Abstract
Numerous studies have investigated the prognostic role of brain and acute leukemia, cytoplasmic (BAALC) gene expression in adult patients with acute myeloid leukemia (AML); however, the results are inconclusive. A meta-analysis was conducted to provide a comprehensive evaluation of the prognostic role of BAALC gene expression in AML. Eligible studies were searched through PubMed, Embase, the Cochrane Library, the China National Knowledge Infrastructure and the China Biology Medicine Disc. Correlations between the BAALC gene expression and clinicopathological features and prognosis were analyzed. A total of 15 studies were examined. The pooled results suggest that high BAALC expression had an unfavorable outcome in AML. The combined hazard ratio (HR) for overall survival (OS) was 1.53 and the summary HR for the disease-free survival rate was 1.64. In addition, subgroup analyses considering cytogenetic and survival analysis were also conducted. High BAALC gene expression appeared to be an adverse prognostic indicator in patients with cytogenetically normal AML (HR for OS, 1.43) and in subgroups of survival analysis with multivariate analysis (HR for OS, 2.35). These results indicate that high BAALC gene expression served as an independent poor prognostic indicator in adult patients with AML.
Keywords: brain and acute leukemia cytoplasmic gene, acute myeloid leukemia, prognosis, meta-analysis
Introduction
Acute myeloid leukemia (AML), as a clonal malignant hematopoietic disorder in hematopoietic stem cells, is the most common form of acute leukemia among humans (1). In the last decade, the risk of AML was classified through distinct cytogenetic abnormalities; however, 40–50% of patients with AML had normal cytogenetic characterization, albeit varying in terms of prognosis (2). With the development of molecular biology, aberrant transcription and epigenetic dysfunction have been proven to be closely associated with the pathogenesis and progression of hematological malignancies. They are relevant not only to the pathogenesis of the disease, but are also important for predicting the response to therapy and the survival rate. The abnormal expression of genes and epigenetics is closely connected with the occurrence and development of AML and it plays an important role in the pathogenesis and prognosis of AML, particularly in cytogenetically normal-acute myeloid leukemia (CN-AML), such as nucleophosmin (NPM1), Fms-related tyrosine kinase 3 (FLT3) and CCAAT/enhancer binding protein α mutations (3,4). However, gene mutation is still not detected in certain patients with AML.
The brain and acute leukemia, cytoplasmic (BAALC), which maps on chromosome 8 at 8q22.3, was originally observed in the neuroderm and its expression was reported as a hematopoietic precursor, such as the early hematopoietic cells of cluster of differentiation 34+ (5). However, no expression was observed in the peripheral mononuclear cells. The site and cellular mechanism(s) of the effects of the BAALC gene are unclear, thereby possibly resulting in differentiation failure caused by cell shape, motility and adhesion in the association between cells (6). Subsequent research has shown that the highly expressed BAALC gene is associated with minimal residual disease, which leads to a low complete remission (CR) rate and overall survival (OS). BAALC is regarded as a superior independent prognostic indicator. However, certain studies have also argued that it is not an effective prognostic indicator in patients with AML. The present study conducted a meta-analysis of the existing literature to gain insight into the correlated characteristics of the BAALC gene expression in AML and to evaluate its prognostic values.
Materials and methods
Retrieval strategy, and inclusion and exclusion criteria
A search of the relevant studies in the literature using PubMed, Embase, the Cochrane Library, the China National Knowledge Infrastructure and the China Biology Medicine Disc was conducted with the retrieval time starting from the beginning of the establishment of each of the databases until September 2013. The English search terms included ‘AML’, ‘acute myeloid leukemia’, ‘acute myeloid leukeamia’, ‘BAALC’ and ‘Brain and acute myeloid leukemia cytoplasmic.’ The search coverage was restricted to ‘human’ and the language was limited to ‘Chinese and English’. The Chinese search terms contained ‘BAALC’ and ‘acute myeloid leukemia’. Repetitive literature was removed by reading the titles and abstracts, and the following inclusion criteria were used: i) Included research objectives were retrospective and prospective cohort studies of acute leukemia; ii) only adult patients with AML, including de novo AML, secondary AML and therapy-related AML, were considered; iii) association between the BAALC gene expression level and survival was investigated; iv) prognostic follow-up time was ≥2 years; and v) survival information data, including OS and disease-free survival rate (DFS), were provided. The following exclusion criteria were used: i) Absence of key information, such as hazard ratio (HR), and graphs could not be obtained by other means; and ii) studies only concentrate on pediatric AML or acute promyelocytic leukemia.
Quality evaluation
Two investigators (S.J. Xiao and J.Z. Shen) conducted an independent quality analysis of the included studies. The Newcastle-Ottawa Scale (NOS) evaluation was adopted to grade the included studies with respect to three dimensions (selection, comparability and exposure) and they contained four, one and three items, respectively. A star system was used to semi-quantitatively assess the quality of the studies. Studies with a grade ≥7 were rated as high-quality, those with a grade of 3–6 were rated as intermediate-quality and those with a grade of ≤2 were rated as low-quality studies. A third investigator (J.L. Huang) was involved in the quality evaluation process when differences occurred in the evaluations that were made by the other two investigators. The studies that shared the same author or adopted the same sample were ranked as the studies with the highest score.
Data extraction
The two investigators that evaluated the studies independently integrated the literature that met the above-mentioned standards and they reached a consensus on all the items. For each research study, the author, year of publication, country, number of cases, age and gender of patients, the methods of detecting BAALC expression, the cut-off value, French-American-British (FAB) classification, survival analysis method (multivariate or univariate) and survival data, including CR, HRs and 95% confidence intervals (CIs) for OS and DFS, were obtained. When the author reported univariate and multivariate analyses to obtain HR, the multivariate analysis results, including other variables, was preferably considered as it could be more accurate. When the study did not directly report this information, the investigators who conducted the studies were contacted to provide the data. However, if the author did not provide the data, we attempted to obtain the appropriate data; the P-value was extracted from the log-rank test to estimate HR and 95% CI. When only Kaplan-Meier curves were available, the time-to-event data were extracted from the graphical survival plots. Kaplan-Meier curves were read by the Engauge Digitizer version 4.1 (free software downloaded from http://sourceforge.net) and the method by Tierney et al (7) was used.
Data analysis
Stata 12.0 (StataCorp, College Station, TX, USA) was used to calculate the pooled survival effects of the BAALC gene expression. I2 and Q statistics were also used to test heterogeneity. The statistical heterogeneity of studies was tested with the χ2-based Q-test. In particular, heterogeneity was indicated to be significant when P<0.10. I2 was used for quantifying the inconsistency. I2=0–25, 25–50, 50–75 and >75% indicated no heterogeneity, intermediate heterogeneity, high heterogeneity and extreme heterogeneity, respectively. When I2>25% and the subgroup or sensitivity analysis showed no evident heterogeneity source, the random effects model was used for combined analysis. I2≤25% indicated no heterogeneity; under this circumstance, the fixed effects model was used for the analysis. In addition, sensitivity analysis and Begg's test were also performed to assess the stability of the results and to evaluate the publication bias, respectively.
Results
Search results and characteristics of studies
As shown in Fig. 1, 362 studies were retrieved from the literature, among which, 347 studies were removed as they did not conform to the inclusion criteria. The type of literature that was excluded consisted of repetitive studies, meetings (without complete information) and studies that originated from the same research center. The scores of the remaining studies are shown in Table I. The major deficiency of the excluded studies lies in their failure to provide detailed descriptive analysis of comparability and cohort follow-up. All the included studies reported the prognostic effect for the BAALC gene expression in AML and four studies reported DFS and OS (8–23). Twelve studies provided an introduction to the association between the BAALC gene expression and CR. The majority of the included literature adopted the multivariate analysis to analyze the prognostic significance of the BAALC gene expression, which was conducive in eliminating the confounding bias caused by the disturbance of the confounding factors. The basic features of the included studies are shown in Table II. Thirteen studies adopted the reverse transcription-quantitative polymerase chain reaction (RT-qPCR) method and two studies used the gene chip method to analyze the BAALC gene expression. From the perspective of the cut-off value, 10 studies adopted the median method. The method to set the cut-off value could not be obtained in two of the studies. The NOS was used for assessing the quality of all the studies in the meta-analysis. The NOS results showed that the median overall score was 8 (range, 3–9), which indicated that the methodological quality was high.
Figure 1.
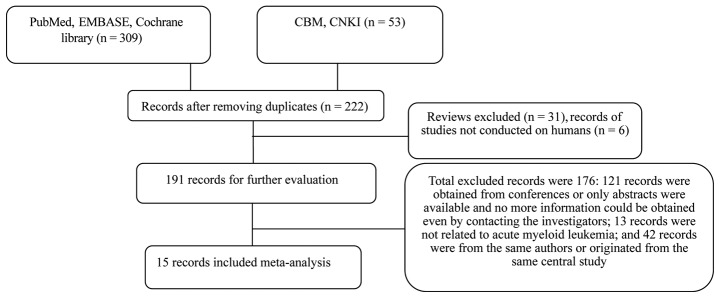
Flow chart of the selection process to identify the eligible studies. CBM, China Biology Medicine Disc; CNKI, China National Knowledge Infrastructure.
Table I.
Newcastle-Ottawa Scale quality assessment of quality cohort studies.
| Selection | Outcome | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| First author, year | Representativeness of the exposed cohort | Selection of the non-exposed cohort | Ascertainment of exposure | Outcome not present at the start | Comparability | Assessment of outcome | Follow-up length | Follow-up adequacy | Score | (Refs.) |
| Yoon, 2013 | * | * | * | * | ** | * | * | * | 9 | (9) |
| Yahya, 2013 | * | * | * | * | ** | * | * | * | 9 | (10) |
| El-Sharnouby, 2010 | * | * | * | * | ** | * | * | * | 9 | (19) |
| Miglino, 2011 | * | * | * | – | – | – | * | – | 4 | (17) |
| Metzeler, 2009 | * | * | * | * | * | * | * | * | 8 | (20) |
| Santamaria, 2010 | * | * | * | * | – | * | * | – | 6 | (18) |
| Thol, 2011 | * | * | * | – | – | – | – | – | 3 | (14) |
| Damiani, 2013 | * | * | * | * | ** | * | * | * | 9 | (11) |
| Langer, 2008 | * | * | * | * | * | * | * | * | 8 | (21) |
| Baldus, 2006 | * | * | * | * | ** | * | * | * | 9 | (22) |
| Bou Samra, 2012 | * | * | * | * | – | * | * | * | 7 | (13) |
| Liu, 2011 | * | * | * | * | * | * | * | * | 8 | (15) |
| Brand, 2013 | – | – | * | * | – | – | * | * | 4 | (12) |
| Bienz, 2005 | * | * | * | * | * | * | * | * | 9 | (23) |
| Weber, 2014 | * | * | * | * | * | * | * | * | 9 | (8) |
Table II.
Main characteristics of the results of eligible studies.
| Age, year (range) | CR | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| First author, year | Country | Character | Case, n (M/F) | Direct method | BAALC high | BAALC low | OS cut-off level | OS calculation method | HR (95% CI) | OR (95% CI) | (Refs.) |
| Yoon, 2014 | Korea | CN-AML | 125 (76/49) | RT-qPCR | 48.51 (18–71) | 49.23 (15–78) | BAALC/ABL=5a | Univariate | 1.94 (0.75–5.03) | 2.45 (1.39–4.32) | (9) |
| Yahya, 2013 | Egypt | CN-AML | 45 (18/27) | RT-qPCR | 45.72 (19–74) | 44.26 (21–73) | Median | Multivariate | 2.63 (1.16–5.88) | 1.56 (2.5–37.2) | (10) |
| El-Sharnouby, 2010 | Egypt | CN-AML | 8 (20/18) | RT-qPCR | 38.75 (17–54) | 35.21 (19–49) | Median | Multivariate | 4.215 (2.31–6.36) | 2.5 (0.67–9.25) | (19) |
| Miglino, 2011 | Italy | AML | 100 | RT-qPCR | (17–84) | (17–84) | Median | Multivariate | 1.51 (0.87–2.61) | 2.6 (1.2–5.82) | (17) |
| Metzeler, 2009 | Germany | CN-AML | 210 (88/122) | RT-qPCR | 62 (18–78) | 57 (17–83) | Median | Univariate | 1.18 (1.02–1.39) | 2.0 (1.11–3.63) | (20) |
| Santamaria, 2010 | Spain | AMLb | 127 (64/63) | RT-qPCR | 53 (15–75) | 56 (20–77) | Median | Multivariate | 3.0 (1.7–5.2) | 4.02 (1.52–10.64) | (18) |
| Thol, 2011 | Germany | CN-AML | 237 | RT-qPCR | NR | NR | NR | Multivariate | 1.95 (1.22–3.13) | 2.5 (1.18–5.26) | (14) |
| Damiani, 2013 | Italy | AML | 175 (84/91) | RT-qPCR | 58 (18–79) | 57 (20–82) | Median | NR | 1.81 (1.12–2.91) | 2.1 (1.01–4.38) | (11) |
| Langer, 2008 | USA | CN-AML | 172 (79/93) | RT-qPCR | 45 (18–59) | 47 (19–59) | Median | Multivariate | 1.84 (1.04–3.28) | 3.7 (1.07–12.5) | (21) |
| Baldus, 2006 | Germany | CN-AML | 307 (147/160) | RT-qPCR | 46 (18–60) | 50 (17–60) | Median | Multivariate | 1.9 (1.3–2.9) | 1.66 (1.03–2.67) | (22) |
| Bou Samra, 2012 | France | CN-AML | 163 | Microarrays | NR | NR | NR | Multivariate | 1.99 (1.32–3) | NR | (31) |
| Liu, 2011 | China | AML | 160 (79/81) | RT-qPCR | 35.2±14 | 39.1±14.8 | Median | Univariate | 1.84 (0.98–3.44) | 1.89 (0.94–3.77) | (15) |
| Brand, 2013 | The Netherlands | AML | 225 | Microarrays | NR | NR | 30th | Multivariate | 1.42 (0.93–2.15) | NR | (12) |
| Weber, 2014 | Germany | CN-AML | 326 (159/167) | RT-qPCR | 52.4 (18.3–64.5) | 53.8 (18.5–64.8) | Median | Multivariate | 1.77 (1.13–2.78) | NR | (8) |
| Bienz, 2005 | Switzerland | AML | 67 (34/33) | RT-qPCR | 46 (18–71) | 53 (19–70) | Median | Multivariate | 3.86 (1.703–8.728) | 2.22 (0.49–10.05) | (23) |
Near median.
Intermediate-risk AML [stratified by molecular score based on ERG, PRAME and EVI1 gene expression, which allows stratification of CN-AML patients in four risk categories; Santamaria et al (35)]. M, male; F, female; BAALC, brain and acute leukemia, cytoplasmic; OS, overall survival; HR, hazard ratio; CI, confidence interval; CR, complete remission; OR, odds ratio; CN-AML, cytogenetically normal-acute myeloid leukemia; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NR, data not reported.
Meta-analysis results
Association of the BAALC gene expression with the FAB subtype
The BAALC expression mainly concentrated on the undifferentiated cells and initial studies suggested that high BAALC gene expression was associated with the M0/M1/M2 subtype (9,10,15,19,21–23). A meta-analysis was conducted on the data from the selected studies. The results are shown in Table III. The results show that BAALC expression tended to occur in poorly differentiated AML, but it was rarely observed in well-differentiated AML.
Table III.
Pooled analysis of the association of BAALC expression with specific FAB subtypes of AML.
| Random effects model overall effect | Fixed effects model overall effect | |||||||
|---|---|---|---|---|---|---|---|---|
| FAB subtype | OR | 95% CI | P-value | Heterogeneity I2, % | OR | 95% CI | P-value | Heterogeneity I2, % |
| M0 | 2.95 | 1.29–6.78 | 0.01 | 0.0 | 2.35 | 0.96–5.77 | 0.06 | 0.0 |
| M1 | 2.20 | 1.58–3.05 | <0.01 | 0.0 | 2.22 | 1.58–3.10 | <0.01 | 0.0 |
| M2 | 1.36 | 1.06–1.77 | 0.02 | 59.1 | 1.38 | 0.87–2.17 | 0.19 | 59.1 |
| M4 | 0.62 | 0.45–0.87 | 0.01 | 43.6 | 0.56 | 0.34–0.95 | 0.03 | 43.6 |
| M5 | 0.27 | 0.15–0.50 | <0.01 | 47.1 | 0.32 | 0.23–0.46 | <0.01 | 47.1 |
| M6 | 0.85 | 0.44–1.65 | 0.63 | 0.0 | 0.83 | 0.40–1.73 | 0.63 | 0.0 |
BAALC, brain and acute leukemia, cytoplasmic; FAB, French-American-British; AML, acute myeloid leukemia; OR, odds ratio; CI, confidence interval.
Clinical characteristics of AML patients with BAALC gene expression
The FLT3-ITD and NPM1 mutations, whose prognoses were recommended by the World Health Organization (WHO) and European Leukemia Net (24,25), were subjected to correlation analysis and the results are shown in Table IV. The meta-analysis of the included data failed to identify a correlation between high BAALC expression and the FLT3-ITD mutation (OR, 1.238, P=0.413); however, high BAALC expression was associated with the NPM1 mutation (OR, 0.166, P<0.001). Seven studies also reported white blood cells (WBC) counts and the percentage of bone marrow at the time of the diagnosis of AML (8–11,18,21,22). A high level of the BAALC expression was not associated with the WBC count and the percentage of bone marrow blasts at the time of diagnosis of AML. High BAALC gene expression had no significant correlation with other clinical characteristics, such as the gender, age, the higher percentage of peripheral blood blasts and lower platelet count in the included studies.
Table IV.
Pooled analysis of the association of BAALC expression with the FLT3-ITD and NPM1 mutations in AML.
| Random effects model overall effect | Fixed effects model overall effect | |||||||
|---|---|---|---|---|---|---|---|---|
| Mutation status subtype | OR | 95% CI | P-value | Heterogeneity I2, % | OR | 95% CI | P-value | Heterogeneity I2, % |
| FLT3-ITD | 1.24 | 0.98–1.57 | 0.41 | 65.4 | 1.24 | 0.98–1.57 | 0.08 | 65.4 |
| NPM1 | 0.17 | 0.12–0.23 | <0.01 | 24.2 | 0.17 | 0.12–0.22 | <0.01 | 24.2 |
BAALC, brain and acute leukemia, cytoplasmic; FLT3, Fms-related tyrosine kinase 3; ITD, internal tandem duplication; NPM1, nucleophosmin; AML, acute myeloid leukemia; OR, odds ratio; CI, confidence interval.
Prognostic impact of the BAALC gene expression in patients with AML
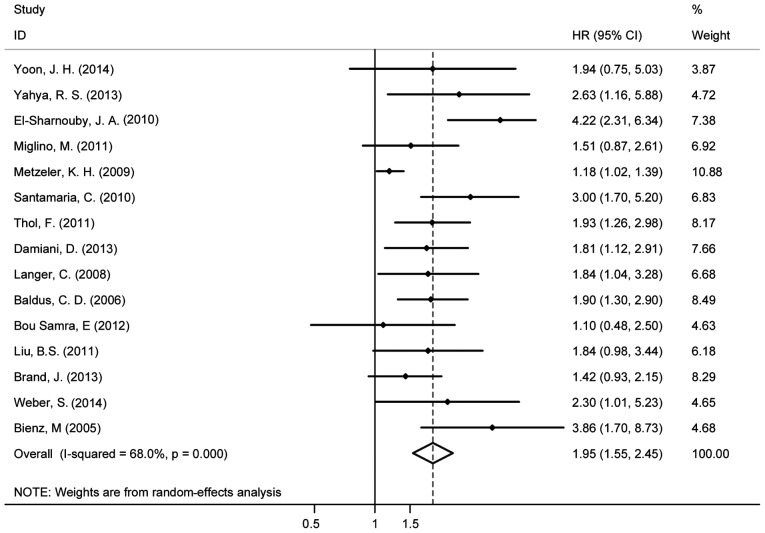
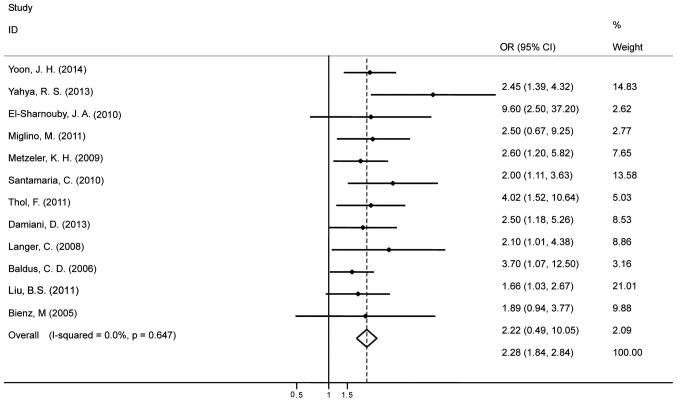
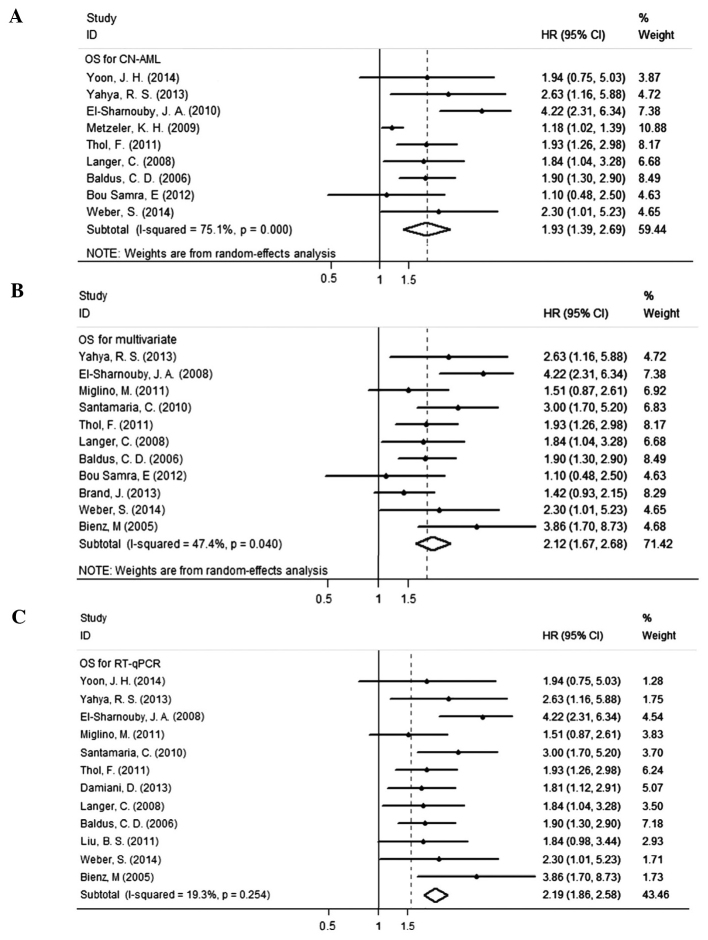
Fifteen studies evaluated the association between the BAALC gene expression and OS for AML and it indicated high heterogeneity (I2=68%). Therefore, the random effects model was employed to perform a meta-analysis (Fig. 2). The OS of patients with the highly expressed BAALC gene was noticeably lower than that of the patients with low BAALC expression with an HR of 1.95 (95% CI, 1.55–2.45). The pool of OR was conducted (Fig. 3) and the results revealed that the highly expressed BAALC gene in AML has a lower CR rate after the patient received chemotherapy [OR, 2.28 (95% CI, 1.84–2.84)]. To obtain the source of heterogeneity and the difference in the survival rates for different groups in the 15 included studies, subgroup analyses were conducted according to AML cytogenetic, the method for detecting the BAALC expression and survival analysis (multivariate or univariate). As shown in Fig. 4, the CN-AML subgroup had an HR of 1.93, (95% CI, 1.39–2.69) with I2=75.1%. In line with the multivariate analysis of the subgroup, the values for patients with high BAALC expression were as follows: HR, 2.12, (95% CI, 1.67–2.68) and I2=47.4%. Using a different detection method, the subgroup analysis of BAALC showed low heterogeneity by RT-qPCR, as evidenced by the following findings: HR, 2.19 (95% CI, 1.86–2.58) and I2=19.3%. In addition, among the 15 studies, four studies also reported the prognostic effect of the BAALC gene expression for DFS. Therefore, a meta-analysis was conducted and the results are shown in Fig. 5. The summary HR for DFS was HR, 2.63 (95% CI, 1.95–3.54) for patients with high BAALC expression compared to those with low BAALC gene expression.
Figure 2.
Forest plot showing the meta-analysis of hazard ratio (HR) and 95% confidence interval (CI) estimates for overall survival rate in patients.
Figure 3.
Forest plot showing the meta-analysis of odds ratio (OR) and 95% confidence interval (CI) estimates for complete remission in patients.
Figure 4.
Forest plots of pooled hazard ratios (HRs) and 95% confidence intervals (CIs) for overall survival (OS) for the prognostic significance of brain and acute leukemia, cytoplasmic (BAALC) expression in defined subgroups of patients with AML. (A) Cytogenetic subgroup; (B) BAALC gene cut-off subgroup; and (C) BAALC detection method subgroup. CN-AML, cytogenetically normal-acute myeloid leukemia; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Figure 5.
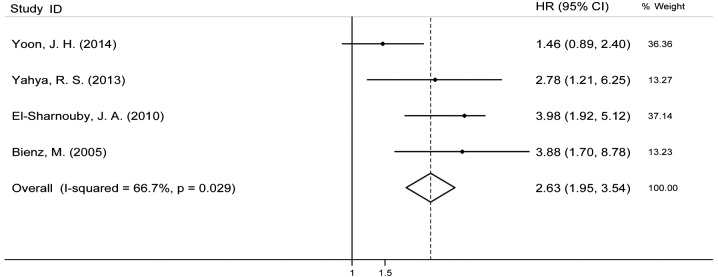
Forest plot showing the meta-analysis of hazard ratio (HR) and 95% confidence interval (CI) estimates for disease-free survival rate in patients.
Sensitivity analysis and publishing bias analysis
Sensitivity analysis showed that the pooled HRs were similar to the overall pooled HR when one study was removed and the remaining studies were analyzed. As shown in Fig. 4, no individual study had a dominant influence on the overall HR. The included studies were also subjected to publishing bias analysis by using the Begg's test. The P-value was 0.921, which indicated no publication bias.
Discussion
The roles of genetic abnormalities, such as gene mutations and chromosome abnormalities, in the occurrence, development and prognosis of AML have been determined. According to the cytogenetic analysis, such as the FLT3-ITD and NPM1 mutation analysis, CN-AML patients were subjected to prognostic stratification (26); the prognostic cytogenetic analysis effectively predicted the different prognoses and formulated corresponding therapeutic schemes for patients (27). However, the prognostic value of the BAALC gene expression in AML remains unclear. The restricted sample size in each of the research studies included in this evaluation made it difficult to detect the corresponding conclusion. Meta-analysis may identify the indicators and their significance. Combining the relevant studies increases statistical power, thereby making it possible to detect the effects that may be missed by individual studies. Conclusively, the present meta-analysis indicated a trend of association between the BAALC gene overexpression and worse prognosis among the entire cohort of patients with AML. The meta-analysis also indicated that high expression of the BAALC gene had a low CR rate [OR, 2.28, (95% CI, 1.84–2.84)], meaning that high expression of the BAALC gene has a poor effect on inducing release and it is prone to induce drug resistance. The present study demonstrated that the worse prognostic impact of the BAALC gene overexpression was observed in patients with AML, with a pooled HR of 1.95 for OS (95% CI, 1.55–2.45), particularly in CN-AML (HR, 1.93; 95% CI, 1.39–2.69). In subgroup analysis of OS, the results also indicated that high BAALC expression is significantly associated with unfavorable OS in multivariate analysis (HR, 2.12; 95% CI, 1.67–2.68). Another meta-analysis showed that FLT3-ITD mutation can predict an unfavourable prognosis with a HR of 1.86 (28); the FLT3-ITD mutation was recommended by the WHO as a prognosis factor in AML. This result indicates that the BAALC gene in AML could be used as a prognostic indicator, as well as to evaluate risk stratification.
The high expression of the BAALC gene usually occurs in acute leukemia, including patients with chronic myeloid leukemia in blast crisis. A percentage of CN-AML patients (65.7%) were found to have high expression of the BAALC gene (23). The highly expressed BAALC had an evidently lower CR rate in allogeneic hematopoietic stem cell transplantation (allo-HSCT) compared to autologous hematopoietic stem cell transplantation. Even if the compound FLT3-ITD mutation or another molecular biology marker had received allo-HSCT, there was no clear difference in the CR rate. Of note, subsequent to receiving allo-HSCT, patients had a lower disease reoccurrence rate compared to those that underwent other types of hematopoietic stem cell transplantation procedures (21). Damm et al (29) established that an integrative prognostic risk evaluation system for CN-AML by a biological marker includes the BAALC expression. The result showed that high-risk patients have a longer OS and DFS if they received hematopoietic stem cell transplantation from a related donor compared to patients who received stem cells from a non-related donor. However, intermediate-risk patients have a lower OS and relapse-free survival if they received hematopoietic stem cell transplantation from a related donor compared to patients who received stem cells from a non-related donor (29). The detection of the molecular biology marker, including the BAALC gene in AML patients, was significant to guiding the patient's prognosis and treatment.
The BAALC gene was originally found in the neuroderm. Subsequent research has showed that it is also expressed in immature bone marrow stem cell progenitors, but not in mature cells. The functions of the BAALC gene remain unclear. The present study also conducted research on the correlation between the BAALC gene expression and FAB classification and observed that FAB classification was prone to occur due to poor differentiation, such as M0/M1/M2, but it was seldom observed in M4/M5 when the BAALC gene expression was high. No correlation was observed between the BAALC gene expression and the number of leukocytes and the percentage of bone marrow blasts during diagnosis; owing to the lack of available data, the association was not evaluated by a meta-analysis. A certain degree of correlation was also observed between high BAALC expression and the NPM1 mutation but not the FLT3-ITD mutation, the mechanism of which is unclear, as it is known that the NPM1 mutation expresses as an independent favorable prognostic biomarker in AML. The pathogenetic impact of BAALC expression and its association with different adverse prognostic indicators in the evolution of leukemia remains elusive. Xu et al (30) induced the knockout of the BAALC gene in small hairpin RNA, which resulted in an evident decrease of cell proliferation and increase of apoptosis, and it demonstrated the role of the BAALC gene in causing leukemia in immature acute leukemic cells. Certain studies also highlighted that the BAALC gene expression blocked the differentiation of the myeloid cells and induced the occurrence of leukemia when combined with HOXA9 (31), which promoted the self-renewal of cells. In addition, the mechanism associated with the BAALC gene expression is under investigation; certain hypotheses have been proposed and tested. Franzoni et al (32) proposed and demonstrated that modification following histone translation affected the BAALC gene expression. In another study, Heesch et al (33) identified that an insulin-like growth factor combined with protein 7 (IGFBP7) was strongly correlated with the BAALC gene, that IGFBP7 inhibited the proliferation and transformation of leukemia and that demethylation drugs enhanced the expression of IGFBP7. The gene sequencing of the binding domain of the BAALC gene promoter identified a gene polymorphism (SNP rs62527607), the locus change of which was conducive to the combination of the RUNX1 transcription factor and the continuous expression of the BAALC gene (34). As further studies will be conducted to investigate the relevant mechanism of the BAALC gene, the detection of BAALC may facilitate the risk evaluation of AML patients and enable targeted therapy of the corresponding genes.
In the present study, the meta-analysis showed an evident heterogeneity (I2=68%); the subgroup analysis of the included studies also suggested that the BAALC expression analysis based on the gene chip had the greatest influence on heterogeneity, among which the research results of Metzeler et al (20) had the largest weight. However, that study was not regarded to be among the low-quality studies, as determined by the NOS scale, which was used to evaluate the studies. The other studies included mainly used the RT-qPCR method to detect the BAALC gene expression. The different types of methodologies used in the included studies are believed to be the major source of the heterogeneity. In addition, in the study of Metzeler et al (20) the meta-analysis used suggested that BAALC high expression was correlated with MN1 gene expression; however, that finding was not subjected to multivariate prognostic analysis. The majority of the included studies were subjected to multivariate analysis, which is regarded as one of the sources of heterogeneity. Further analysis is required to determine the cause(s) of the evident heterogeneity.
The meta-analysis adopted in the present study has certain limitations. The majority of the included studies only adopted the retrospective study method rather than the prospective. In addition, the included studies adopted different methods to detect BAALC gene expression with a non-unified cut-off value. As the median was mostly adopted by the included studies, the findings of the different research studies saliently differed from each other in terms of the cut-off value, thus having a substantial bearing on the heterogeneity of the studies. Of note, the differences in the cut-off value would also impact the results. In view of the fact that a small number of studies used a set cut-off value to analysis the prognosis, subgroup analysis was not conducted.
In conclusion, the meta-analysis results in the present study showed that high expression of the BAALC gene served as an independent poor prognostic indicator in AML. Thus, it could be applied to the clinical prognostic evaluation and guide the treatment of patients with AML. Identification of a more effective prognostic indicator and risk evaluation system could be used for the risk evaluation rating of AML and the formulation of risk-adapted therapeutic strategies for patients.
Acknowledgements
The present study was financially supported by the National and Fujian Provincial Key Clincial Specialty Discipline Construction Program (China), the Major Research National Natural Science Foundation of China-Young Scientist Program (grant no. 81300428) and the Surface Project of National Natural Science Foundation of China (grant no. 81370629).
References
- 1.Estey EH. Acute myeloid leukemia: 2012 update on diagnosis, risk stratification, and management. Am J Hematol. 2012;87:89–99. doi: 10.1002/ajh.22246. [DOI] [PubMed] [Google Scholar]
- 2.Shen Y, Zhu YM, Fan X, et al. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood. 2011;118:5593–5603. doi: 10.1182/blood-2011-03-343988. [DOI] [PubMed] [Google Scholar]
- 3.Mrozek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood? Blood. 2007;109:431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scholl S, Fricke HJ, Sayer HG, Hoffken K. Clinical implications of molecular genetic aberrations in acute myeloid leukemia. J Cancer Res Clin Oncol. 2009;135:491–505. doi: 10.1007/s00432-008-0524-x. [DOI] [PubMed] [Google Scholar]
- 5.Baldus CD, Tanner SM, Kusewitt DF, et al. BAALC, a novel marker of human hematopoietic progenitor cells. Exp Hematol. 2003;31:1051–1056. doi: 10.1016/S0301-472X(03)00263-7. [DOI] [PubMed] [Google Scholar]
- 6.Tanner SM, Austin JL, Leone G, et al. BAALC, the human member of a novel mammalian neuroectoderm gene lineage, is implicated in hematopoiesis and acute leukemia. Proc Natl Acad Sci USA. 2001;98:13901–13906. doi: 10.1073/pnas.241525498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber S, Alpermann T, Dicker F, et al. BAALC expression: a suitable marker for prognostic risk stratification and detection of residual disease in cytogenetically normal acute myeloid leukemia. Blood Cancer J. 2014;4:e173. doi: 10.1038/bcj.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon JH, Kim HJ, Shin SH, et al. Implication of higher BAALC expression in combination with other gene mutations in adult cytogenetically normal acute myeloid leukemia. Leuk Lymphoma. 2014;55:110–120. doi: 10.3109/10428194.2013.800869. [DOI] [PubMed] [Google Scholar]
- 10.Yahya RS, Sofan MA, Abdelmasseih HM, Saudy N, Sharaf-Eldein MA. Prognostic implication of BAALC gene expression in adult acute myeloid leukemia. Clin Lab. 2013;59:621–628. doi: 10.7754/clin.lab.2012.120604. [DOI] [PubMed] [Google Scholar]
- 11.Damiani D, Tiribelli M, Franzoni A, et al. BAALC overexpression retains its negative prognostic role across all cytogenetic risk groups in acute myeloid leukemia patients. Am J Hematol. 2013;88:848–852. doi: 10.1002/ajh.23516. [DOI] [PubMed] [Google Scholar]
- 12.Brand J, van Vliet MH, de Best L, et al. A standardized microarray assay for the independent gene expression markers in AML: EVI1 and BAALC. Exp Hematol Oncol. 2013;2:7. doi: 10.1186/2162-3619-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bou Samra E, Klein B, Commes T, Moreaux J. Development of gene expression-based risk score in cytogenetically normal acute myeloid leukemia patients. Oncotarget. 2012;3:824–832. doi: 10.18632/oncotarget.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thol F, Damm F, Ludeking A, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29:2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 15.Liu BS, Guo XT, Zha J, et al. Simultaneous detection of MDR1 and BAALC gene expression in acute myeloid leukemias and its clinical implication. Chin J Cancer Prev Treat. 2011;18:1691–1695. [Google Scholar]
- 16.Rockova V, Abbas S, Wouters BJ, et al. Risk stratification of intermediate-risk acute myeloid leukemia: integrative analysis of a multitude of gene mutation and gene expression markers. Blood. 2011;118:1069–1076. doi: 10.1182/blood-2011-02-334748. [DOI] [PubMed] [Google Scholar]
- 17.Miglino M, Colombo N, Pica G, et al. WT1 overexpression at diagnosis may predict favorable outcome in patients with de novo non-M3 acute myeloid leukemia. Leuk Lymphoma. 2011;52:1961–1969. doi: 10.3109/10428194.2011.585673. [DOI] [PubMed] [Google Scholar]
- 18.Santamaria C, Chillon MC, Garcia-Sanz R, et al. BAALC is an important predictor of refractoriness to chemotherapy and poor survival in intermediate-risk acute myeloid leukemia (AML) Ann Hematol. 2010;89:453–458. doi: 10.1007/s00277-009-0864-x. [DOI] [PubMed] [Google Scholar]
- 19.El-Sharnouby JA, Ahmed LM, Taha AM, Kamal O. Prognostic significance of CEBPA mutations and BAALC expression in acute myeloid leukemia Egyptian patients with normal karyotype. Egypt J Immunol. 2010;15:131–143. [PubMed] [Google Scholar]
- 20.Metzeler KH, Dufour A, Benthaus T, et al. ERG expression is an independent prognostic factor and allows refined risk stratification in cytogenetically normal acute myeloid leukemia: a comprehensive analysis of ERG, MN1, and BAALC transcript levels using oligonucleotide microarrays. J Clin Oncol. 2009;27:5031–5038. doi: 10.1200/JCO.2008.20.5328. [DOI] [PubMed] [Google Scholar]
- 21.Langer C, Radmacher MD, Ruppert AS, et al. High BAALC expression associates with other molecular prognostic markers, poor outcome, and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: a Cancer and Leukemia Group B (CALGB) study. Blood. 2008;111:5371–5379. doi: 10.1182/blood-2007-11-124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldus CD, Thiede C, Soucek S, Bloomfield CD, Thiel E, Ehninger G. BAALC expression and FLT3 internal tandem duplication mutations in acute myeloid leukemia patients with normal cytogenetics: prognostic implications. J Clin Oncol. 2006;24:790–797. doi: 10.1200/JCO.2005.01.6253. [DOI] [PubMed] [Google Scholar]
- 23.Bienz M, Ludwig M, Leibundgut EO, et al. Risk assessment in patients with acute myeloid leukemia and a normal karyotype. Clin Cancer Res. 2005;11:1416–1424. doi: 10.1158/1078-0432.CCR-04-1552. [DOI] [PubMed] [Google Scholar]
- 24.Sabattini E, Bacci F, Sagramoso C, Pileri SA. WHO classification of tumours of haematopoietic and lymphoid tissues in 2008: an overview. Pathologica. 2010;102:83–87. [PubMed] [Google Scholar]
- 25.Dohner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 26.Schlenk RF, Dohner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 27.Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Port M, Bottcher M, Thol F, et al. Prognostic significance of FLT3 internal tandem duplication, nucleophosmin 1 and CEBPA gene mutations for acute myeloid leukemia patients with normal karyotype, and younger than 60 years: a systematic review and meta-analysis. Ann Hematol. 2014;93:1279–1286. doi: 10.1007/s00277-014-2072-6. [DOI] [PubMed] [Google Scholar]
- 29.Damm F, Heuser M, Morgan M, et al. Integrative prognostic risk score in acute myeloid leukemia with normal karyotype. Blood. 2011;117:4561–4568. doi: 10.1182/blood-2010-08-303479. [DOI] [PubMed] [Google Scholar]
- 30.Xu B, Chen G, Shi P, et al. shRNA-mediated BAALC knockdown affects proliferation and apoptosis in human acute myeloid leukemia cells. Hematology. 2012;17:35–40. doi: 10.1179/102453312X13221316477499. [DOI] [PubMed] [Google Scholar]
- 31.Heuser M, Berg T, Kuchenbauer F, et al. Functional role of BAALC in leukemogenesis. Leukemia. 2012;26:532–536. doi: 10.1038/leu.2011.228. [DOI] [PubMed] [Google Scholar]
- 32.Franzoni A, Passon N, Fabbro D, Tiribelli M, Damiani D, Damante G. Histone post-translational modifications associated to BAALC expression in leukemic cells. Biochem Biophys Res Commun. 2012;417:721–725. doi: 10.1016/j.bbrc.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Heesch S, Bartram I, Neumann M, et al. Expression of IGFBP7 in acute leukemia is regulated by DNA methylation. Cancer Sci. 2011;102:253–259. doi: 10.1111/j.1349-7006.2010.01760.x. [DOI] [PubMed] [Google Scholar]
- 34.Eisfeld AK, Marcucci G, Liyanarachchi S, et al. Heritable polymorphism predisposes to high BAALC expression in acute myeloid leukemia. Proc Natl Acad Sci USA. 2012;109:6668–6673. doi: 10.1073/pnas.1203756109. [DOI] [PMC free article] [PubMed] [Google Scholar]