Abstract
Recently, several studies regarding the association between the pri-miR-124-1 rs531564 polymorphism and cancer susceptibility were explored. Owing to inconsistent results of these studies, a meta-analysis was conducted to determine the association of this polymorphism with cancer risk. Relevant studies were identified by a systematic search of PubMed, EMBASE, Web of Science and CNKI on-line databases. Odds ratios (ORs) and 95% confidence intervals (CIs) from eligible studies were pooled, and heterogeneity and publication bias were also evaluated. A total of five studies with 2,253 cases and 2,510 controls were included in the meta-analysis. Overall, the results showed that the pri-miR-124-1 rs531564 polymorphism was significantly associated with a reduced cancer risk (G vs. C: OR, 0.86; 95% CI, 0.77–0.96; GG vs. CC: OR, 0.52; 95% CI, 0.34–0.79; GG vs. CG/CC: OR, 0.54; 95% CI, 0.36–0.81). Furthermore, in the subgroup analysis by cancer sites, a statistical association was identified between the rs531564 polymorphism and a decreased esophageal squamous cell carcinoma (ESCC) risk (G vs. C: OR, 0.87; 95% CI, 0.77–0.98; GG vs. CC: OR, 0.54; 95% CI, 0.34–0.84). These findings suggested that the genetic variant of rs531564 may have a potential value in decreasing cancer risk, particularly in ESCC patients.
Keywords: pri-miR-124-1 rs531564, microRNA, polymorphism, cancer risk, meta-analysis
Introduction
MicroRNAs (miRNAs) are well-conserved non-coding RNAs that regulate gene expression by base-pairing to the 3′-untranslated regions of mRNA targets, thus resulting in post-transcriptional gene repression or target degradation (1,2). Emerging evidence indicates that miRNAs can act as tumor suppressors or oncogenes and participate in a wide variety of cellular processes, such as cell growth, differentiation, inflammation and apoptosis (3,4). It has also been observed that miRNAs could have played important roles in tumor development, diagnosis and prognosis (5). Studies have demonstrated that the single-nucleotide polymorphisms (SNPs) or mutations in miRNA-coding genes functionally affected the expression and biogenesis of mature miRNAs and their target genes, consequently contributing to cancer susceptibility (6–8).
Among the variant SNPs, an important G/C polymorphism, designated as rs531564, was identified in pri-miR-124-1 and the G allele was reported to alter the process and expression of mature miR-124 in the nervous system (9). Previously, Yang et al (10) and Ye et al (11) studied the role of this polymorphism in the cancer risk among Caucasian populations. Additionally, several studies suggested that there was no evident association between the variant genotype of the pri-miR-124-1 rs531564 polymorphism and cancer risk in Chinese populations (12–14). However, Xiong et al (15) identified that the G allele of rs531564 was less frequent in the cervical cancer patients when compared with healthy subjects (P=0.014) and the polymorphism was clearly associated with a reduced risk of cervical cancer in Chinese women. In addition, You (16) and Zhang et al (17) reported that the rs531564 genotype was significantly associated with the risk of esophageal squamous cell carcinoma (ESCC) in Chinese populations. The results were contradictory and inconclusive, therefore a meta-analysis was performed in the present study to evaluate the association between the rs531564 polymorphism and cancer susceptibility in the Chinese population.
Materials and methods
Literature search
A literature search for eligible studies that explored the association between the pri-miR-124-1 rs531564 polymorphism and cancer risk was carried out using PubMed, EMBASE, Web of Science and CNKI databases (until October 1, 2014). Keywords used in the searches included: (‘miR-124’ or ‘miRNA-124’ or ‘rs531564’ or ‘pri-miR-124-1’) and (‘tumor’ or ‘cancer’ or ‘carcinoma’) and (‘polymorphism’ or ‘SNP’ or ‘allele’ or ‘variation’). The search was performed without restriction on language or publication years. The reference lists of retrieved studies were also checked for additional literature.
Selection criteria
Studies were identified as eligible if they met the following criteria: i) Evaluated the association of the pri-miR-124-1 rs531564 polymorphism and cancer risks; ii) case-control studies; and iii) detailed genotype data for estimating odds ratios (ORs) and 95% confidence intervals (CIs). Studies were excluded based on the following criteria: i) Reviews or animal studies; ii) lack of sufficient data for meta-analysis; and iii) repeated outcomes from the same samples. When overlapping data of the same case series were included in more than one study, only the most recent or complete study was used in the meta-analysis.
Data extraction
The information extracted from each study was as follows: First author, publication year, country origin, ethnicity, cancer types, diagnostic methods for cancer, source of controls, genotyping methods, the numbers of genotyped cases and controls and Hardy-Weinberg equilibrium (HWE) for control groups. Quality assessment of the included studies was evaluated according to the Newcastle-Ottawa Quality Assessment Scale (NOS) (18). The information was extracted by two investigators (Fang and Zeng) in duplicate and any disagreements were discussed by group discussions to achieve a consensus.
Statistical analysis
The pooled ORs and 95% CIs were summarized to assess the strength of the association between the pri-miR-124-1 rs531564 polymorphism and cancer susceptibility with five genetic models: Allele contrasts (G vs. C), and homozygote (GG vs. CC), heterozygote (CG vs. CC), dominant (GG/CG vs. CC) and recessive models (GG vs. CG/CC), separately. Heterogeneity was examined with the χ2 and I2 test. When heterogeneity was absent (P>0.10, I2<50), a fixed effect model was used for secondary analysis. Otherwise, a random effect model was employed (19,20). HWE was measured with the Pearson's goodness-of-fit test in control groups and P<0.05 was deemed not to conform to HWE. Sensitivity analysis was conducted to assess the stability of the combined results by the omission of every single study each time or excluding studies with disrupted HWE. Funnel plots were also used to estimate the possible publication bias. All the analyses were operated using Cochrane Collaboration's Review Manager Software 5.2 and P<0.05 was considered to indicate a statistically significant difference.
Results
Characteristics of studies
Based on the selection criteria above, five case-control studies (13–17) were finally enrolled in the meta-analysis with publication years that ranged from 2011 to 2014. The detailed literature retrieval process is presented in Fig. 1. A total of 4,763 subjects were involved in the meta-analysis, including 2,253 cancer cases and 2,510 healthy controls. Multiple cancer types were analyzed in the studies, including triple-negative breast cancer (13), cervical cancer (15) and ESCC (14,16–17). The diagnoses of these tumors were confirmed by histological or histopathological examinations. In addition, three studies used hospital-based controls (13–15), one used population-based controls (17) and one used population- and hospital-based controls (16). Polymerase chain reaction-based ligase detection reaction was performed in two studies (14,15) and three used other methods. The genotype frequencies of the controls were all fitted in HWE, except for one study by You (16). The NOS scores of the selected studies were >5 (moderate-to-high quality). The main characteristics and methodological quality of the included studies are summarized in Table I.
Figure 1.
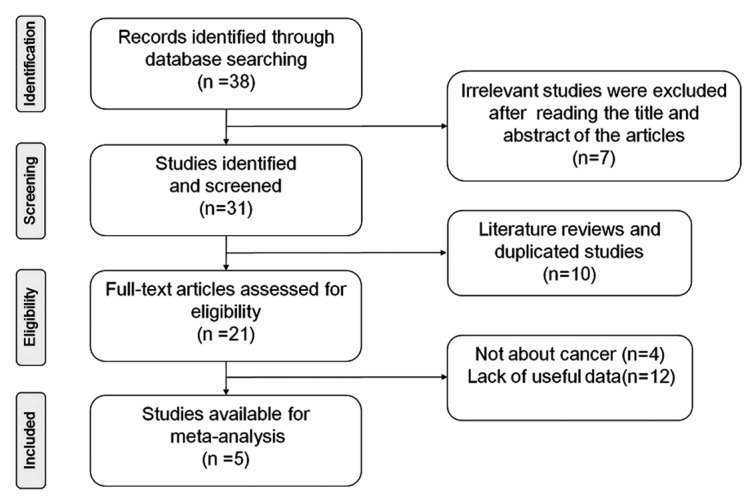
Flow diagram of the study selection process.
Table I.
Characteristics of the included studies in the meta-analysis.
| Case genotypes | Control genotypes | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First author, year | Country | Ethnicity | Cancer type | Diagnostic method | Source of controls | Genotyping | CC | CG | GG | CC | CG | GG | PHWE | NOS score | (Refs.) |
| Ma, 2013 | China | Asian | TNBC | Histopathological | HB | MassARRAY | 126 | 52 | 4 | 136 | 45 | 8 | 0.098 | 7 | (13) |
| Xiong, 2014 | China | Asian | Cervical | Histopathological | HB | PCR-LDR | 91 | 15 | 1 | 151 | 51 | 6 | 0.507 | 6 | (15) |
| You, 2011 | China | Asian | ESCC | Histopathological | PB/HB | MALDI-TOF-MS | 166 | 69 | 9 | 105 | 72 | 4 | 0.037 | 6 | (16) |
| Yin, 2013 | China | Asian | ESCC | Histopathological | HB | PCR-LDR | 454 | 146 | 11 | 470 | 168 | 19 | 0.400 | 7 | (14) |
| Zhang, 2014 | China | Asian | ESCC | Histological | PB | SNaPshot | 803 | 295 | 11 | 910 | 331 | 34 | 0.555 | 8 | (17) |
TNBC, triple-negative breast cancer; ESCC, esophageal squamous cell carcinoma; HB, hospital-based; PB, population-based; HWE, Hardy-Weinberg equilibrium; PCR-LDR, polymerase chain reaction-based ligase detection reaction; NOS, Newcastle-Ottawa Quality Assessment Scale.
Quantitative synthesis
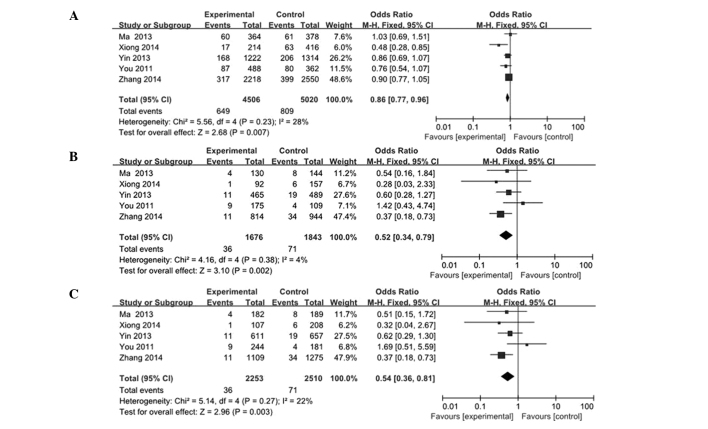
In the present analysis, data from five case-control studies were collected and analyzed to investigate the association between the pri-miR-124-1 rs531564 polymorphism and cancer risk. As a result, the meta-analysis of overall studies revealed that a significantly decreased cancer risk was observed in the G vs. C, GG vs. CC and GG vs. CG/CC models tested (G vs. C: OR, 0.86; 95% CI, 0.77–0.96; GG vs. CC: OR, 0.52; 95% CI, 0.34–0.79; GG vs. CG/CC: OR, 0.54; 95% CI, 0.36–0.81; Fig. 2). In the subgroup analysis by cancer types, the results showed that the rs531564 genotype was markedly associated with a decreased risk of ESCC (G vs. C: OR, 0.87; 95% CI, 0.77–0.98; GG vs CC: OR, 0.54; 95% CI, 0.34–0.84; Fig. 3). The main results are summarized in Table II.
Figure 2.
Overall meta-analysis of the association between pri-miR-124-1 rs531564 and cancer risk. (A) G vs. C; (B) GG vs. CC; (C) GG vs. CG/CC. The squares and horizontal lines correspond to the odds ratio (OR) and 95% confidence interval (CI). The area of the squares reflects the study specific weight (inverse of the variance). The diamond represents the pooled OR and 95% CI.
Figure 3.
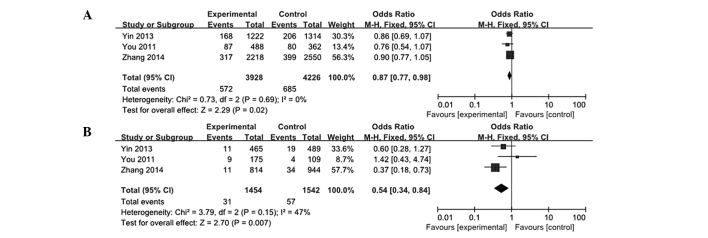
Subgroup analysis of the association between pri-miR-124-1 rs531564 and esophageal squamous cell carcinoma risk. (A) G vs. C; (B) GG vs. CC. Odds ratio and 95% confidence interval (CI) were pooled. The area of the squares reflects the study specific weight (inverse of the variance). The diamond represents the pooled OR and 95% CI.
Table II.
Meta-analysis of the rs531564 polymorphism with the cancer risk.
| Test of association | Test of heterogeneity | |||||
|---|---|---|---|---|---|---|
| Study groups | OR (95% CI) | Z | P-value | Model | Ph | I2, % |
| Overall | ||||||
| G vs. C | 0.86 (0.77–0.96) | 2.68 | 0.007 | F | 0.23 | 28 |
| GG vs. CC | 0.52 (0.34–0.79) | 3.10 | 0.002 | F | 0.38 | 4 |
| CG vs. CC | 0.86 (0.67–1.10) | 1.24 | 0.220 | R | 0.03 | 62 |
| GG/CG vs. CC | 0.84 (0.67–1.03) | 1.65 | 0.100 | R | 0.08 | 52 |
| GG vs. CG/CC | 0.54 (0.36–0.81) | 2.96 | 0.003 | F | 0.27 | 22 |
| ESCC | ||||||
| G vs. C | 0.87 (0.77–0.98) | 2.29 | 0.020 | F | 0.69 | 0 |
| GG vs. CC | 0.54 (0.34–0.84) | 2.70 | 0.007 | F | 0.15 | 47 |
| CG vs. CC | 0.87 (0.68–1.11) | 1.14 | 0.260 | R | 0.08 | 60 |
| GG/CG vs. CC | 0.88 (0.77–1.01) | 1.76 | 0.080 | F | 0.23 | 32 |
| GG vs. CG/CC | 0.64 (0.30–1.37) | 1.15 | 0.250 | R | 0.09 | 59 |
OR, odds ratio; CI, confidence interval; ESCC, esophageal squamous cell carcinoma; vs, versus; R, random effect model; F, fixed effect model; Ph, P-value of Q test for heterogeneity test.
Moderate heterogeneity was observed in the CG vs. CC and GG/CG vs. CC models tested [CG vs. CC: P=0.22, P-value of Q test for heterogeneity test (Ph)=0.03, I2=62%; GG/CG vs. CC: P=0.10, Ph=0.08, I2=52%; Table II]. Therefore, the random effect models were used and sensitivity analyses were performed. When the study by Xiong et al (15) was removed, the heterogeneity in the GG/CG vs. CC model was largely reduced, but the result remained insignificant (P=0.13, Ph=0.25, I2=26%). When the study with disrupted HWE by You (16) was removed, the heterogeneity in the GG/CG vs. CC model was reduced and the result was consistently insignificant (P=0.15, Ph=0.12, I2=49%). Sensitivity analysis indicated that the pooled ORs were not altered by deleting one study at a time (data not shown).
Assessment of publication bias
Funnel plots were performed to access the publication bias of eligible studies, in which the standard error of log (OR) for each study was plotted against its log (OR). The graphical funnel plots (Fig. 4) appeared symmetrical and no clear evidence of publication bias was observed among all the genetic models.
Figure 4.
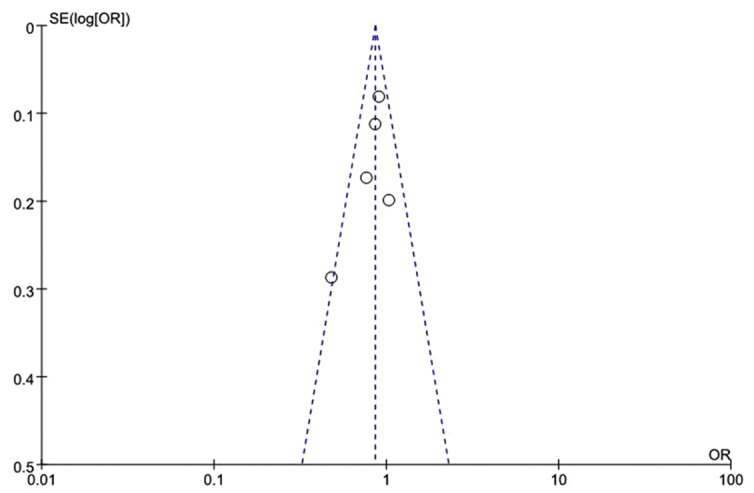
A funnel plot for evaluating publication bias using the allele model. The funnel plot was almost symmetrical. OR, odds ratio, SE, standard error.
Discussion
miRNAs are emerging as crucial regulators of diverse biological pathways and have been indicated in the etiology of human tumors through modulating approximately one third of the human genome (21,22). Disruption of miRNA-dependent regulation by SNPs in either miRNAs genes or miRNA-binding sites resulted in a myriad of diseases including human malignancy (23,24). However, the effects of genetic variants in miRNA-related genes on cancer susceptibility remain largely unknown.
To the best of our knowledge, this is the first meta-analysis studied on the association between the pri-miR-124-1 rs531564 polymorphism and cancer risk in Chinese populations. According to the study, the rs531564 polymorphism was significantly correlated with a reduced cancer risk in the G vs. C, GG vs. CC and GG vs. CG/CC models tested when all the studies were pooled into analysis. However, heterogeneity existed in the heterozygous (CG vs. CC: Ph=0.03, I2=62%) and dominant models (GG/CG vs. CC: Ph=0.08, I2=52%) (Table II). Following the removal of the study by Xiong et al (15), the heterogeneity in the dominant model was largely reduced, suggesting that the rs531564 polymorphism may have the opposite effects in different tumors and causing the heterogeneity, yet the limitation of cancer types inhibited further verification. In the stratified analysis by cancer sites, the results confirmed that the rs531564 variant carriers had decreased risks for ESCC in the G vs. C and GG vs. CC models tested, indicating the polymorphism may have a potentially protective value in ESCC patients (Fig. 3).
Recently, the role of miR-124 in cancer development has received increasing attention. Shi et al (25) demonstrated that miR-124 was commonly reduced in prostate cancer, which directly targets the androgen receptor and subsequently induces an increased expression of the tumor suppressive gene, p53. Furuta et al (26) suggested cyclin-dependent kinase 6 (CDK6) as a possible target for miR-124, which was a key modulator of cell cycle and differentiation. The study also indicated that the restoration of miR-124 expression impairs the growth of hepatocellular carcinoma cells. In addition, decreased expression of miR-124 was also detected in glioblastoma multiforme (GBM) and transfection of miR-124 induced G0/G1 cell cycle arrest in GBM cell lines through inhibition of CDK6 and phosphorylated retinoblastoma proteins (27). Therefore, miR-124 may act as a tumor suppressor and have significant effects on the cancer risk by modulating these targets.
However, several limitations to the present meta-analysis should be considered. Firstly, the number of studies for each site-specific cancer was so limited that further stratified analysis on breast and cervical cancer could not be performed. Secondly, lack of sufficient data and detailed individual information restricted further evaluation on the effects of potential interactions between the genetic variation and epidemiological factors such as age, gender and lifestyle-related factors. Thirdly, the cumulative effects of miRNA-related genetic variants were also not investigated. Fourthly, there was not enough study on the Chinese population and no study on other populations in the meta-analysis. Therefore, larger sample sizes with different ethnic groups are required for further study. Finally, only published studies in English or Chinese were enrolled in this analysis, and a potential language bias may exist.
In conclusion, the meta-analysis suggested a significant association between the pri-miR-124-1 rs531564 polymorphism and decreased cancer susceptibility in Chinese population, which may be a potential biomarker for the early diagnosis of ESCC. However, investigations of diverse ethnic populations and various types of cancer are of great value to verify these findings.
Acknowledgements
The present study was sponsored by the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry, and was supported by grants from the Natural Science Foundation of China (nos. 30873044 and 81272372).
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 3.Ruan K, Fang X, Ouyang G. MicroRNAs: Novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285:116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 4.Chen T, Li Z, Yan J, Yang X, Salminen W. MicroRNA expression profiles distinguish the carcinogenic effects of riddelliine in rat liver. Mutagenesis. 2012;27:59–66. doi: 10.1093/mutage/ger060. [DOI] [PubMed] [Google Scholar]
- 5.Barbarotto E, Schmittgen TD, Calin GA. MicroRNAs and cancer: profile, profile, profile. Int J Cancer. 2008;122:969–977. doi: 10.1002/ijc.23343. [DOI] [PubMed] [Google Scholar]
- 6.Hu Z, Liang J, Wang Z, Tian T, Zhou X, Chen J, Miao R, Wang Y, Wang X, Shen H. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009;30:79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 7.Wang F, Sun G, Zou Y, Li Y, Hao L, Pan F. Association of microRNA-499 rs3746444 polymorphism with cancer risk: Evidence from 7188 cases and 8548 controls. PLoS One. 2012;7:e45042. doi: 10.1371/journal.pone.0045042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy R, De Sarkar N, Ghose S, Paul RR, Pal M, Bhattacharya C, Chowdhury SK, Ghosh S, Roy B. Genetic variations at microRNA and processing genes and risk of oral cancer. Tumour Biol. 2014;35:3409–3414. doi: 10.1007/s13277-013-1450-3. [DOI] [PubMed] [Google Scholar]
- 9.Qi L, Hu Y, Zhan Y, Wang J, Wang BB, Xia HF, Ma X. A SNP site in pri-miR-124 changes mature miR-124 expression but no contribution to Alzheimer's disease in a Mongolian population. Neurosci Lett. 2012;515:1–6. doi: 10.1016/j.neulet.2012.02.061. [DOI] [PubMed] [Google Scholar]
- 10.Yang H, Dinney CP, Ye Y, Zhu Y, Grossman HB, Wu X. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008;68:2530–2537. doi: 10.1158/0008-5472.CAN-07-5991. [DOI] [PubMed] [Google Scholar]
- 11.Ye Y, Wang KK, Gu J, Yang H, Lin J, Ajani JA, Wu X. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila) 2008;1:460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y, Du WD, Chen G, Ruan J, Xu S, Zhou FS, Zuo XB, Lv ZJ, Zhang XJ. Association analysis of genetic variants in microRNA networks and gastric cancer risk in a Chinese Han population. J Cancer Res Clin Oncol. 2012;138:939–945. doi: 10.1007/s00432-012-1164-8. [DOI] [PubMed] [Google Scholar]
- 13.Ma F, Zhang P, Lin D, Yu D, Yuan P, Wang J, Fan Y, Xu B. There is no association between microRNA gene polymorphisms and risk of triple negative breast cancer in a Chinese Han population. PLoS One. 2013;8:e60195. doi: 10.1371/journal.pone.0060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin J, Wang X, Zheng L, et al. Hsa-miR-34b/c rs4938723 T>C and hsa-miR-423 rs6505162 C>A polymorphisms are associated with the risk of esophageal cancer in a Chinese population. PLoS One. 2013;8:e80570. doi: 10.1371/journal.pone.0080570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong X, Cheng J, Liu X, Tang S, Luo X. Correlation analysis between miR-124 rs531564 polymorphisms and susceptibility to cervical cancer. Nan Fang Yi Ke Da Xue Xue Bao. 2014;34:210, 213. (In Chinese) [PubMed] [Google Scholar]
- 16.You WY. A Case Control study on the Association between Polymorphisms of microRNA Genes and Susceptibility for Kazakh's Esophageal Cancer (unpublished PhD thesis) 2011. (In Chinese) [Google Scholar]
- 17.Zhang J, Huang X, Xiao J, et al. Pri-miR-124 rs531564 and pri-miR-34b/c rs4938723 polymorphisms are associated with decreased risk of esophageal squamous cell carcinoma in Chinese populations. PLoS One. 2014;9:e100055. doi: 10.1371/journal.pone.0100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 19.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: An update. Contemp Clin Trials. 2007;28:105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Xu W, Xu J, Liu S, Chen B, Wang X, Li Y, Qian Y, Zhao W, Wu J. Effects of common polymorphisms rs11614913 in miR-196a2 and rs2910164 in miR-146a on cancer susceptibility: A meta-analysis. PLoS One. 2011;6:e20471. doi: 10.1371/journal.pone.0020471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radojicic J, Zaravinos A, Vrekoussis T, Kafousi M, Spandidos DA, Stathopoulos EN. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle. 2011;10:507–517. doi: 10.4161/cc.10.3.14754. [DOI] [PubMed] [Google Scholar]
- 22.Salzman DW, Weidhaas JB. SNPing cancer in the bud: microRNA and microRNA-target site polymorphisms as diagnostic and prognostic biomarkers in cancer. Pharmacol Ther. 2013;137:55–63. doi: 10.1016/j.pharmthera.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicoloso MS, Sun H, Spizzo R, et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010;70:2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin Y, Lee CG. Single Nucleotide Polymorphisms Associated with microRNA Regulation. Biomolecules. 2013;3:287–302. doi: 10.3390/biom3020287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi XB, Xue L, Ma AH, Tepper CG, Gandour-Edwards R, Kung HJ, deVere White RW. Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene. 2013;32:4130–4138. doi: 10.1038/onc.2012.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 27.Silber J, Lim DA, Petritsch C, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]