Abstract
The ABCC multidrug resistance associated proteins (ABCC-MRP), a subclass of ABC transporters are involved in multiple physiological processes that include cellular homeostasis, metal detoxification, and transport of glutathione-conjugates. Although they are well-studied in humans, yeast, and Arabidopsis, limited efforts have been made to address their possible role in crop like wheat. In the present work, 18 wheat ABCC-MRP proteins were identified that showed the uniform distribution with sub-families from rice and Arabidopsis. Organ-specific quantitative expression analysis of wheat ABCC genes indicated significantly higher accumulation in roots (TaABCC2, TaABCC3, and TaABCC11 and TaABCC12), stem (TaABCC1), leaves (TaABCC16 and TaABCC17), flag leaf (TaABCC14 and TaABCC15), and seeds (TaABCC6, TaABCC8, TaABCC12, TaABCC13, and TaABCC17) implicating their role in the respective tissues. Differential transcript expression patterns were observed for TaABCC genes during grain maturation speculating their role during seed development. Hormone treatment experiments indicated that some of the ABCC genes could be transcriptionally regulated during seed development. In the presence of Cd or hydrogen peroxide, distinct molecular expression of wheat ABCC genes was observed in the wheat seedlings, suggesting their possible role during heavy metal generated oxidative stress. Functional characterization of the wheat transporter, TaABCC13 a homolog of maize LPA1 confirms its role in glutathione-mediated detoxification pathway and is able to utilize adenine biosynthetic intermediates as a substrate. This is the first comprehensive inventory of wheat ABCC-MRP gene subfamily.
Keywords: ABCC-MRP proteins, cadmium stress, Triticum aestivum, detoxification, yeast complementation, seed development
Introduction
ATP-binding cassette transporter (ABC) proteins are found in all living organisms and constitute one of the largest known superfamily (Henikoff et al., 1997; Rea, 1999). They are involved in multitude functions in animals and plants for the transport of broad substrates ranging from glutathione-conjugates, xenobiotic compounds, intermediate metabolites and hormones (Rea, 1999; Bakos et al., 2000; Augustine et al., 2005; Kang et al., 2010; Klaassen and Aleksunes, 2010). The ABCC subfamily of these transporters classically referred as multi drug resistance associated proteins (MRPs) have been best characterized in Arabidopsis. Each of these MRP proteins contains at least one highly conserved ATPase domain as an energy provider (~200 aa residues long) also referred as nucleotide binding domains (NBD). This ATPase domain comprises of a Walker motif A and Walker motif B on either end of an ABC signature motif (Klein et al., 2006). MRP proteins usually contain two repeats of transmembrane (TMD) and NBD represented as TMD1-NBD1-TMD2-NBD2 (Klein et al., 2006).
Fifteen MRP transporters (AtMRP1-15) are reported from model plant Arabidopsis (Sanchez-Fernandez et al., 2001; Kolukisaoglu et al., 2002). Genome wide identification and expression analysis of ABC transporters including MRP subclass has also been explored from crop plants like rice (OsABCC1-17) (Jasinski et al., 2003; Garcia et al., 2004) maize (ZmABCC1-13) (Swarbreck et al., 2003; Pang et al., 2013) and Vitis vinifera (VvABCC1-26) (Cakier and Kilickaya, 2013). ABCC-MRP from Arabidopsis were shown to be involved in functions that included vacuolar sequestration of metabolites, cellular signaling (Martinoia et al., 1993), hormone transport (Ko et al., 2014), pathogen response (Ji et al., 2014; Walter et al., 2008) development of plant tissues (Wu et al., 2014), and secondary metabolite transport (Jasiñski et al., 2001). Earlier, MRP proteins were considered as the classical GSH-S conjugate pumps since evidence for other physiological roles were lacking (Ishikawa et al., 1997). Subsequently, new MRPs from Arabidopsis were characterized for their roles in hormonal regulation and physiological process (Gaedeke et al., 2001). Forward and reverse genetic approaches, showed their importance in the transport of glutathione and glucuronides conjugates (Liu et al., 2001), heavy metal tolerance (Gaillard et al., 2008; Park et al., 2012), chlorophyll catabolite transport (Lu et al., 1998; Tommasini et al., 1998; Frelet-Barrand et al., 2008), guard cell signaling and phytic acid transport (Nagy et al., 2009) and herbicide transport (Frelet-Barrand et al., 2008).
Studies indicated that ABCC-MRP transporters could be potential targets for trait development in higher crops (Xu et al., 2009; Panzeri et al., 2011). Embryo-specific silencing of ABC-MRP like gene resulted in the reduced phytic acid content of maize and soybean seeds (Shi et al., 2007). Recently, ABCC1 from grape berry showed their ability to transport anthocyanidin 3-O-glucosides in-vitro in an ATP- and GSH-dependent manner with high affinity (Francisco et al., 2013). A single partial wheat ABCC-MRP gene (AAL47686.1, partial CDS) have been speculated for xenobiotic detoxification (Theodoulou et al., 2003). Recently, another wheat MRP transporter, TaABCC3 was shown to be involved in grain development and providing resistance against secreted mycotoxin from Fusarium (Walter et al., 2015).
In yeast, six MRP proteins referred as Ycf1, Ybt1, Bpt1, Vmr1, Yor1, and Nft1 (Paumi et al., 2009) are present. Yeast MRPs are characterized for their possible role in the vacuolar transport of certain secondary metabolites and heavy metal sequestration (Li et al., 1996; Sharma et al., 2002). Yeast mutants defective in one or multiple MRP genes have been utilized as an important resource to address the functional activity of MRP orthologs across the kingdom (Tommasini et al., 1996, 1998). MRPs from plants complemented YCF1 function in yeast, thus speculating their potential involvement as a glutathione transporter (Tommasini et al., 1998; Wang and Wu, 2006).
Allohexaploid wheat (Triticum aestivum L.), is an important cereal crop, consumed as a staple food by a large population in the developing countries. Although, plant ABCC-MRP transporters were identified from model crops, limited attempts were made to characterize this subfamily from wheat. In this study, to address the importance of ABCC proteins for their diverse biological and physiological processes in wheat, systematic analysis and molecular function of ABCC genes was performed. Molecular gene structure and chromosomal locations of the putative wheat ABCC genes were predicted by both genome-wide analysis and ESTs searches. Eighteen wheat ABCC genes were identified and subjected to phylogenetic analyses along with Arabidopsis and rice counterparts. Their expression using qRT-PCR was studied to reveal their transcript accumulation in different tissues i.e., roots, stem, leaves, flag leaf, developing wheat grains, and in roots exposed to Cd stress. In addition, we demonstrated that TaABCC13 (previously reported as TaMRP3, Bhati et al., 2014) has the ability to utilize adenine biosynthetic intermediates as a substrate in yeast. This is the first report providing a detailed inventory with molecular characterization of wheat ABCC-MRP transporters along with the possible functional evidence for the transport of glutathione-conjugated substrate.
Materials and methods
Plant materials, growth conditions, and treatments
Bread wheat cv. C306, a good processing quality Indian variety (Singh et al., 2014) was used for this study. For tissue sample collection, plants were grown in growth chambers under a 12 h photoperiod at 400 μmol m−2 s−1, 70 percent relative humidity and 25°C/18°C (day/night). Healthy plants with 14 days after anthesis (DAA) mature seeds were used to collect different vegetative tissues i.e., roots, stem, leaves, flag leaf, and seeds. Tissue samples were collected under random bulk up experiment method from different plants and snap frozen. Collected samples were stored at −80°C before RNA extraction. In total three biological replicates were used for the tissue samples. To study gene expression through seed maturation, the main individual spikes of the biological replicates were tagged at the first day after anthesis (DAA). The tagged spikes were harvested at subsequent developing days at 7, 14, 21, and 28 DAA and frozen in liquid nitrogen for RNA extraction.
For treatments, surface sterilized seeds were soaked in half strength Hoagland's solution in Petri dishes for germination. Germinating seedlings were transferred to a hydroponic growing system with half-strength Hoagland's solution. Seedlings of 7 days old (with one fully expanded leaf) were treated with Hoagland's solution supplemented with the 50 μM Cd (CdCl2) or 10 μM H2O2 for 24 H separately. After 24 h of stress, leaves and root samples were snapped frozen in liquid nitrogen and stored at −80°C for further use. Seedlings without stress treatment were used as the experimental control. The CdCl2 treated seedlings were subjected to estimate relative metal accumulation. Roots and leaves were collected and dried overnight at 60°C. The equal weight of treatment and control tissues (roots and leaves) was microwave digested in HNO3 (Suprapur® Merck, Germany). These digested samples were used for metal analysis by using inductively coupled plasma mass spectrometry (ICP-MS; 7700×Agilent Technologies, Santa Clara, CA).
To study the effect of hormones, abscisic acid (ABA) and gibberellic acid (GA3) were used as suggested earlier with minor modifications (Hwang et al., 2003; Eastmond and Jones, 2005). Six to seven spikes of 14 DAA stage were used to collect seeds and were subjected to incubation with either ABA (100 μM) or GA3 (60 μM) containing 20 mM CaCl2. Control seeds imbibed in CaCl2buffer were used. Three replicates containing 22–25 seeds per plates with their respective hormonal treatments were used for the experiment. After 60 min of incubation seeds were collected and rapidly chilled before storage at −80°C. RNA was extracted from the respective treatments and was subjected for qRT-PCR. Five technical replicates from biological replicates were used for qRT-PCR analysis.
Bioinformatics analysis
To identify new ABCC-MRP subclass members from wheat, orthologous sequences from Arabidopsis and rice were used as query in two independent approaches. First these queries were used against the wheat EST database in tBLASTn algorithm at NCBI. The EST hits with a threshold BLAST score of 400 were considered as significant. These EST sequences were screened for presence of Walker A, B, and ABCC-MRP like ATPase domains as defined in the NCBI CDD database. The EST sequences containing typical domains were used for blast analysis on the wheat genome to map their unique position hit at different chromosome regions.
In the second approach the orthologous query sequences were individually mapped over wheat genome assemblies available at full length information for each individual member was derived from wheat genome contigs available at International wheat genome sequencing consortia (IWGSC, www.wheatgenome.org) and further these annotated ORFs were qualitatively checked with the gene structure database of wheat reported by Mayer et al. (2014). The full length amino acid sequences for each TaABCC were used to predict the domain topology and arrangement using PROSITE tool available on the ExPASy protein server. MEGA6 based alignment file prepared from all TaABCC that was used to develop an unrooted phylogenetic tree and identification of conserved amino acid patterns within TaABCC proteins. Domain prediction was performed by using Conserved Domain Database (CDD) (Marchler-Bauer et al., 2011). WebLogo3 generated logo presentations were analyzed for presence of representative ABCC-MRP domains. These identified full length sequences were named as per nomenclature guidelines and inventory of plant ABC proteins (Verrier et al., 2008). Most of these TaABCC proteins were named firstly on the basis of phylogeny and their recovery from wheat genome.
RNA isolation and quantitative real time PCR (qRT-PCR)
RNA was extracted from different tissues like root, stem, leaf, flag leaf, and developing seeds (7, 14, 21, and 28 DAA-days after anthesis) of wheat plants. Total RNA was extracted using the RNeasy Plant MiniKit (Qiagen, Valencia, CA, USA), following manufacturer's instructions. Genomic DNA contamination was removed by DNase-I treatment (RNase free kit, Ambion, USA). Transcriptor First Strand cDNA Synthesis Kit RT-PCR (Roche, USA) was used for cDNA preparation from two micrograms of RNA. Reverse transcription was performed using random hexamer primers following the manufacturer's guidelines. Gene specific primers (Table S1) were used with QuantiTect SYBR Green RT-PCR Master mix (Qiagen, USA) based qRT-PCR reactions up to 45 cycles on ABI 7500 Fast System (Applied Biosystems, Foster City, CA, USA). Five replicates from each biological sample were used to perform qRT-PCR analysis. For PCR reaction 4–5 replicates for each gene were amplified from 3 independent cDNA preparations arising from different biological replicates. Relative expression level was quantified using 2−ΔΔCt method after normalizing Ct values against 18 s rRNA expression (Schmitteng, 2001). The data are expressed as mean ± standard deviation. Statistical analysis was performed using Origin 6.0 software (Origin Lab Corporation, MA, USA). One-Way analysis of variance (ANOVA) followed by Turkey's multiple comparison test was applied to check the level of significance. In all the tests, statistical significance was checked at p < 0.01 and/or p < 0.05.
Phenotypic complementation of yeast adenine biosynthetic mutants and quantitation of pigmentation
Yeast strain, YPH499 (ABC154) defective in adenine biosynthesis pathway, with a genotype of MATα, ura3–52; leu2- Δ 1; lys2–801; his3 Δ 200; trp1 Δ 63; ade2–101 was used as a positive control. Yeast mutant for ycf1 (ABC 470) with genotype of MATα, Dycf1::KanMX2; ura3–52; leu2-D1; lys2–801; his3D200; trp1D63; ade2–101 prepared in the same auxotrophic background was also used for the current study (37). For complementation experiment, TaABCC13 + pYES263 harboring in yeast Δycf1 mutant was used. Pre-culture of wild type, mutant and transformed yeast colonies were grown overnight in YPD broth and brought to the same OD600. The colonies were subsequently streaked on 0.5% YPD media for 4 days along with negative controls. The red pigmentation produced by the yeast colonies that includes ade2 mutants was quantified spectrophotometrically as described earlier (Chaudhuri et al., 1997). Briefly, yeast cells were grown to the saturation in half strength YPD to an OD of 2.0. Equal amount of cells were further lysed using glass beads in 5% sulfosalicyclic acid. The amount of pigmentation was measured at the absorbance of 530 nm.
Results
Inventory, molecular structure, and phylogenetic analysis of wheat ABCC proteins
EST databases and genome assembly of wheat were used to identify candidate ABCC-MRP genes. In total ~75 wheat ESTs was screened from dbEST BLAST results. ESTs were assembled to identify possible partial or full ORF with conserved domains and were designated as new TaABCC members. The result of BLAST analysis identified AAL47686.1 and AAQ10074.1 as previously annotated partial ABCC proteins from wheat. In addition to that, TaMRP3 (AIK23242.1) was also identified previously and characterized in wheat tissues (Bhati et al., 2014). Subsequently, the information generated from the recently released wheat genome was also utilized to identify more wheat ABCC-MRPs (Mayer et al., 2014). Our refined searches resulted in identification of 18 ABCC subclass proteins from wheat, hereafter referred as TaABCC1–TaABCC18. The nomenclature was based on the guidelines those are widely accepted for plant ABC protein (Verrier et al., 2008). Following this, previously reported TaMRP3 gene (Bhati et al., 2014) has now been referred as TaABCC13.
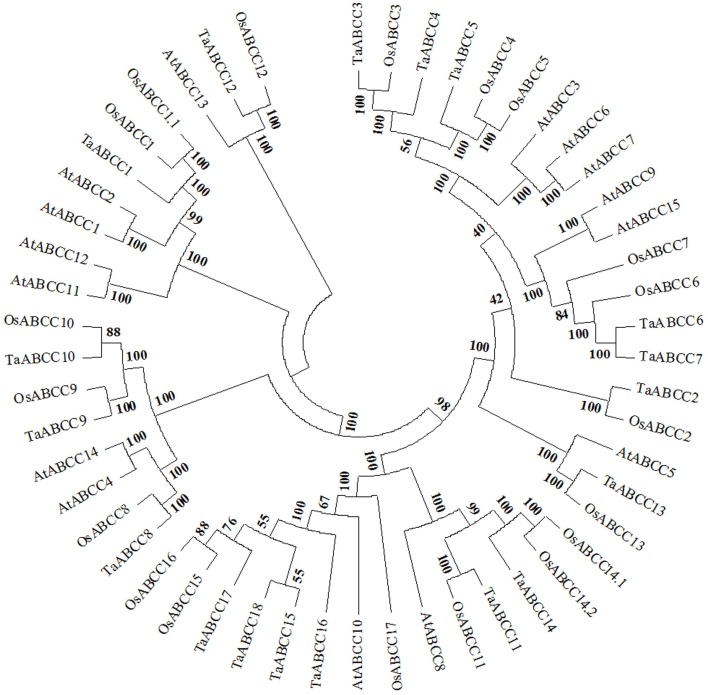
To assign the possible functional clues to wheat ABCC proteins, the identified wheat ABCC protein sequences were subjected to phylogenetic analysis along with ABCC-MRP members from Arabidopsis and rice. Based on our analysis each of the cluster consisted of multiple ABCC from Arabidopsis and rice suggesting a wide distribution of wheat ABCC proteins (Figure 1). Among wheat ABCC, TaABCC15 showed highest homology with TaABCC17 with percentage identity of 88.2. On the contrary, highest divergence was observed between TaABCC2 and TaABCC3 with percentage identity of 29.8. When a cross species comparison was done, the highest percentage identity of 75.1 was observed for TaABCC5 and AtMRP5. Similarly with rice, the highest percentage identity of 87.6 was observed for TaABCC6 with OsMRP6. Interestingly, TaABCC3 and TaABCC4 belong to the group that included a Cd inducible AtMRP3 (Bovet et al., 2003). Low phytic acid homolog genes TaABCC13 (previously reported as TaMRP3, Bhati et al., 2014) and AtMRP5 are grouped together along with OsMRP13 (Figure 1).
Figure 1.
Phylogeny alignment of plant ABCC-MRP proteins. Phylogenetic tree analysis of different MRP genes identified from wheat (TaABCC1-18), Arabidopsis (AtABCC1-15), rice (OsABCC1-17) (Updated sequence accessions with the new systematic names were used from Verrier et al., 2008).
Translated amino acid sequences for each TaABCC confirms presence of Signature motif and functional domains like transmembrane (TM) and ABCC-MRP (NBD) like domain those are conserved ubiquitously among members of ABCC-MRP transporter subclass. The predicted full length amino acid sequences from all TaABCC proteins were analyzed on CDD and Expasy PROSITE. The analysis revealed specific signatures of TM-NBD-TM-NBD with the possible location on membrane except for TaABCC members, without any mapped EST (Figure 2A). A ClustalW based sequence alignment was used to create logo representation for signature motif and each conserved domain. Logo representation strongly suggests more conserved amino acid in ABCC-MRP domains as compared to transmembrane domains (Figure 2B). ABCC-MRP1 domain in all wheat ABCC proteins has conserved NBD represented by K-S/T-S/T amino acids. ABC signature motif (SGGQKQR) was conserved and present among all the wheat ABCC proteins in the NBD domain of ABCC-MRP transporters (Figure 2B).
Figure 2.
Phylogenetic tree for TaABCC proteins based on the neighbor joining method, schematic diagram of the domain arrangement in different TaABCC and logo representation of amino acid conserved in different domains. (A) The un-rooted phylogenetic tree (left side) from TaABCC amino acid sequences was developed using the NJ method on MEGA6 software and domain representations (right side) were prepared using PROSITE server, TM- Transmembrane domain, MRP- nucleotide binding domain from Multidrug Resistant associated Protein). (B) Multiple alignment was analyzed on WebLogo 3 server from University of California, Berkeley for logo representation of TM, ABCC_MRP domains and signature motif; the Y-axis represents conservation of amino acid at that position (height).
Genomic distribution of ABCC genes on wheat genomes
Genomic evolution of modern wheat is contributed by three different diploid parents i.e., T. urartu, Ae. speltoides, and Ae. tauschii (contributed A, B, and D genome, respectively) (Mayer et al., 2014). The isolated ORFs for TaABCC genes were used to map their representative chromosomal locations based on contig assembly at IWGSC BLAST server (http://wheat-urgi.versailles.inra.fr). The CDS sequences for selective wheat ABCC genes were further confirmed by 5′ or 3′ RACE sequencing and subsequently refined by using IWGSC for their chromosomal locations (Table 1). Each TaABCC genes was mapped on all three homoeologous wheat chromosome but with varying similarity score. Genomic coordinates for each wheat ABCC genes and their homoeologous are mentioned in Table S2. All the wheat ABCC genes were also analyzed for their genomic structure using IWGSC sequences. Result suggested the presence of multiple introns (ranging from 2 to 27) for wheat ABCC genes (Figure S1).
Table 1.
Inventory and chromosomal location of ABCC genes from wheat.
| Gene | Representative ESTs | Chromosomal Location |
|---|---|---|
| TaABCC1* | HX108911.1, DR735499.1, BJ224413.1, CJ805925 | 2AL, 2BL, 2DL |
| TaABCC2 | CJ691783.1, CJ585444.1 | 3AL, 3BL, 3DL |
| TaABCC3* | CJ699639.1, HX193542.1, CV762434.1 | 3AS, 3DS, 4AS |
| TaABCC4* | CJ956417.1, CD877047.1, CJ944377.1, | 3AL, 3BL, 3DL |
| TaABCC5 | None Found with significant similarity score | 1AL, 1BL, 1DL |
| TaABCC6* | CJ859948.1, HX191927.1, CJ668043.1, CJ856118.1, | 2AL, 2BL, 2DL |
| TaABCC7 | None Found with significant similarity score | 2AL, 2BL, 2DL |
| TaABCC8* | CJ668035.1, HX159331.1, AL822018.1, BQ620567.1 | 5BL, 5DL, 5AL |
| TaABCC9 | BJ309016.1, CJ649956.1, CJ541775.1 | 2AS, 2BS, 2DS |
| TaABCC10 | None found with significant similarity score | 2DS, 2AS, 2BS |
| TaABCC11* | CD884377.1, HX070437.1, CD865647.1, CJ690695.1 | 7AL, 7BL, 7DL |
| TaABCC12 | HX143310.1 | 7AS, 7BS, 7DS |
| TaABCC13* | CD910572.1, CD910572.1, CA730883.1, HX147655.1 | 5AL, 4BL, 4DL |
| TaABCC14 | CJ667633.1, CJ560395.1, BJ303226.1 | 3AS, 3BS, 3DS |
| TaABCC15* | HX159319.1, CJ656810.1, CJ689124.1 | 7AS, 7BS, 7DS, |
| TaABCC16* | CV760609.1, CJ656810.1, HX159319.1, CJ689124.1 | 7AS, 7BS, 7DS, |
| TaABCC17 | CJ667633.1, CJ560395.1, BJ303226.1 | 3A, 3B, 3D |
| TaABCC18 | None found with significant similarity score | 7AS, 7BS, 7DS |
Indicates the ABCC genes for which sequence information was confirmed by either 5′ or 3′ RACE. Underline indicates the primers amplifying the transcript arising from these genomes.
Differential expression patterns of ABCC genes in wheat
In order to gain insight into the transcript accumulation of wheat ABCC transporter in different tissues, qRT-PCR analysis was performed. Expression studies were planned for those genes which are either covered by multiple reliable EST or present in our in-house transcriptome assembly from multiple wheat tissues. No unique representative ESTs (dbEST) was observed for TaABCC5, TaABCC7, TaABCC10, and TaABCC18; hence they were not selected for the expression study. The expression pattern of the remaining wheat ABCC genes was studied in roots, stem, leaves, and flag leaf that could help in speculating their site of molecular function. Analysis of the transcript abundance revealed that all the genes displayed differential expression patterns across the set of wheat tissue samples. Expression data, suggested the induction of wheat ABCC genes in leaves, root, stem, and flag leaf at different folds (Figure 3).
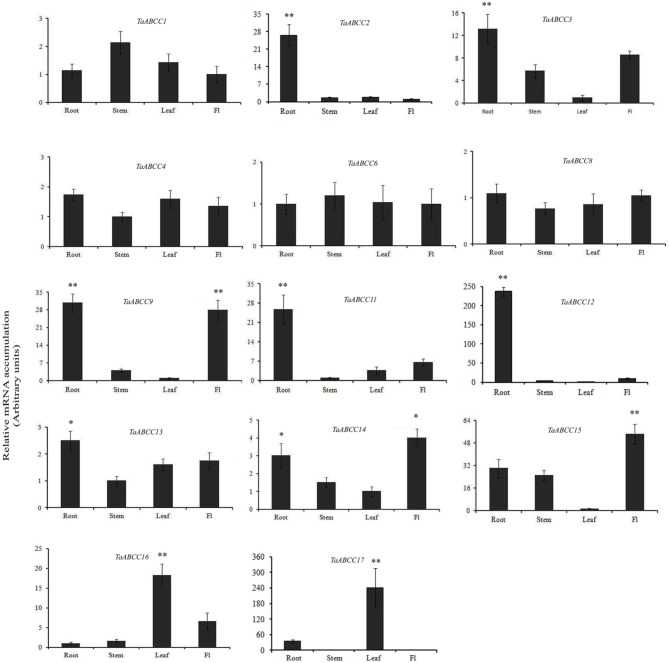
Figure 3.
Expression analysis of wheat ABCC genes in different parts of wheat plants. qRT-PCR analysis was performed on the cDNA templates prepared from 2 μg of DNase free RNA isolated from roots (R), stem (S), leaves (L), and flag leaf (FL) of 14 DAA wheat plants. Relative transcript accumulation of the genes was calculated. Each bar indicates the mean of five replicates with the indicated standard deviation of the mean. ** indicates significant difference at p < 0.01. * indicates significant difference at p < 0.05.
The transcript levels for TaABCC8 and TaABCC6 showed no significant change in expression levels and are ubiquitously expressed in all the tissue studied. Remaining TaABCC genes showed differential expression responses. Interestingly, expression of TaABCC4 and TaABCC13 were observed in similar patterns for all the tissues, although at different folds. In roots transcript accumulation was observed highest for TaABCC12 (~220-fold) followed by TaABCC2, TaABCC11 (~25-fold), and TaABCC3 (~12-fold) (Figure 3). TaABCC9 and TaABCC14 showed predominant expression in both roots and flag leaf. TaABCC16 and TaABCC17 are expressed at significantly higher level in leaves compared other tissues (Figure 3). Overall, expression data suggested differential and overlapping expression patterns implicating their function in wheat.
Temporal expression of wheat ABCC genes in seeds
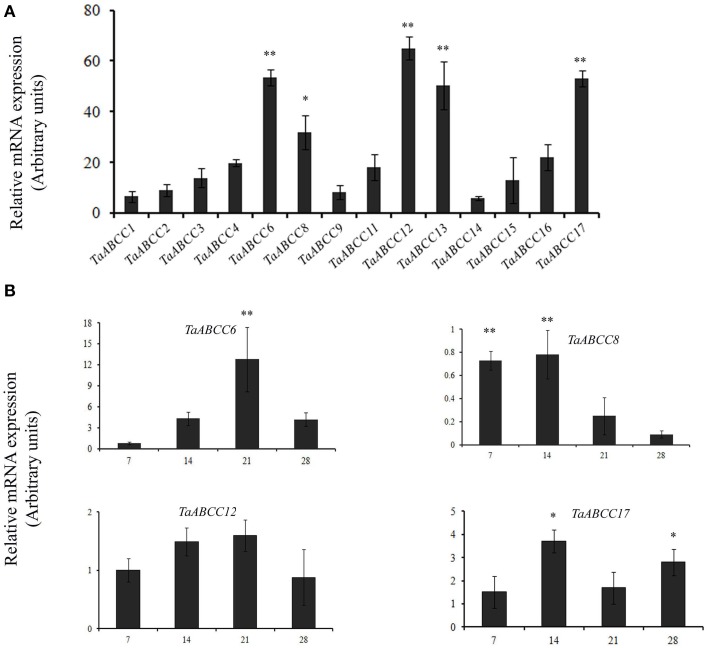
In order to characterize TaABCC genes in seeds, quantitative expression analyses were performed in 14 DAA wheat seeds. This time point represents a complete differentiation of outer aleurone layer and endosperm tissue of wheat seed. Expression data suggested significantly higher expression for TaABCC6, TaABCC8, TaABCC12, TaABCC13, and TaABCC17 in wheat seeds (Figure 4A). The relative transcript accumulation of these transcripts ranges ~30− to ~65-fold. Among the expressed wheat ABCC genes, relative transcript abundance of TaABCC12 was highest in seed (Figure 4A).
Figure 4.
Quantitative expression of wheat ABCC genes in wheat seeds. (A) The cDNA templates were prepared from 2 μg of DNase free RNA isolated from wheat seeds of 14 DAA. Each bar indicates the mean of 4–5 replicates with the indicated standard deviation of the mean. (B) qRT-PCR analysis during wheat seed development. The cDNA templates were prepared from 2 μg of DNAse free RNA isolated from different time point of seed maturation at 7, 14, 21, and 28 DAA. Each bar indicates the mean of five replicates with the indicated standard deviation of the mean. ** indicates significant difference at p < 0.01. * indicates significant difference at p < 0.05.
Temporal expression analysis of highly expressed ABCC genes in wheat was performed at different stages of grain filling (i.e., 7, 14, 21, and 28 DAA). TaABCC13 analysis was excluded since it was previously reported (previously reported as TaMRP3, Bhati et al., 2014). Only TaABCC8 was highly expressed at the early time points of grain filling compared to other wheat ABCC genes (Figure 4B). The expression of TaABCC8 decreased further with the maturation of the wheat seeds. TaABCC6 was significantly induced at 21 DAA compared to that of other time points. Overall, expression analysis suggested that all the TaABCC genes responds differentially during the development of wheat seed.
Hormonal regulation of ABCC genes
The selected ABCC genes those are highly expressed during wheat seed development were further studied for their response to exogenous treatment of GA3 and abscisic acid (ABA). In the current study, we chose GA3 and ABA, since these are the two major hormones that control the dynamics of seed maturation and grain filling (Sreenivasulu et al., 2006; Thiel et al., 2008). Expression analysis of genes in 14 DAA seeds when exposed to ABA showed no significant changes in the transcript accumulation of TaABCC8 (Figure 5A). The transcript abundance of TaABCC6 and TaABCC13 was slightly repressed. Only TaABCC12 and TaABCC17 were induced by ABA and GA3. In contrast, upon GA3 treatment, transcript accumulation of TaABCC6, TaABCC8, TaABCC12 TaABCC13, and TaABCC17 was significantly increased with respect to the control (Figure 5B). These results suggested that wheat ABCC genes are preferentially induced in seeds; when exogenously treated with GA3.
Figure 5.
Hormonal regulation of wheat ABCC genes. Fourteen DAA old wheat seeds were treated with either ABA (100 μM) or GA3 (60 μM) for 60 min. Seeds were collected and were subjected for RNA isolation. For quantification, cDNA templates were prepared from 2 μg of DNase free RNA isolated from wheat tissue. Each bar indicates the mean of five replicates with the indicated standard deviation of the mean. ** indicates significant difference at p < 0.01 with respect to control.
Expression patterns of ABCC genes under cadmium stress
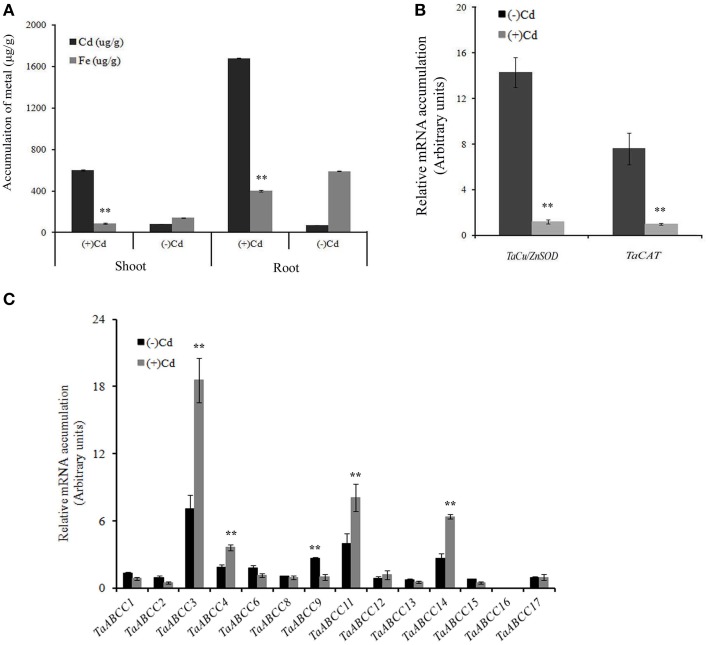
Heavy metal (HM) toxicity is one of the major abiotic stresses leading to hazardous effects in plants. Multiple plant ABCC-MRPs were speculated for their functional role for vacuolar sequestration, thus reducing the cellular metal toxicity (Bovet et al., 2003; Gaillard et al., 2008). In order to characterize the TaABCC against the metal stress, we exposed the wheat seedlings to Cd. After exposure, Cd accumulation was measured in roots and shoots of wheat seedlings. Results showed significantly higher accumulation of Cd in the plants exposed to metal compared to their respective controls (Figure 6A). In roots, Cd accumulation was 2.5-fold higher when compared to shoots (Figure 6A). Previously, it has been suggested that the plants exposed to Cd are a poor accumulator of iron (Meda et al., 2007). During our study, we also measured the iron accumulation in roots and shoots of Cd treated wheat seedlings. Our result suggested a significant decrease in the uptake of iron in roots treated with Cd (Figure 6A). No significant changes in the accumulation of iron in shoots were observed, suggesting the possibility that in this study most of the Cd gets retained in the roots rather being transported in the shoots within the time frame of the experiment.
Figure 6.
Effect CdCl2 on expression of wheat ABCC genes. (A) Cd accumulation in shoots and roots of 3 weeks old wheat seedlings when exposed to CdCl2. (B) qRT-PCR analysis of TaCu/ZnSOD and TaCAT when exposed to roots of wheat seedlings. (C) qRT-PCR analysis of all the identified wheat ABCC genes in roots of wheat seedlings exposed to CdCl2. For quantification, cDNA templates were prepared from 2 μg of DNAse free RNA isolated from wheat roots. Each bar indicates the mean of five replicates with the indicated standard deviation of the mean. ** indicates significant difference at p < 0.01 with respect their control.
The highest concentration of Cd exposure to the plants is usually accompanied with the interference in the expression of TaCu/ZnSOD and catalase transcripts (TaCAT) (Qiu et al., 2013). Thus, quantitative real time PCR (qRT-PCR) expression analysis of TaCu/ZnSOD transcripts was checked. Down-regulation of TaCu/ZnSOD and TaCAT transcript was observed in presence of Cd suggesting its toxic effect (Figure 6B). To study molecular responses to Cd, wheat ABCC genes were studied for their relative expression analysis in roots. Based on the expression of TaABCC genes, they could be classified into two categories; those which are differentially regulated and others where no significant change in the transcript accumulation was observed. The relative transcript levels among wheat ABCC genes suggested an enhanced expression of four genes i.e., TaABCC3, TaABCC4, TaABCC11, and TaABCC14 when exposed to Cd (Figure 6C). The highest transcript accumulation was observed for TaABCC3 that showed an increased transcript abundance of two- to three-folds when compared to control roots. Surprisingly, some of the wheat ABCC genes showed suppression in the expression. This suppression was observed consistently among the biological replicates. These slightly suppressed genes included TaABCC1, TaABCC6, and TaABCC9. No significant changes in the expression levels were observed for TaABCC2, TaABCC8, TaABCC12, TaABCC13, TaABCC16, and TaABCC17 (Figure 6C).
Expression patterns of ABCC genes in presence of H2O2
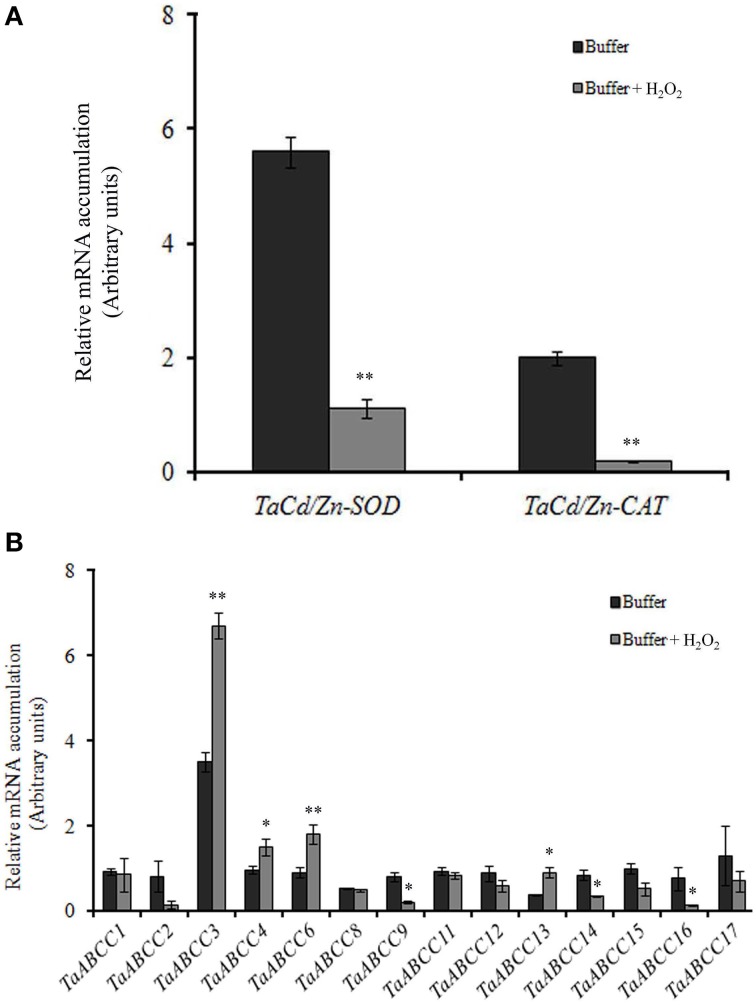
Previous reports suggested that exposure of plants to abiotic stress such as Cd causes accumulation of H2O2 (Cho and Seo, 2005). The induction of TaABCC transcripts when exposed to Cd could be also possibly due to generation of oxidative stress. To address this, wheat seedlings were exposed to oxidative stress (H2O2). Results showed down-regulation of TaCu/ZnSOD and TaCAT transcript when wheat seedlings were exposed to H2O2 (Figure 7A). Expression data suggested that few of the TaABCC genes were highly up-regulated in presence of H2O2. qRT-PCR analysis indicated an increased transcript accumulation of TaABCC3, TaABCC4, TaABCC6, and TaABCC13 in roots when exposed to H2O2 (Figure 7B). Upon H2O2treatment, transcript accumulation was highest for TaABCC3 with the cumulative abundance of two-fold higher when compared to control roots. On the contrary, no significant changes in the transcript mRNA accumulation were observed for TaABCC1, TaABCC8, TaABCC11, TaABCC12, and TaABCC17. Interestingly, transcript expression of TaABCC2, TaABCC9, TaABCC14, and TaABCC16 were slightly reduced (Figure 7B). These results suggested that Cd induced accumulation of H2O2 might be responsible for enhanced transcript abundance of certain wheat ABCC genes.
Figure 7.
Effect of H2O2 on expression of wheat ABCC genes. (A) qRT-PCR analysis of TaCu/ZnSOD and TaCAT when the roots of wheat seedlings were exposed to H2O2. (B) qRT-PCR analysis of all the identified wheat ABCC genes in wheat seedlings exposed to H2O2. The cDNA templates were prepared from 2 μg of DNA-free RNA isolated from the roots of the wheat seedlings. Each bar indicates the mean of 4–5 replicates with the indicated standard deviation of the mean. ** indicates significant difference at p < 0.01 with respect to control. * indicates significant difference at p < 0.05 with respect to control.
TaABCC13 is involved in transport of glutathione-conjugates in yeast
Previously, TaABCC13 (earlierTaMRP3, Bhati et al., 2014) was shown to complement yeast Δycf1 mutant for its Cd sensitivity. Yeast MRP proteins, Ycf1p and Bpt1p (Li et al., 1996; Petrovic et al., 2000) are the two members that are known to be involved in transport of glutathione-conjugates and have different degree of sensitivity toward Cd (Klein et al., 2000; Sharma et al., 2002). Based on these studies we suspected that TaABCC13 should also has the ability to detoxify glutathione-conjugates. To ascertain the role in glutathione-mediated detoxification by MRP, phenotypic assays for pigment accumulation in adenine biosynthetic mutants has been utilized (Sharma et al., 2003). Yeast Δycf1 when complemented with TaABCC13 could rescue the red pigmentation under adenine limiting conditions (Figure 8A). In order to calculate the extent of complementation, assays were performed to measure the intensity of pigmentation in yeast (Sharma et al., 2003). Our data suggested a significant accumulation (~2.5-folds) of the pigmentation in the wild type and yeast Δycf1 complemented with TaABCC13 compared to control (Figure 8B). These results confirm the ability of TaABCC13 to transport toxic adenine biosynthetic intermediates (phosphoribosyl-amino-imidazole and phosphoribosyl-amino-imidazole carboxylate) in Δycf1 those are conjugated with glutathione.
Figure 8.
Phenotypic complementation of yeast YCF1 mutant with TaABCC13 under adenine-limiting condition. (A) Parent strain YPH299 and its ScΔycf1 were used to complement with TaABCC13. All the cultures were used to complement with TaABCC13. All the cultures were grown to the OD of 0.5 and subsequently streaked on YPD plates with 0.5% of yeast extract. For control, ScΔycf1 transformed with empty plasmid pYES263 was used. All the strain used was isogeneic for nutritional marker. Pictures were taken 4 days post-incubation at 30°C. (B) Quantification of red pigmentation for mutant and ScΔYCF1 complemented with TaABCC13 was performed on the cell culture grown under adenine limiting conditions as described earlier. Each bar indicates the mean of four replicates with the indicated standard deviation of the mean. * indicates significant difference at p < 0.01 with respect to control.
Discussion
In the past, there have been significant efforts to understand the multiple functions of ABC transporters in yeast, humans and recently in plants. Although their roles in model plants have been explored extensively, but such evidences are still largely obscure from crops like wheat. Recent reports from rice, maize, and grapes conferred valuable roles to ABC transporters and offers initiative for further characterizing their biological implications. With advancement in understanding the functions of ABCC-MRP subclass, it became evident that they act as major detoxifiers and sequester metal-chelators into the plant vacuoles (Martinoia et al., 1993). Role of wheat ABCC-MRP proteins were not largely addressed till date. This work led to the identification of 18 wheat MRPs that were characterized in detail for their gene expression.
Gene structure and genomic distribution of wheat ABCC
ABCC-MRP subclass represents one of the important groups of ABC transporters that contains N-terminal extension TMD domain. This study identified 18 full length ABCC proteins. The largest identified protein was TaABCC1 with 1630 aa whereas the smallest one TaABCC18 contains 1024 aa (Table 1). Similar size distribution was also observed for rice and Arabidopsis (Verrier et al., 2008). Wheat being a three genome system, it was anticipated to have three homoeologous of representative genes (Table 1). All identified genes were located on either of the chromosome1, 2, 3, 4, 5, and 7 (Table 1). We failed to recover the full length sequence for few homoeologous due to limited wheat genome contig information. Nonetheless, considering the presence of homoeologous in wheat, this sums to a total of 54 wheat ABCC genes. The recent draft of wheat genome has also annotated putative members of ABCC-MRP subfamily (Mayer et al., 2014). Thus, analysis of the draft annotation sequences for ABCC-MRP like transporters was done and summarized along with their sequence identifier (Table S3). Interestingly, performing in-depth analysis of the above data resulted in the multiple annotations for the same protein sequences of varying lengths. Alternatively, utilizing EST and transcriptome based strategies to identify the expressed genes can be one of the reliable approaches for a complex and challenging genome like wheat. The IWGSC genome analysis suggested maximal wheat ABCC annotated sequences mapped to chromosome set 7, chromosome set 2, and chromosome set 3 of wheat genome. Four of the wheat ABCC genes are located on the chromosome 7 along with their respective homoeologous, showing high density region for wheat ABCC. This represents the comparatively high density of ABC-MRP annotation on chromosome 7, which can be as a result of intra-chromosomal duplication events during evolution of hexaploid wheat genome (Mayer et al., 2014). Our analysis of chromosomal location matches with the IWGSC annotation report. Contrarily, the current study was based on manual gene annotation using the ESTs and a genome contig database which takes into account the expression of the genes and addresses their functional roles in wheat. Furthermore, upon careful examination, no ESTs were mapped from NCBI for TaABCC5, TaABCC7, TaABCC10, and TaABCC18. Based on our previous observation, it seems some of these wheat ABCC genes may not be expressed, although careful experiments are required to confirm the expression of these transcripts.
In a few cases, this work reflects a more comprehensive observation compared to that of IWGSC annotations and gene structure predictions. During our analysis varying lengths of ABCC proteins were ascertained, but truncated TaABCC18 was identified as pseudogene. Such annotated truncated or pseudo genes were previously reported in case of Arabidopsis (Sanchez-Fernandez et al., 2001) and maize (Pang et al., 2013).
Wheat ABCC genes showed differential expression patterns
Studying the expression patterns in specific tissues and organs suggests the molecular clues for their role and help to address their functionality in plants. The preferential expression patterns suggested their specificities for the respective tissue/organs. For example, TaABCC2 and TaABCC3 showed highest expression in the roots whereas TaABCC1 and TaABCC16 are preferentially expressed in the stem and leaves. Phylogenetic analysis suggested that AtABCC6 and TaABCC3 are present in the same cluster. AtMRP6 is highly expressed at the initiation point of secondary roots, especially in xylem-opposite peri-cycle cells where lateral roots initiate (Gaillard et al., 2008). Growing roots are active sites for the synthesis of auxins (Ljung et al., 2005). TaABCC3 is also a close match for AtMRP3 and both the genes were found to be induced by HMs (Brunetti et al., 2015). Further TaABCC3 was induced by the presence of GA3, indicating its role in root architecture development, although further functional studies are required for confirmation. Further, examining the promoter of TaABCC3 derived from Chromosome 3B, resulted in identification of regions those are associated with root specific expression (GSH transporter and DRE), GA3 (GA-Myb) regulated domains and phosphate responsive domain (P1BS) (data not shown). This suggests that TaABCC3 can be a prime candidate for plant genetic engineering to enhance tolerance against heavy-metal accumulation.
TaABCC14 and TaABCC15 are highly abundant in flag leaf and both are clustered together in the phylogenetic tree. Flag leaf is the important organ for the biosynthesis of transitory starches, micronutrients and other metabolites that contribute to the seed maturation during the senescence (Ali et al., 2010). These observations suggested an important role of specific ABC transporters in flag leaf. TaABCC1 forms a distinct cluster with OsABCC1 and showed high expression in stem, suggesting its preferred site of function. OsABCC1 was recently shown to be involved in arsenic tolerance by sequestering it into the node cell vacuoles (Song et al., 2014). Both TaABCC1 and OsABCC1 are not induced by the presence of Cd. Similar expression profiles and close clustering on the phylogeny tree might indicate TaABCC1 may also perform such roles in plant tissue.
The early expression of these genes during seed development suggested their possible roles for seed trait development. Seed maturation is a result of controlled flux of hormones (GA3, ABA, and ethylene) and nutrients between the seed tissues (Sreenivasulu et al., 2006; Thiel et al., 2008). Thus, possible impact of the GA3 and ABA was tested on the transcript accumulation of wheat ABCC genes that are highly expressed in seeds. Earlier a PDR1-like gene was shown to be repressed by ABA treatment (Zhang et al., 2013) likewise in our case TaABCC6 and TaABCC13 were also repressed (Figure 5). Most of the wheat ABCC genes were highly induced by GA3, which is supported by previous expression pattern observed for AtABCC13/ABCC11 for this hormone (Guizani et al., 2014). The differential expression response of wheat ABCC genes for hormone treatment will help in assimilating clues regarding their possible roles in integrated pathways that are involved in multiple abiotic stresses. TaABCC13 is the closest functional ortholog for the OsMRP13 and AtMRP5 those are involved in phytic acid transport. TaABCC13 is a functional protein that could be also possibly involved in PA transport, especially in the aleurone tissue of the seed (Bhati et al., 2014). Thus, targeting TaABCC13 by recently developed genome editing tools will be a viable strategy to generate low phytate crop in wheat. On the similar grounds, the functionality of some of the other wheat ABCC genes could be assessed by using yeast or Arabidopsis mutants.
In this study, response of wheat ABCC genes for Cd mediated response was also addressed. During our experiments, Cd accumulation was found in roots and aerial parts of wheat plants, thus transcriptional regulation of genes involved in Cd transport and detoxification is expected. Down regulation of Cu/ZnSOD transcripts upon Cd stress is in accordance with the response observed previously in wheat, and Arabidopsis (Cuypers et al., 2011; Qiu et al., 2013). On the contrary, induction of Cu/ZnSOD transcripts was observed in soybean and perennial rye-grass suggesting that regulation of antioxidant activities those are induced by Cd is dependent on the plant species (Clemens, 2001). Uptake of other micronutrient especially Fe gets perturbed when plant roots were exposed to HMs like Cd (Astolfi et al., 2014). Similarly, in our study, uptake of iron was decreased in the presence of Cd (Figure 6A). The reduced support for the uptake of Fe and other micronutrient is explained due to “inducible deficiency” or because of Cd induced regulation of Fe-homeostasis (Cuypers et al., 2011). The expression of wheat ABCC genes was observed when exposed to either Cd or H2O2. Based on these results, it is suggested that the regulation of wheat ABCC could be either due to the direct effect of Cd or through the H2O2 generated by toxicity. Interestingly, TaABCC11 was only specifically induced by Cd and no significant change in expression was observed when exposed to H2O2. For the remaining wheat ABCC genes it seems that H2O2 generated by the Cd toxicity might be responsible for differential gene expression.
Conservation of functional activity of wheat ABCC
Yeast defective in ycf1 (yeast cadmium factor) an MRP transporter has been classically utilized for confirming the functional role of ABCC-MRP transporters from plants (Tommasini et al., 1998; Wang and Wu, 2006; Bhati et al., 2014). Earlier we reported that, TaABCC13 (earlier referred as TaMRP3) rescued Δycf1 sensitivity toward cadmium but its functional activity in utilizing glutathione-conjugates were not studied (Bhati et al., 2014). Under adenine limiting conditions, the toxic intermediates phosphoribosyl-amino-imidazole and phosphoribosyl-amino-imidazole carboxylate gets accumulated in these WT (ade1/ade2: YPH499) mutant. These toxic metabolites are conjugated with glutathione and transported inside the vacuole primarily by YCF1 (Sharma et al., 2003) showing red pigmentation. Therefore, defective YCF1 showed significant decrease in phenotypic pigmentation. Based on the pigmentation assays, the current study confirms the ability of TaABCC13 in transport of toxic adenine biosynthetic intermediates by the accumulation of red-pigmentation in YCF1 yeast mutant (Figure 8). Based on the PSort analysis, TaABCC13 reliability score for membrane localization was similar to that of ScYCF1, ZmLpa1, AtMRP1, AtMRP2, AtMRP5 and PvMRP1 (data not shown). AtMRP1 and AtMRP2 complements yeast YCF1 mutants as well as actively influx glutathione-conjugated substrate into the plant vacuoles (Martinoia et al., 1993; Liu et al., 2001). Similarly, ABCC1 from grape berry is also able to transport glutathione dependent substrates (Francisco et al., 2013). Based on these observations, it is evident that TaABCC13 could detoxify Cd and is also involved in the membrane transport of glutathione-conjugated substrates by sequestration. Thus, TaABCC13 is involved in a conserved function that a classical ABCC-MRP are known to perform. Surprisingly, no significant change in the expression of TaABCC13 was observed when exposed to Cd at 24 h. Thus, it could be important to study the expression of TaABCC13 when exposed to different doses of Cd. In roots, TaABCC3 was highly up-regulated when exposed to Cd toxicity. It would be valuable to screen remaining wheat ABCC genes for the functional rescue of Δycf1 sensitivity for Cd.
The current study provided the spatial-temporal characterization of 18 wheat ABCC genes. Wheat ABCC genes showed differential expression in the presence of Cd or H2O2 suggesting their importance in abiotic stresses. Additionally, TaABCC13 was able to rescue the functional activity of yeast mutant by utilizing glutathione-conjugated substrates. The insight provided herein will be a much needed foundation regarding the wheat ABCC protein function that could be directed for cereal crop improvement.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Executive Director, NABI for facilities and support. We wish to thank Prof. Anand K. Bacchawat (IISER, Mohali) for providing ycf1 mutant and help in the pigmentation assays. The research was supported by NABI intramural funds and partly by DBT sponsored project (Grant No: BT/PR5989/AGII/106/867/2012). KB acknowledges DBT, Govt. of India for fellowships. SA was supported by ICMR, Govt. of India for fellowship. MK was supported by UGC-CSIR, Govt. of India for Fellowship. SS and VS were supported by grant to AP. Technical assistance by Atul Kumar is greatly appreciated for helping in ICPMS.
Glossary
Abbreviations
- ABC-MRP
ATP binding cassette multi drug resistance protein
- DAA
day after anthesis
- PA
phytic acid
- lpa
low phytic acid.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00488
Exon/intron structures of wheat ABCC genes are shown. Black boxes represent exons and black lines represent introns. Total number of introns for each gene are represented on the right side.
References
- Ali M. A., Hussain M., Khan M. I., Ali Z., Zulkiffal M., Anwar J., et al. (2010). Source-sink relationship between photosynthetic organs and grain yield attributes during grain filling stage in spring wheat (Triticum aestivum). Int. J. Agric. Biol. 12, 509–515. [Google Scholar]
- Astolfi S., Ortolani M. R., Catarcione G., Paolacci A. R., Cesco S., Pinton R., et al. (2014). Cadmium exposure affects iron acquisition in barley (Hordeum vulgare) seedlings. Physiol. Plant. 152, 646–659. 10.1111/ppl.12207 [DOI] [PubMed] [Google Scholar]
- Augustine L. M., Markelewicz R., Jr., Boekelheide K., Cherrington N. J. (2005). Xenobiotic and endobiotic transporter mRNA expression in the blood–testis barrier. Drug Metab. Dispos. 33, 182–189. 10.1124/dmd.104.001024 [DOI] [PubMed] [Google Scholar]
- Bakos E., Evers R., Sinko E., Varadi A., Borst P., Sarkadi B. (2000). Interactions of the human multidrug resistance proteins MRP1 and MRP2 with organic anions. Mol. Pharmacol. 57, 760–768. 10.1124/mol.57.4.760 [DOI] [PubMed] [Google Scholar]
- Bhati K. K., Aggarwal S., Sharma S., Mantri S., Singh S. P., Bhalla S., et al. (2014). Differential expression of structural genes for the late phase of phytic acid biosynthesis in developing seeds of wheat (Triticum aestivum L.). Plant Sci. 224, 74–85. 10.1016/j.plantsci.2014.04.009 [DOI] [PubMed] [Google Scholar]
- Bovet L., Eggmann T., Meylan-Bettex M., Polier J., Kammer P., Marin E., et al. (2003). Transcript levels of AtMRPs after cadmium treatment: induction of AtMRP3. Plant Cell Environ. 26, 371–381. 10.1046/j.1365-3040.2003.00968.x [DOI] [Google Scholar]
- Brunetti P., Zanella L., De Paolis A., Di Litta D., Cecchetti V., Falasca G., et al. (2015). Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J. Exp. Bot. 66, 3815–3829. 10.1093/jxb/erv185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakier B., Kilickaya O. (2013). Whole-genome survey of the putative ATP-binding cassette transporter family genes in Vitis vinifera. PLoS ONE 8:e78860. 10.1371/journal.pone.0078860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri B., Ingavale S., Bachhawat A. K. (1997). apd1±, a gene required for red pigment formation in the ade6 mutants of Schizosaccharomyces pombe encodes an enzyme required for glutathione biosynthesis: a role for glutathione and a glutathione-conjugate pump. Genetics 145, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho U., Seo N. (2005). Oxidative stress in A. thaliana exposed to cadmium is due to hydrogen peroxide accumulation. Plant Sci. 168, 113–120. 10.1016/j.plantsci.2004.07.021 [DOI] [Google Scholar]
- Clemens S. (2001). Molecular mechanism of plant metal tolerance and homeostasis. Planta 212, 475–486. 10.1007/s004250000458 [DOI] [PubMed] [Google Scholar]
- Cuypers A., Smeets K., Ruytinx J., Opdenakker K., Keunen E., Remans T., et al. (2011). The cellular redox state as a modulator in cadmium and copper responses in Arabidopsis thaliana seedlings. J. Plant Physiol. 168, 309–316. 10.1016/j.jplph.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Eastmond P. J., Jones R. L. (2005). Hormonal regulation of gluconeogenesis in cereal aleurone is strongly cultivar-dependent and gibberellin action involves SLENDER1 but not GAMYB. Plant J. 44, 483–493. 10.1111/j.1365-313X.2005.02544.x [DOI] [PubMed] [Google Scholar]
- Francisco R. M., Regalado A., Ageorges A., Burla B. J., Bassin B., Eisenach C., et al. (2013). ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell 25, 1840–1854. 10.1105/tpc.112.102152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frelet-Barrand A., Kolukisaoglu H. U., Plaza S., Ruffer M., Azevedo L., Hortensteiner S., et al. (2008). Comparative mutant analysis of Arabidopsis ABCC-type ABC transporters: AtMRP2 contributes to detoxification, vacuolar organic anion transport and chlorophyll degradation. Plant Cell Physiol. 49, 557–569. 10.1093/pcp/pcn034 [DOI] [PubMed] [Google Scholar]
- Gaedeke N., Klein M., Kolukisaoglu U., Forestier C., Müller A., Ansorge M., et al. (2001). The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. EMBO J. 20, 1875–1887. 10.1093/emboj/20.8.1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard S., Jacquet H., Vavasseur A., Leonhardt N., Forestier C. (2008). AtMRP6/AtABCC6, an ATP-Binding Cassette transporter gene expressed during early steps of seedling development and up-regulated by cadmium in Arabidopsis thaliana. BMC Plant Biol. 8:22. 10.1186/1471-2229-8-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia O., Bouige P., Forestier C., Dassa E. (2004). Inventory and comparative analysis of rice and Arabidopsis ATP-binding cassette (ABC) systems. J. Mol. Biol. 343, 249–265. 10.1016/j.jmb.2004.07.093 [DOI] [PubMed] [Google Scholar]
- Guizani T. E., Blanc N., Triki S., St-Pierre B., Ducos E. (2014). Expression pattern of AtABCC13/MRP11 reveals developmental, hormonal, and nutritional regulations. Biol. Plant. 58, 231–240. 10.1007/s10535-013-0387-0 [DOI] [Google Scholar]
- Henikoff S., Greene E. A., Pietrokovski S., Bork P., Attwood T. K., Hood L. (1997). Gene families: the taxonomy of protein paralogs and chimeras. Science 278, 609–614. 10.1126/science.278.5338.609 [DOI] [PubMed] [Google Scholar]
- Hwang Y. S., Bethke P. C., Gubler F., Jones R. L. (2003). cPrG-HCl a potential H+/Cl−symporter prevents acidification of storage vacuoles in aleurone cells and inhibits GA-dependent hydrolysis of storage protein and phytate. Plant J. 35, 154–163. 10.1046/j.1365-313X.2003.01789.x [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Li Z. S., Lu Y. P., Rea P. A. (1997). The GS-X pump in plant, yeast, and animal cells: structure, function, and gene expression. Biosci. Rep. 17, 189–207. 10.1023/A:1027385513483 [DOI] [PubMed] [Google Scholar]
- Jasinski M., Ducos E., Martinoia E., Boutry M. (2003). The ATP-binding cassette transporters: structure, function, and gene family comparison between rice and Arabidopsis. Plant Physiol. 131, 1169–1177. 10.1104/pp.102.014720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasiñski M., Stukkens Y., Degand H., Purnelle B., Marchand-Brynaert J., Boutry M. (2001). A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell 13, 1095–1107. 10.1105/tpc.13.5.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Peng Y., Meckes N., Allen S., Stewart N., Traw M. B. (2014). ABC transporter AtABCG16 increases plant tolerance to abscisic acid and assists in basal resistance against Pseudomonas syringae DC3000. Plant Physiol. 166, 879–888. 10.1104/pp.114.248153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Hwang J. U., Lee M., Kim Y. Y., Assmann S. M., Martinoia E., et al. (2010). PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107, 2355–2360. 10.1073/pnas.0909222107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen C. D., Aleksunes L. M. (2010). Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol. Rev. 62, 1–96. 10.1124/pr.109.002014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M., Burla B., Martinoia E. (2006). The multidrug resistance-associated protein (MRP/ABCC) subfamily of ATP-binding cassette transporters in plants. FEBS Lett. 580, 1112–1122. 10.1016/j.febslet.2005.11.056 [DOI] [PubMed] [Google Scholar]
- Klein M., Martinoia E., Hoffmann-Thoma G., Weissenbock G. (2000). A membrane-potential dependent ABC-like transporter mediates the vacoular uptake of rye flavones glucuronidase: regulation of glucuronide uptake by glutathione and its conjugates. Plant J. 21, 289–304. 10.1046/j.1365-313x.2000.00684.x [DOI] [PubMed] [Google Scholar]
- Ko D., Kang J., Kiba T., Park J., Kojima M., Do J., et al. (2014). Arabidopsis ABCG14 is essential for the root-to- shoot translocation of cytokinin. Proc. Natl. Acad. Sci. U.S.A. 19, 7150–7155. 10.1073/pnas.1321519111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolukisaoglu H. U., Bovet L., Klein M., Eggmann T., Geisler M., Wanke D., et al. (2002). Family business: the multidrug-resistance related protein (MRP) ABC transporter genes in Arabidopsis thaliana. Planta 216, 107–119. 10.1007/s00425-002-0890-6 [DOI] [PubMed] [Google Scholar]
- Li Z. S., Szczypka M., Lu Y. P., Thiele D. J., Rea P. A. (1996). The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. J. Biol. Chem. 271, 6509–6517. [DOI] [PubMed] [Google Scholar]
- Liu G., Sanchez-Fernandez R., Li Z. S., Rea P. A. (2001). Enhanced multi specificity of Arabidopsis vacuolar multidrug resistance-associated protein-type ATP-binding cassette transporter, AtMRP2. J. Biol. Chem. 276, 8648–8656. 10.1074/jbc.M009690200 [DOI] [PubMed] [Google Scholar]
- Ljung K., Hull A. K., Celenza J., Yamada M., Estelle M., Normanly J., et al. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17, 1090–1104. 10.1105/tpc.104.029272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. P., Li Z. S., Drozdowicz Y. M., Hortensteiner S., Martinoia E., Rea P. A. (1998). AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: functional comparisons with AtMRP1. Plant Cell 10, 267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., et al. (2011). CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39, 225–229. 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia E., Grill E., Tommasini R., Kreuz K., Amrhein N. (1993). An ATP-dependent glutathione-S-conjugate ‘export’ pump in the vacuolar membrane of plants. Nature 364, 247–249. 10.1038/364247a0 [DOI] [Google Scholar]
- Mayer K. F., Rogers K. F., Doležel J., Pozniak C., Eversole K., Feuillet C., et al. (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. International Wheat Genome Sequencing Consortium (IWGSC). Science 345:1251788 10.1126/science.1251788 [DOI] [PubMed] [Google Scholar]
- Meda A. R., Scheuermann E. B., Prechsl U. E., Erenoglu B., Schaaf G., Hayen H., et al. (2007). Iron acquisition by phytosiderophores contributes to cadmium tolerance. Plant Physiol. 143, 1761–1773. 10.1104/pp.106.094474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy R., Grob H., Weder B., Green P., Klein M., Frelet-Barrand A., et al. (2009). The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 Is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J. Biol. Chem. 284, 33614–33622. 10.1074/jbc.M109.030247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang K., Li Y., Liu M., Meng Z., Yu Y. (2013). Inventory and general analysis of the ATP-binding cassette (ABC) gene superfamily in maize (Zea mays L.). Gene 526, 411–428. 10.1016/j.gene.2013.05.051 [DOI] [PubMed] [Google Scholar]
- Panzeri D., Cassani E., Doria E., Tagliabue G., Forti L., Campion B., et al. (2011). A defective ABC transporter of the MRP family, responsible for the bean lpa1 mutation, affects the regulation of the phytic acid pathway, reduces seed myo-inositol and alters ABA sensitivity. New Phytol. 191, 70–83. 10.1111/j.1469-8137.2011.03666.x [DOI] [PubMed] [Google Scholar]
- Park J., Song W., Ko D., Eom Y., Hansen T. H., Schiller M., et al. (2012). The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 69, 278–288. 10.1111/j.1365-313X.2011.04789.x [DOI] [PubMed] [Google Scholar]
- Paumi C. M., Chuk M., Snider J., Stagljar I., Michaelis S. (2009). ABC transporters in Saccharomyces cerevisiae and their interactors: new technologies advances the biology of the ABCC (MRP) subfamily. Microbiol. Mol. Biol. Rev. 73, 577–593. 10.1128/MMBR.00020-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic S., Pascolo L., Gallo R., Cupelli F., Ostrow J. D., Goffeau A., et al. (2000). The products of YCF1 and YLL015w (BPT1) cooperate for the ATP-dependent vacuolar transport of unconjugated bilirubin in Saccharomyces cerevisiae. Yeast 16, 561–571. [DOI] [PubMed] [Google Scholar]
- Qiu Z., Li J., Zhang M., Bi Z., Li Z. (2013). He-Ne laser pretreatment protects wheat seedlings against cadmium-induced oxidative stress. Ecotoxicol. Environ. Saf. 88, 135–141. 10.1016/j.ecoenv.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Rea P. A. (1999). MRP subfamily ABC transporters from plants and yeast. J. Exp. Bot. 50, 895–913. 10.1093/jxb/50.Special_Issue.895 [DOI] [Google Scholar]
- Sanchez-Fernandez R., Davies T. G. E., Coleman J., Rea P. A. (2001). The Arabidopsis thaliana ABC protein subfamily, a complete inventory. J. Biol. Chem. 276, 30231–30244. 10.1074/jbc.M103104200 [DOI] [PubMed] [Google Scholar]
- Schmitteng L. (2001). Analysis of relative gene expression data using a real time quantitative PCRand 2-(Delta Delta C (T)) method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Sharma K. G., Kaur R., Bachhawat A. K. (2003). The glutathione-mediated detoxification pathway in yeast: an analysis using the red pigment that accumulates in certain adenine biosynthetic mutants of yeasts reveals the involvement of novel genes. Arch. Microbiol. 180, 108–117. 10.1007/s00203-003-0566-z [DOI] [PubMed] [Google Scholar]
- Sharma K. G., Mason D. L., Liu G., Rea P. A., Bachhawat A. K., Michaelis S. (2002). Localization, regulation, and substrate transport properties of Bpt1p, a Saccharomyces cerevisiae MRP-type ABC transporter. Eukaryotic Cell 1, 391–400. 10.1128/EC.1.3.391-400.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wang W., Schellin K., Li B., Faller M., Styoop J. M., et al. (2007). Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 25, 930–937. 10.1038/nbt1322 [DOI] [PubMed] [Google Scholar]
- Singh A., Mantri S., Sharma M., Chaudhury A., Tuli R., Roy J. (2014). Genome-wide transcriptome study in wheat identified candidate genes related to processing quality, majority of them showing interaction (quality × development) and having temporal and spatial distributions. BMC Genomics 15:29. 10.1186/1471-2164-15-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W. Y., Yamaki T., Yamaji N., Ko D., Jung K. H., Fujii-Kashino M., et al. (2014). A rice ABC transporter, OsABCC1, reduces arsenic accumulation in the grain. Proc. Natl. Acad. Sci. U.S.A. 111, 15699–15704. 10.1073/pnas.1414968111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N., Radchuk V., Strickert M., Miersch O., Weschke W., Wobus U. (2006). Gene expression patterns reveal tissue-specific signalling networks controlling programmed cell death and ABA-regulated maturation in developing barley grains. Plant J. 47, 310–327. 10.1111/j.1365-313X.2006.02789.x [DOI] [PubMed] [Google Scholar]
- Swarbreck D., Ripoll P. J., Brown D. A., Edwards K. J., Theodoulou F. (2003). Isolation and characterisation of two multidrug resistance associated protein genes from maize. Gene 315, 153–164. 10.1016/S0378-1119(03)00734-0 [DOI] [PubMed] [Google Scholar]
- Theodoulou F. L., Clark I. M., He X.-L., Pallett K. E., Cole D. J., Hallahan D. L. (2003). Co-induction of glutathione-S-transferases and multidrug resistance associated protein by xenobiotics in wheat. Pest Manag. Sci. 59, 202–214. 10.1002/ps.576 [DOI] [PubMed] [Google Scholar]
- Thiel J., Weier D., Sreenivasulu N., Strickert M., Weichert N., Melzer M., et al. (2008). Different hormonal regulation of cellular differentiation and function in nucellar projection and endosperm transfer cells: a microdissection-based transcriptome study of young barley grains. Plant Physiol. 148, 1436–1452. 10.1104/pp.108.127001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasini R., Evers R., Vogt E., Mornet C., Zaman G., Schinkel A. H., et al. (1996). The human multidrug resistance-associated protein functionally complements the yeast cadmium resistance factor-1. Proc. Natl. Acad. Sci. U.S.A. 93, 6743–6748. 10.1073/pnas.93.13.6743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasini R., Vogt E., Fromenteau M., Hoertensteiner S., Matile P., Amrhein N., et al. (1998). An ABC−transporter of Arabidopsis thaliana has both glutathione−conjugate and chlorophyll catabolite transport activity. Plant J. 13, 773–780. 10.1046/j.1365-313X.1998.00076.x [DOI] [PubMed] [Google Scholar]
- Verrier P. J., Bird D., Burla B., Dassa E., Forestier C., Geisler M., et al. (2008). Plant ABC proteins – a unified nomenclature and updated inventory. Trends Plant Sci. 13, 151–159. 10.1016/j.tplants.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Walter S., Brennan J., Arunachalam C., Ansari K., Hu X., Khan M. R., et al. (2008). Components of the gene network associated with genotype-dependent response of wheat to the Fusarium mycotoxin deoxynivalenol. Funct. Integr. Genomics 8, 421–427. 10.1007/s10142-008-0089-4 [DOI] [PubMed] [Google Scholar]
- Walter S., Kahla A., Arunachalam C., Perochon A., Khan M. R., Scofield S. R., et al. (2015). A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J. Exp. Bot. 66, 2583–2593. 10.1093/jxb/erv048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Wu M. (2006). An ATP-binding cassette transporter related to yeast vacuolar ScYCF1 is important for Cd sequestration in Chlamydomonas reinhardtii. Plant Cell Environ. 10, 1901–1912. 10.1111/j.1365-3040.2006.01566.x [DOI] [PubMed] [Google Scholar]
- Wu L., Guan Y., Wu Z., Yang K., Lv J., Converse R., et al. (2014). OsABCG15 encodes a membrane protein that plays an important role in anther cuticle and pollen exine formation in rice. Plant Cell Rep. 11, 1881–1899. 10.1007/s00299-014-1666-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. H., Zhao H. J., Liu Q. L., Frank T., Engel K. H., An G., et al. (2009). Mutations of the multi-drug resistance associated protein ABC transporter gene 5 result in reduction of phytic acid in rice seeds. Theor. Appl. Genet. 119, 75–83. 10.1007/s00122-009-1018-1 [DOI] [PubMed] [Google Scholar]
- Zhang R., Zhu J., Cao H. Z., An Y. R., Huang J. J., Chen X. H., et al. (2013). Molecular cloning and expression analysis of PDR1-like gene in ginseng subjected to salt and cold stresses or hormonal treatment. Plant Physiol. Biochem. 71, 203–211. 10.1016/j.plaphy.2013.07.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Exon/intron structures of wheat ABCC genes are shown. Black boxes represent exons and black lines represent introns. Total number of introns for each gene are represented on the right side.