Abstract
MicroRNA-29b (miR-29b) targets numerous important genes that mediate carcinogenesis and tumor development in breast cancer in vitro and in vivo. The aim of the present study was to determine the clinical significance of miR-29b expression in primary breast cancer patients. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) of miR-29b and certain target genes of miR-29b, such as DNA methyltransferase 3A (DNMT3A), ten-eleven translocation 1 (TET1) and thymine DNA glycosylase (TDG), was performed in 94 primary breast cancer samples. Low expression of miR-29b in primary tumors was significantly associated with poorer disease-free survival (DFS) (P=0.0075) and overall survival (OS) (p=0.0012). Multivariate analysis indicated that miR-29b expression was an independent prognostic factor for OS [relative risk=15.6 (2.33–348), P=0.0026]. In addition, a significant inverse correlation was identified between the expression levels of DNMT3A and miR-29b in estrogen receptor-positive breast cancer patients (P=0.027). To the best of our knowledge, this is the first study to investigate the clinicopathological significance of miR-29b in breast cancer cases and miR-29b is shown to act as a tumor suppressive microRNA in breast cancer and as a potential marker for recurrence and metastasis in breast cancer patients.
Keywords: breast cancer, prognosis, microRNA-29b, DNA methyltransferase 3A, clinicopathological significance
Introduction
Breast cancer is a leading cause of cancer-related mortality among women in industrialized countries. Despite advances in the technologies used for its diagnosis and treatment, recurrence and metastasis of breast cancer remain serious clinical issues. Therefore, there is an urgent need to identify biomarkers or techniques to be used for the early detection of carcinogenesis or recurrence following curative surgery using minimally invasive tests.
MicroRNAs (miRs) are small non-coding RNAs consisting of 20–22 nucleotides. Changes in the levels of miRs are involved in the initiation and progression of human cancers due to the altered translation of various target genes (1). The recent increase in miR interest is attributed to the breakthrough discovery of their role in numerous pathological processes, including malignant transformation (2). In fact, miRs have been reported as potential biomarkers of various malignancies (3,4).
MicroRNA-29b (miR-29b) regulates a number of important genes that mediate carcinogenesis and tumor development in breast cancer (5–8). For example, miR-29b targets a network of pro-metastatic regulators involved in angiogenesis, collagen remodeling and proteolysis, thereby inhibiting metastasis (5). Furthermore, miR-29b directly targets DNA methyltransferase 3A (DNMT3A), DNMT3B, ten-eleven translocation 1 (TET1) and thymine DNA glycosylase (TDG), all of which play crucial roles in the progression and metastasis of various cancers by altering the DNA methylation status (9–16). Although almost all the studies investigating the numerous important roles of miR-29b in breast cancer have been experimental studies conducted in vitro and in vivo (5–8,16), the clinicopathological significance of miR-29b in breast cancer cases has not been determined clinically. The present study evaluated the importance of miR-29b in breast cancer cases and additionally showed the associations between miR-29b and several target genes of miR-29b indicated in the regulation of DNA methylation status in clinical samples.
Materials and methods
Patients
Breast cancer patients (n=94) who underwent surgical treatment at several hospitals [National Hospital Organization Kyushu Cancer Center (Fukuoka, Fukuoka) Kyushu University Beppu Hospital (Beppu, Oita), Oita Prefectural Hospital (Yufu, Oita) and Takada-Chuo Hospital (Yokohama, Kanagawa), all in Japan)] between 1990 and 1999 were enrolled in the study. Prior to sample acquisition, each patient provided written informed consent at the respective hospital. The study was approved by the ethics committees of Kyushu University. Patients were excluded who had been diagnosed with ductal carcinoma in situ. Three patients who had distant metastasis at first diagnosis received no neo-adjuvant chemotherapy. Post-operative adjuvant chemotherapy and endocrine therapy were performed according to the St. Gallen Consensus Conference guidelines (17). Among the 94 patients, 63 were estrogen receptor (ER)-positive. The expression levels of the HER2 protein could not be confirmed in the cases, as the measurements used for HER2 expression were not common when the surgeries were performed. The mean observation period ranged from 1 to 124 months (median, 54 months). Among the 94 patients, only 30, 81 and 57 patients were examined for TET1, TDG and DNMT3A expression, respectively, due to the deficiency of samples for quantification of each cDNA by reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Total RNA extraction and first-strand cDNA synthesis
The resected tumor tissue specimens were frozen immediately in liquid nitrogen and stored at −80°C until analysis. The total RNA extraction from the primary tumors was performed according to the ISOGEN-LS (Nippon Gene Co., Ltd., Tokyo, Japan) manufacturer's instructions. The reverse transcription reactions and first-strand cDNA synthesis were performed as described previously (18).
RT-qPCR for miR-29b, TET1, TDG and DNMT3A
Quantitative analysis was performed of miR-29b- and RNU6B (internal control)-specific cDNAs derived from total RNA extracted from resected tumors using gene-specific primers, according to the TaqMan MicroRNA Assay protocol (Assay IDs: 000413 for hsa-miR-29b-3p and 001093 for RNU6B; Applied Biosystems, Carslbad, California, USA). The procedures were as described previously (18). The raw miR expression levels were normalized to RNU6B expression for calculation of the relative miR expression values. To determine the relative expression levels of TET1, TDG and DNMT3A, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control. RT-qPCR was performed using the LightCycler® 480 system and the LightCycler® 480 Probes Master kit (Roche Applied Science, Penzberg, Germany). The sequences of the primers for TET1, TDG and DNMT3A were as follows: TET1 sense, 5′-TCTGTTGTTGTGCCTCTGGA-3′ and antisense, 5′-GCCTTTAAAACTTTGGGCTTC-3′; TDG sense, 5′-ATGCAGCAGTGAACCTTGTG-3′ and antisense, 5′-GTCATCCACTGCCCATTAGG-3′; and DNMT3A sense, 5′-AAGGAGGAGCGCCAAGAG-3′ and antisense, 5′-ATCACCGCAGGGTCCTTT-3′. The expression of DNMT3B was not detected in the samples.
Statistical analysis
For miR-29b analysis, differences between clinicopathological factors were analyzed using χ2 tests for categorical variables. Disease-free survival (DFS) and overall survival (OS) times were measured from the time of the first surgery until the date of mortality or last follow-up. Survival curves were determined by the Kaplan-Meier method and statistical significance between groups was assessed using the Wilcoxon test. Multivariate analysis was performed to assess the relative influence of prognostic factors on OS using the Cox proportional hazards model with a forward stepwise procedure. Statistical analysis was performed by JMP® Pro version 9.0.2 for Mac OS (SAS Institute Japan, Tokyo, Japan).P<0.05 was considered to indicate a statistically significant difference.
Results
Low miR-29b expression in primary tumor tissues is a prognostic factor for breast cancer patients
miR-29b expression was assessed in primary tumor tissues from 94 breast cancer patients. Patients were divided into miR-29b high and low expression groups according to the median value of miR-29b expression. Clinicopathological factors were subsequently analyzed in association with miR-29b levels. The miR-29b low expression group exhibited a significantly larger tumor size and more advanced clinical stages compared to the miR-29b high expression group (Table I). In terms of DFS and OS, the miR-29b low expression group showed a significantly poorer prognosis than that of the miR-29b high expression group (Fig. 1). Among the ER-positive cases, the low miR-29b expression group had significantly poorer DFS and OS compared to the high miR-29b expression group (Fig. 2). Among the ER-negative cases, low miR-29b expression correlated with a poorer OS only (Fig. 3).
Table I.
miR-29b expression and clinicopathological factors.
| miR-29b expression | |||
|---|---|---|---|
| Factors | Low (n=47), no. (%) | High (n=47), no. (%) | P-value |
| Age, mean years ± SD | 55±11 | 54±11 | |
| ER | |||
| Positive | 31 (66) | 32 (68) | 0.59 |
| Negative | 15 (32) | 12 (26) | |
| Progesterone receptor | |||
| Positive | 26 (55) | 31 (66) | 0.24 |
| Negative | 18 (38) | 13 (28) | |
| T factors | |||
| T1 | 13 (28) | 25 (53) | 0.01 |
| T2–4 | 34 (72) | 22 (47) | |
| Lymph node metastasis | |||
| Absent | 22 (47) | 27 (57) | 0.31 |
| Present | 25 (53) | 20 (43) | |
| Lymphatic invasion | |||
| Absent | 18 (38) | 16 (34) | 0.73 |
| Present | 23 (49) | 24 (51) | |
| Venous invasion | |||
| Absent | 33 (70) | 34 (72) | 0.60 |
| Present | 7 (15) | 6 (13) | |
| Stage | |||
| Stage I | 7 (15) | 17 (36) | 0.02 |
| Stages II–IV | 40 (85) | 30 (64) | |
miR-29b, microRNA-29b; SD; standard deviation, ER, estrogen receptor.
Figure 1.
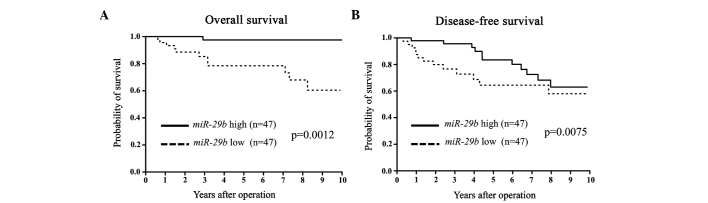
(A) Overall survival (OS) and (B) disease-free survival (DFS) curves for breast cancer patients according to the expression levels of microRNA-29b (miR-29b) in primary tumors. The differences in OS and DFS were significant (P=0.0012 and 0.0075, respectively).
Figure 2.
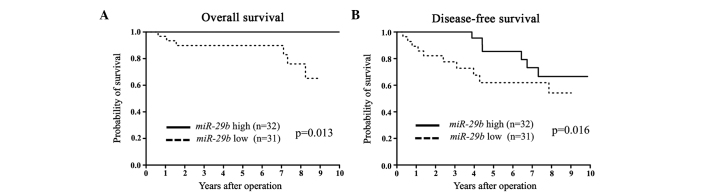
(A) Overall survival (OS) and (B) disease-free survival (DFS) curves for breast cancer patients according to the expression levels of microRNA-29b (miR-29b) in estrogen receptor (ER)-positive primary tumors. The differences in OS and DFS between the miR-29b low and high expression levels were significant (P=0.013 and 0.016, respectively).
Figure 3.
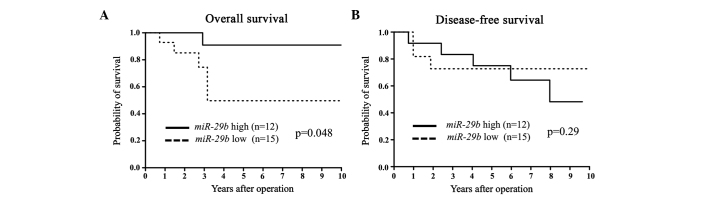
(A) Kaplan-Meier overall survival (OS) and (B) disease-free survival (DFS) curves for breast cancer patients according to the expression level of microRNA-29b (miR-29b) in estrogen receptor (ER)-negative primary tumors. The difference in OS between the miR-29b low and high expression levels was significant (P=0.048).
Multivariate analysis of OS showed that the low level of miR-29b expression was an independent prognostic predictor in all patients (Table II).
Table II.
Results of multivariate analysis of clinicopathological factors for overall survival (Cox proportional hazards model).
| Multivariate analysis | ||
|---|---|---|
| Factors | RR (95% CI) | P-value |
| T factor (T1/2–4) | 3.14 (0.39–19.5) | 0.250 |
| Lymph node metastasis | 1.15 (0.13–24.9) | 0.910 |
| Lymphatic invasion | 11.4 (0.61–743) | 0.120 |
| Venous invasion | 2.59 (0.56–12.7) | 0.220 |
| Stage (I/II–IV) | 2.57 (0.04–211) | 0.650 |
| miR-29b expression | 15.6 (2.33–348) | 0.003 |
RR, relative risk; CI, confidence interval; miR-29b, microRNA-29b.
Evaluation of TET1, TDG and DNMT3A expression levels and their comparison with miR-29b levels in breast cancer patients
Additionally, the expression levels of TET1, TDG and DNMT3A were examined in breast cancer primary tumor tissues. These levels were subsequently compared between the miR-29b high and low expression groups. There were no significant differences in any of the patients between the miR-29b high and low expression groups (Fig. 4). However, in analyses of the ER-positive patients, DNMT3A showed significantly higher expression in the miR-29b low expression compared to the high expression group (P=0.027; Fig. 5).
Figure 4.
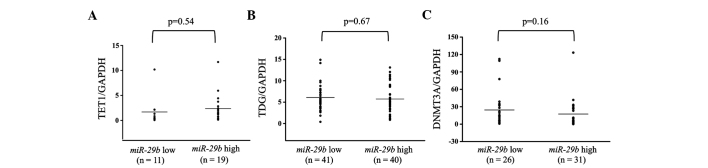
MicroRNA (miR) expression of (A) TET1, (B) TDG and (C) DNMT3A in miR-29b low- and high-expressing primary breast cancer tumors. The high miR-29b expression level was above and the low miR-29b expression level was below the mean expression value of all the samples (n=94). The horizontal line in the graph represents the mean of each group. There were no significant differences in any of the samples. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Figure 5.
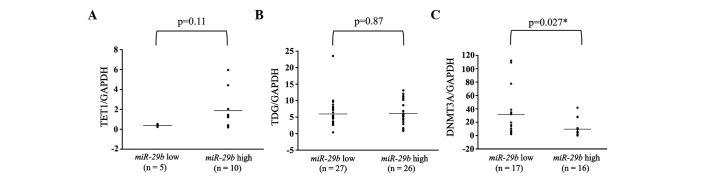
MicroRNA (miR) expression of (A) TET1, (B) TDG and (C) DNMT3A in miR-29b low- and high-expressing primary estrogen receptor (ER)-positive breast cancer tumors. The high miR-29b expression level was above and the low miR-29b expression level was below the mean expression value of all the samples. The horizontal line in the graph represents the mean of each group. The difference in DNMT3A expression was significant (*P=0.027). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Discussion
In the present study, low miR-29b expression in primary breast tumors correlated significantly with poor DFS and OS in breast cancer patients. This was consistent with previous in vitro and in vivo findings that miR-29b acts as a tumor suppressive miR (5–7). For example, Chou et al (5) showed that miR-29b was induced by GATA3 and inhibited metastasis by targeting various genes (ANGPTL4, LOX, MMP and VEGFA) involved in modifying the tumor microenvironment.
With respect to the clinicopathological factors, larger tumor sizes and more advanced stages were detected in the miR-29b low expression compared to the high expression group. This finding suggested that the suppression of miR-29b is associated with tumor progression. To clarify how miR-29b contributed to breast cancer progression, the study focused on candidate target genes of miR-29b according to TarBase 6.0 (19). Among 103 genes, we were interested in those that regulate epigenetic status, such as TET1, TDG and DNMT3A. The direct interactions between miR-29b and TET1, TDG and DNMT3A were confirmed by luciferase assays and western blot analysis (16). Although there are numerous pathways that regulate the levels of TET1 (13,20), TDG (21) and DNMT3A (22) in breast cancer, significant inverse correlations were identified between the expression levels of miR-29b and DNMT3A in ER-positive patients. The overexpression of DNMT3A correlates with a poor prognosis in numerous cancers, including breast cancer (10,23). Starlard-Davenport et al (24) demonstrated that transfection of pre-miR-29b into breast cancer cell lines inhibited cell proliferation, decreased DNMT3A and DNMT3B mRNA levels and decreased the promoter methylation status of several tumor suppressor genes.
With respect to breast cancer subtypes, the present results showed a significant correlation between low miR-29b expression and poor OS, independent of the ER status. According to the results of the multivariate analysis, miR-29b is a powerful biomarker for predicting patient outcomes in all the subtypes of breast cancer.
In conclusion, miR-29b expression in breast cancer primary tumors was an independent prognostic factor for OS. Low miR-29b expression in primary tumors may predict poor OS and DFS in breast cancer patients. Additionally, in ER-positive cases, a significant inverse correlation between the expression levels of miR-29b and DNMT3A was identified.
Acknowledgements
The authors would like to thank Ms. K. Oda, Ms. Kasagi, Ms. S. Kono and Ms. T. Kawano for their technical assistance. The present study was supported in part by the Japan Society for the Promotion of Science (grant no. 25830102).
References
- 1.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 2.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 3.Yang H, Yu J, Wang L, Ding D, Zhang L, Chu C, Chen Q, Xu Z, Zou Q, Liu X. miR-320a is an independent prognostic biomarker for invasive breast cancer. Oncol Lett. 2014;8:1043–1050. doi: 10.3892/ol.2014.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tejero R, Navarro A, Campayo M, Viñolas N, Marrades RM, Cordeiro A, Ruíz-Martínez M, Santasusagna S, Molins L, Ramirez J, et al. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PLoS One. 2014;9:e101899. doi: 10.1371/journal.pone.0101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou J, Lin JH, Brenot A, Kim JW, Provot S, Werb Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol. 2013;15:201–213. doi: 10.1038/ncb2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang T, Liang Y, Lin Q, Liu J, Luo F, Li X, Zhou H, Zhuang S, Zhang H. miR-29 mediates TGFβ1-induced extracellular matrix synthesis through activation of PI3K-AKT pathway in human lung fibroblasts. J Cell Biochem. 2013;114:1336–1342. doi: 10.1002/jcb.24474. [DOI] [PubMed] [Google Scholar]
- 7.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 8.Pandey M, Sultana S, Gupta KP. Involvement of epigenetics and microRNA-29b in the urethane induced inception and establishment of mouse lung tumors. Exp Mol Pathol. 2014;96:61–70. doi: 10.1016/j.yexmp.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Cao XY, Ma HX, Shang YH, Jin MS, Kong F, Jia ZF, Cao DH, Wang YP, Suo J, Jiang J. DNA methyltransferase3a expression is an independent poor prognostic indicator in gastric cancer. World J Gastroenterol. 2014;20:8201–8208. doi: 10.3748/wjg.v20.i25.8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibrahem L, Mahfouz R, Elhelw L, Abdsalam EM, Soliman R. Prognostic significance of DNMT3A mutations in patients with acute myeloid leukemia. Blood Cells Mol Dis. 2015;54:84–89. doi: 10.1016/j.bcmd.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 11.Niederwieser C, Kohlschmidt J, Volinia S, Whitman SP, Metzeler KH, Eisfeld AK, Maharry K, Yan P, Frankhouser D, Becker H, et al. Prognostic and biologic significance of DNMT3B expression in older patients with cytogenetically normal primary acute myeloid leukemia. Leukemia. 2015;29:567–575. doi: 10.1038/leu.2014.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu CH, Peng KL, Kang ML, Chen YR, Yang YC, Tsai CH, Chu CS, Jeng YM, Chen YT, Lin FM, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Reports. 2012;2:568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- 13.Sun M, Song CX, Huang H, Frankenberger CA, Sankarasharma D, Gomes S, Chen P, Chen J, Chada KK, He C, et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci USA. 2013;110:9920–9925. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu HG, Zhan W, Yan L, Qin RY, Yan YP, Yang ZJ, Liu GC, Li GQ, Wang HF, Li XL, et al. TET1 partially mediates HDAC inhibitor-induced suppression of breast cancer invasion. Mol Med Rep. 2014;10:2595–2600. doi: 10.3892/mmr.2014.2517. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Yu T, Shi J, Chen X, Zhang W, Lin T, Liu Z, Wang Y, Zeng Z, Wang C, et al. Thymine DNA glycosylase is a positive regulator of Wnt signaling in colorectal cancer. J Biol Chem. 2014;289:8881–8890. doi: 10.1074/jbc.M113.538835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morita S, Horii T, Kimura M, Ochiya T, Tajima S, Hatada I. miR-29 represses the activities of DNA methyltransferases and DNA demethylases. Int J Mol Sci. 2013;14:14647–14658. doi: 10.3390/ijms140714647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldhirsch A, Wood WC, Senn HJ, Glick JH, Gelber RD. International Consensus Panel on the Treatment of Primary Breast Cancer: Fifth International Conference on Adjuvant Therapy of Breast Cancer, St Gallen, March 1995. Eur J Cancer. 1995;31A:1754–1759. doi: 10.1016/0959-8049(95)00479-3. [DOI] [PubMed] [Google Scholar]
- 18.Ota D, Mimori K, Yokobori T, Iwatsuki M, Kataoka A, Masuda N, Ishii H, Ohno S, Mori M. Identification of recurrence-related microRNAs in the bone marrow of breast cancer patients. Int J Oncol. 2011;38:955–962. doi: 10.3892/ijo.2011.926. [DOI] [PubMed] [Google Scholar]
- 19.Vergoulis T, Vlachos IS, Alexiou P, Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N, Dalamagas T, Hatzigeorgiou AG. TarBase 6.0: Capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012;40:D222–D229. doi: 10.1093/nar/gkr1161. (D1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.da Costa NM, Hautefeuille A, Cros MP, Melendez ME, Waters T, Swann P, Hainaut P, Pinto LF. Transcriptional regulation of thymine DNA glycosylase (TDG) by the tumor suppressor protein p53. Cell Cycle. 2012;11:4570–4578. doi: 10.4161/cc.22843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng EK, Li R, Shin VY, Siu JM, Ma ES, Kwong A. MicroRNA-143 is downregulated in breast cancer and regulates DNA methyltransferases 3A in breast cancer cells. Tumour Biol. 2014;35:2591–2598. doi: 10.1007/s13277-013-1341-7. [DOI] [PubMed] [Google Scholar]
- 23.Yu Z, Xiao Q, Zhao L, Ren J, Bai X, Sun M, Wu H, Liu X, Song Z, Yan Y, et al. DNA methyltransferase 1/3a overexpression in sporadic breast cancer is associated with reduced expression of estrogen receptor-alpha/breast cancer susceptibility gene 1 and poor prognosis. Mol Carcinog. 2014 Jan 25; doi: 10.1002/mc.22133. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 24.Starlard-Davenport A, Kutanzi K, Tryndyak V, Word B, Lyn-Cook B. Restoration of the methylation status of hypermethylated gene promoters by microRNA-29b in human breast cancer: A novel epigenetic therapeutic approach. J Carcinog. 2013;12:15. doi: 10.4103/1477-3163.115720. [DOI] [PMC free article] [PubMed] [Google Scholar]


