasture. Sci. 61, 721–730. 10.1071/CP10121 [DOI] [Google Scholar]
- Eagles H. A., Cane K., Trevaskis B. (2011). Veery wheats carry an allele of Vrn-A1 that has implications for freezing tolerance in winter wheats. Plant Breed. 130, 413–418. 10.1111/j.1439-0523.2011.01856.x [DOI] [Google Scholar]
- Fu D. L., Szucs P., Yan L. L., Helguera M., Skinner J. S., Zitzewitz J., et al. (2005). Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol. Genet. Genom. 273, 54–65. 10.1007/s00438-004-1095-4 [DOI] [PubMed] [Google Scholar]
- Galiba G., Quarrie S. A., Sutka J., Morgounov A. (1995). RFLP mapping of the vernalization (Vrn1) and frost resistance (Fr1) genes on chromosome 5A of wheat. Theor. Appl. Genet. 90, 1174–1179. 10.1007/bf00222940 [DOI] [PubMed] [Google Scholar]
- Guo Z. A., Song Y. X., Zhou R., Ren Z. L., Jia J. Z. (2010). Discovery, evaluation and distribution of haplotypes of the wheat Ppd-D1 gene. New Phytol. 185, 841–851. 10.1111/j.1469-8137.2009.03099.x [DOI] [PubMed] [Google Scholar]
- Han J., Zhang L. S., Li J. T., Shi L. J., Xie C. J., You M. S., et al. (2009). Wheat Backbone parents “triumph/Yanda 1817” hybrid progenies derived genetic analysis of composition. Acta. Agronomica. Sinica. 5, 1395–1404. 10.3724/SP.J.1006.2009.01395 [DOI] [Google Scholar]
- Iwaki K., Nishida J., Yanagisawa T., Yoshida H., Kato K. (2002). Genetic analysis of Vrn-B1 for vernalization requirement by using linked dCAPS markers in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 104, 571–576. 10.1007/s00122-001-0769-0 [DOI] [PubMed] [Google Scholar]
- Law C. N., Worland A. J. (1997). Genetic analysis of some flowering time and adaptive traits in wheat. New Phytol. 137, 19–28. 10.1046/j.1469-8137.1997.00814.x [DOI] [Google Scholar]
- Law C. N., Worland A. J., Giorgi B. (1975). The genetic control of ear-emergence time by chromosomes 5A and 5D of wheat. Heredity 36, 49–584. 10.1038/hdy.1976.5 [DOI] [Google Scholar]
- Milec Z., Tomková L., Sumíková T., Pánková K. (2012). A new multiplex PCR test forthe determination of Vrn-B1 alleles in bread wheat (Triticum aestivum L.). Mol. Breed. 30, 317–323. 10.1007/s11032-011-9621-7 [DOI] [Google Scholar]
- Muterko A., Balashova I., Cockram J., Kalendar R., Sivolap Y. (2014). The new wheat vernalization response allele Vrn-D1s is caused by DNA transposon insertion in the first intron. Plant Mol. Biol. Rep. 33, 294–303. 10.1007/s11105-014-0750-0 [DOI] [Google Scholar]
- Santra D. K., Santra M., Allan R. E., Campbell K. G., Kidwell K. K. (2009). Genetic and molecular characterization of vernalization genes Vrn-A1, Vrn-B1, and Vrn-D1 in spring wheat germplasm from the Pacific Northwest region of the U.S.A. Plant Breed. 28, 576–584. 10.1111/j.1439-0523.2009.01681.x [DOI] [Google Scholar]
- Shcherban A. B., Efremova T. T., Salina E. A. (2012). Identification of a new Vrn-B1 allele using two near-isogenic wheat lines with difference in heading time. Mol. Breed. 29, 675–685. 10.1007/s11032-011-9581-y [DOI] [Google Scholar]
- Shitsukawa N., Iqari C., Shimada S., Kitagawa S., Sakamoto K., Saito H. (2007). The einkorn wheat (Triticum monococcum) mutant, maintained vegetative phase, is caused by a deletion in the VRN1 gene. Genes. Genet. Syst. 82, 167–170. 10.1266/ggs.82.167 [DOI] [PubMed] [Google Scholar]
- Wang L., Niu J. S., Li Q. Y., Qin Z., Ni Y. J., Xu H. X. (2015). Allelic variance at the vernalization gene locus Vrn-D1 in a group of sister wheat (Triticum aestivum) lines and its effects on development. J. Agricul. Sci. 153, 588–601. 10.1017/S0021859614000409 [DOI] [Google Scholar]
- Worland A. J., Börner A., Korzun V., Li W. M., Petrovíc S., Sayers E. J. (1998). The influence of photoperiod genes on the adaptability of European winter wheats. Euphytica 100, 385–394. 10.1023/A:1018327700985 [DOI] [Google Scholar]
- Yan L., Fu D., Lin C., Blechl A., Tranquilli G., Bonafede M., et al. (2006), The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. U.S.A. 103, 19581–19586. 10.1073/pnas.0607142103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Helguera M., Kato K., Fukuyama S., Sherman J., Dubcovsky J. (2004a). Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor. Appl. Genet. 109, 1677–1686. 10.1007/s00122-004-1796-4 [DOI] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Blechl A., Tranquilli G., Ramakrishna W., SanMiguel P., et al. (2004b). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303, 1640–1644. 10.1126/science.1094305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Tranquilli G., Helguera M., Fahima T., Dubcovsky J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. U.S.A. 100, 6263–6268. 10.1073/pnas.0937399100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang Y., Wu S., Yang J., Liu H., Zhou Y. (2012). A single nucleotide polymorphism at theVrn-D1promoter region in common wheat is associated with vernalization response. Theor. Appl. Genet. 125,1697–1704. 10.1007/s00122-012-1946-z [DOI] [PubMed] [Google Scholar]
- Zhang X. K., Xia X. C., Xiao Y. G., Dubcovsky J., He Z. H. (2008). Allelic variation at the vernalization genes Vrn-A1, Vrn-B1, Vrn-D1 and Vrn-B3 in Chinese common wheat cultivars and their association with growth habit. Crop Sci. 48, 458–470. 10.2135/cropsci2007.06.035522875177 [DOI] [Google Scholar]
- Zhu J., Pearce S., Burke A., See D. R., Skinner D. Z., Dubcovsky J., et al. (2014). Copy number and haplotype variation at the VRN-A1 and central FR-A2 loci are associated with frost tolerance in hexaploid wheat. Theor. Appl. Genet. 127, 1183–1197. 10.1007/s00122-014-2290-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Q. S. (2003). Wheat Improvement and Pedigree Analysis in Chinese Wheat Cultivars. Beijing: China Agriculture Press. [Google Scholar]
- Żmieñko A., Samelak A., Kozłowski P., Figlerowicz M. (2014). Copy number polymorphism in plant genomes. Theor. Appl. Genet. 127, 1–18. 10.1007/s00122-013-2177-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR Primers for detecting vernalization response and photoperiod genes in the Chinese bread wheat cultivars.
Molecular characterization of vernalization and photoperiod response genes in all wheat cultivars surveyed from the Yellow and Huai valley of China.