Abstract
Ralstonia eutropha is a facultative chemolithoautotrophic bacterium that uses the Calvin–Benson–Bassham (CBB) cycle for CO2 fixation. This study showed that R. eutropha strain H16G incorporated 13CO2, emitted by the oxidative decarboxylation of [1-13C1]-glucose, into key metabolites of the CBB cycle and finally into poly(3-hydroxybutyrate) [P(3HB)] with up to 5.6% 13C abundance. The carbon yield of P(3HB) produced from glucose by the strain H16G was 1.2 times higher than that by the CBB cycle-inactivated mutants, in agreement with the possible fixation of CO2 estimated from the balance of energy and reducing equivalents through sugar degradation integrated with the CBB cycle. The results proved that the ‘gratuitously’ functional CBB cycle in R. eutropha under aerobic heterotrophic conditions participated in the reutilization of CO2 emitted during sugar degradation, leading to an advantage expressed as increased carbon yield of the storage compound. This is a new insight into the role of the CBB cycle, and may be applicable for more efficient utilization of biomass resources.
The Calvin–Benson–Bassham (CBB) cycle, employing ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) as a key CO2-fixing enzyme, is used for primary production by most plants, algae and various autotrophic microorganisms1. Because the CBB cycle is a highly energy-consuming pathway dependent on reductive assimilation of CO2, it is strictly repressed by regulation in several plants, algae and cyanobacteria when essential ATP and reducing equivalents are unavailable2,3,4,5. In facultative photoautotrophic purple bacteria, the CBB cycle operates not only during carbon assimilation under photoautotrophic conditions but also for dissipating excess reducing equivalents under photoheterotrophic conditions. Reg/Prr two-component signal transduction systems sense the redox states of cells and regulate global gene expression for various metabolisms including the CBB cycle in the purple non-sulphur bacteria Rhodobacter sphaeroides and Rhodospirillum rubrum6. Synthesis of Rubisco was completely repressed in R. sphaeroides under aerobic chemoheterotrophic conditions6. Algae and some chemolithoautotrophic bacteria grow mixotrophically by simultaneous function of autotrophic and heterotrophic metabolisms, which require light and adequate inorganic electron donors, respectively, along with organic compounds.
A Gram-negative facultative chemolithoautotrophic bacterium, Ralstonia eutropha (Cupriavidus necator) strain H16, can utilize various organic compounds such as sugars, organic acids, fatty acids and plant oils for heterotrophic growth. In autotrophic growth mode, the bacterium can utilize H2 as the energy source and fix CO2 by the CBB cycle7. Two sets of the enzymes in the CBB cycle are encoded in cbbc and cbbp operons in chromosome 2 and megaplasmid pHG1, respectively. The expression of the cbb genes is activated by a common transcriptional regulator, CbbR, encoded in the cbbc operon, when the intracellular concentration of phosphoenolpyruvate (PEP) becomes low under autotrophic conditions8. Interestingly, it has been shown that partial derepression of the cbb genes occurs on some substrates, including fructose and citrate9, given that weak activities of Rubisco and other CBB-cycle enzymes were detected in the late heterotrophic growth phase10. However, to date, the effect of the partially derepressed CBB cycle on heterotrophic metabolism in R. eutropha has not been investigated.
R. eutropha H16 has been also known to accumulate poly(3-hydroxybutylate) [P(3HB)] as a storage compound under unbalanced growth conditions. It has been estimated that the P(3HB) accumulation has a role in survival under stress conditions. Bacterial P(3HB) and related polyhydroxyalkanoates (PHAs) have attracted industrial attention as possible alternatives to petroleum-based polymer materials because they are biodegradable thermoplastics produced from renewable carbon sources. A number of studies has focused on the biosynthesis of PHAs by R. eutropha, particularly in terms of the biosynthetic pathways and enzymes, and on metabolic engineering aimed at efficient production of PHA copolyesters from inexpensive biomass resources11,12,13,14,15. Recent transcriptome analyses of R. eutropha showed that the expression of the cbb genes was upregulated in the wild type strain H16 under nitrogen-deficient P(3HB) accumulation conditions16,17; however, it was downregulated in the PHA-negative mutant strain PHB−4 grown on gluconate18. Metabolome analysis of the H16 strain detected ribulose-1,5-bisphosphate (RuBP), a key metabolite specific to the CBB cycle, in cells of R. eutropha H16 cultured with fructose or octanoate19. Moreover, we detected slight incorporation of 13C atoms into P(3HB) during incubation of R. eutropha H16 in a fructose-containing medium supplemented with NaH13CO3, and confirmed that 13CO2 fixation was mediated by both the Rubiscos17. These observations strongly suggested some role of the CBB cycle in the heterotrophic P(3HB) biosynthesis from sugars.
We assumed that, when the CBB cycle is functional under heterotrophic conditions in the presence of sugars, it may act on fixation and reutilization of CO2 emitted by oxidative decarboxylation during the sugar degradation. Generally, microbial production of value-added compounds from sugars often accompanies marked loss of carbon because of decarboxylation. In particular, this phenomenon is critical for acetyl-CoA-derived compounds such as P(3HB) because one-third of the total carbon atoms in hexoses are lost as CO2 molecules emitted by the oxidative decarboxylation of pyruvate to acetyl-CoA. Considering the costs of harvest, transportation and saccharification of biomass-based polysaccharides, the loss of carbon during microbial sugar degradation cannot be negligible for the purpose of efficient utilization of biomass resources. Therefore, the design of metabolic pathways avoiding carbon loss is expected to be one way to establish more efficient bioprocesses. Chinen et al. have reported a pathway for efficient L-glutamate production from glucose by Corynebacterium glutamicum employing phosphoketolase (PKT) to bypass the CO2-releasing pyruvate dehydrogenase reaction20. However, to date, the reutilization of the decarboxylated carbon for the bioproduction of value-added compounds using the functions of the CBB cycle under heterotrophic conditions has not been studied.
Metabolomic approaches employing stable isotope labelling of metabolites are powerful tools for the analysis of metabolic dynamics21,22,23. Recently, Hasunuma et al. established the dynamic analysis of plant metabolism by isotope tracing of 13C from 13CO2 fed to Nicotiana tabacum leaves24. In this study, we constructed CBB cycle-inactivated mutants of R. eutropha and compared P(3HB) biosynthesis properties and 13C-labelling profiles with [1-13C1]-glucose, in which the 13C atom was emitted as 13CO2 through the Entner–Doudoroff (ED) pathway, with those of the parent strain. The result provided new insight into the function of the CBB cycle. The ‘gratuitously’ activated CBB cycle in R. eutropha under heterotrophic conditions played a role in fixation and reutilization of the carbon, generally wasted during sugar degradation, for biosynthesis of the storage polyester.
Results
Construction of CBB cycle-inactivated strains of R. eutropha
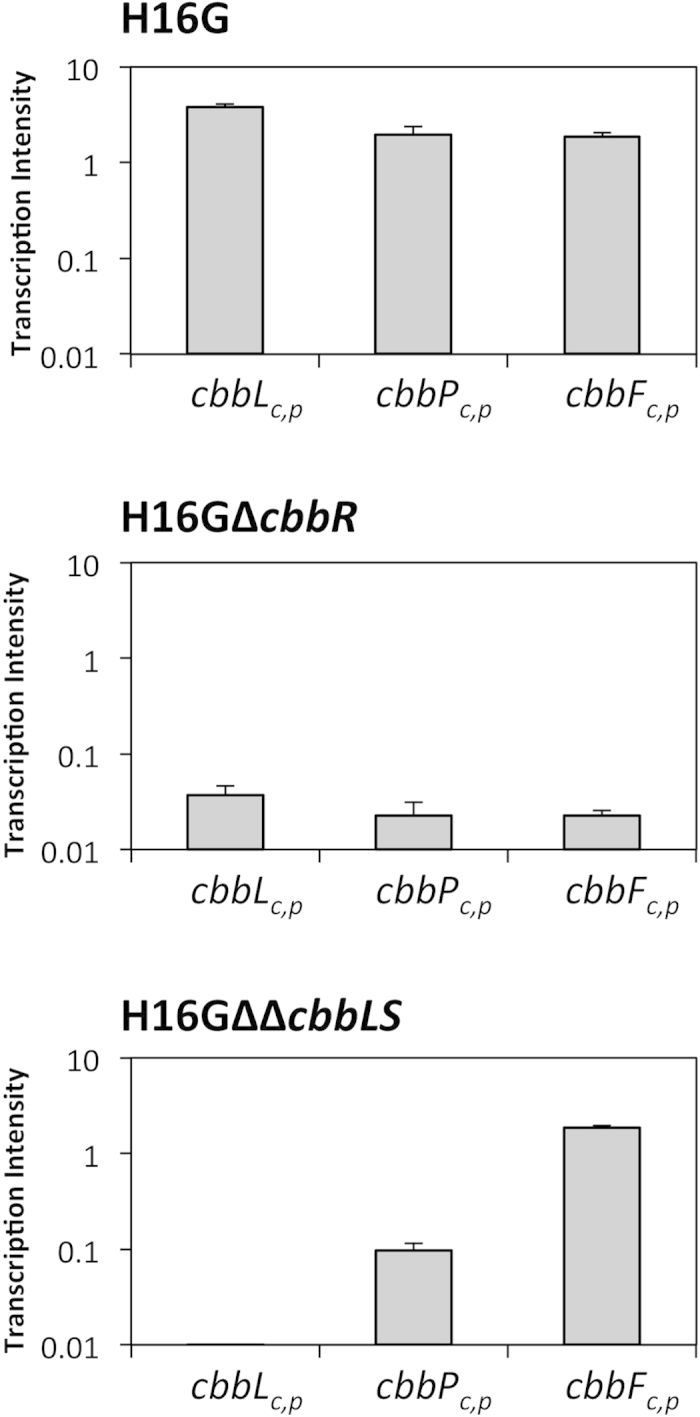
Two CBB cycle-inactivated strains of R. eutropha were constructed using the glucose-assimilating recombinant strain H16G (renamed from the previously constructed strain H16∆nagR_nagE-G793C25) as a host strain. In the mutant strain H16G∆∆cbbLS, both cbbLSc- and cbbLSp-encoding Rubisco enzymes were deleted from chromosome 2 and pHG1, respectively, by homologous recombination. Note that this strain retains other cbb genes involved in regeneration of RuBP in the CBB cycle. Another mutant strain, H16G∆cbbR, was constructed by deletion of cbbR on chromosome 2, encoding a common transcriptional activator for the two cbb operons. It has been reported that a cbbR-deleted strain of R. eutropha was incapable of growing autotrophically owing to insufficient induction of the cbb genes26. Indeed, qRT-PCR analysis demonstrated that expression levels of cbbL (encoding Rubisco large subunit), cbbP (encoding phosphoribulokinase) and cbbF (encoding fructose-1,6-bisphosphatase I/sedoheptulose-1,7-bisphosphatase) in H16G∆cbbR incubated with glucose were approximately one-hundredth compared to those in the parent strain H16G (Fig. 1). H16G∆∆cbbLS showed higher expression of cbbP and cbbF than H16G∆cbbR, as expected.
Figure 1. Relative gene expression levels of cbbL, cbbP and cbbF in R. eutropha strains H16G and the CBB cycle-inactivated strains (H16G∆cbbR and H16G∆∆cbbLS) grown on glucose.
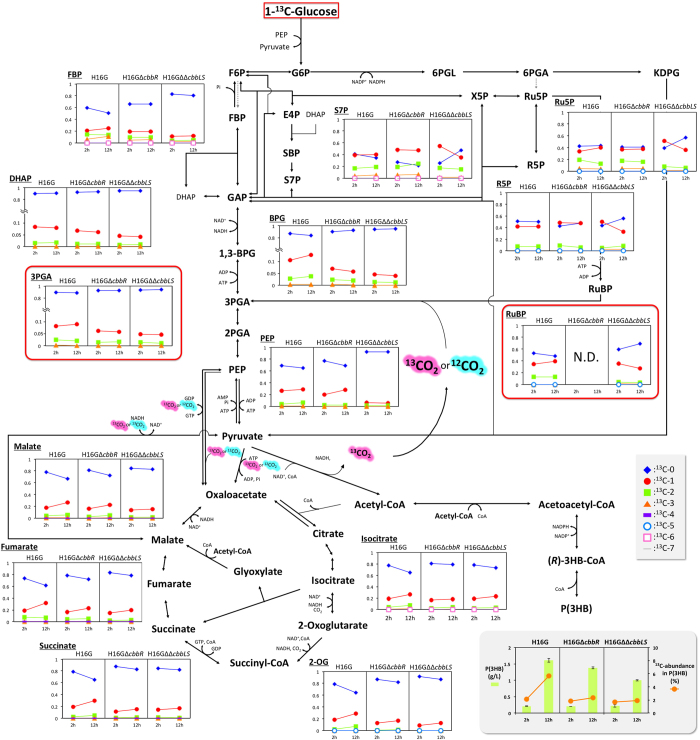
Metabolomics of the R. eutropha strains producing P(3HB) from [1-13C1]-glucose
The R. eutropha strains were first cultivated in a nutrient-rich medium and the grown cells were then incubated in a nitrogen-free mineral salt medium containing [1-13C1]-glucose. The cellular metabolites were extracted from the cells incubated with [1-13C1]-glucose for 2 h and 12 h and subjected to metabolomic analysis to detect the incorporation of 13C into each of the metabolites. Analysis using reversed-phase ion-pair liquid chromatography coupled with triple-quadrupole mass spectrometry (RP-IP-LC/QqQ-MS) was able to determine the 13C-labelling ratios for 31 metabolites including sugar phosphates, organic acids, amino acids and CoA thioesters. Unexpectedly, high incorporation of 13C in free coenzyme A (CoA) was observed during the incubation with [1-13C1]-glucose for all the strains examined, and the profile was nearly the same as those of the detectable CoA-thioesters (acetyl-CoA, butyryl-CoA, succinly-CoA, 3-hydroxybutyryl-CoA and crotonyl-CoA). The 13C abundances in acyl moieties of the CoA thioesters could not be precisely determined, owing to 13C accumulation in the CoA backbone, so that the results for CoA-thioesters were not further used. The mass distributions of 26 metabolites are shown in supplementary Table S2, and those of 14 metabolites in sugar metabolism are shown along with the metabolic pathways in Fig. 2. The changes in abundance of [13C1]-derivatives were useful for evaluating the level of 13C incorporation, given that derivatives multiply labelled with 2 or more 13C atoms were generally present at low abundance (<7%) except for a few metabolites.
Figure 2. Time-dependent changes of mass distribution of metabolites in central metabolisms, as well as production of and 13C abundance in P(3HB) with R. eutropha H16G and H16G∆cbbR, and H16G∆∆cbbLS incubated with [1-13C1]-glucose.
Abbreviations are shown in supplementary Table S1.
The 13C atom was slightly but significantly incorporated into 3-phosphoglycerate (3PGA) (p < 0.01), a product of Rubisco-mediated CO2 fixation, in H16G incubated with [1-13C1]-glucose. In contrast, the abundance of the 13C-containing isotopomers of 3PGA was slightly decreased in H16G∆cbbR and H16G∆∆cbbLS. Similar tendencies were observed for dihydroxyacetone phosphate (DHAP), 1,3-bisphosphoglycerate (1,3-BPG) and fructose-1,6-bisphoaphate (FBP), although [13C1]-phosphoenolpyruvate (PEP) in H16G∆cbbR increased. The abundances of the [13C1]-isotopomer of 3PGA, 1,3-BPG and DHAP in H16G were approximately 10%, which was apparently lower than those of PEP and FBP (~20%). This observation suggested slow turnover of the triose phosphates in R. eutropha under the conditions studied.
Significant and interesting differences among the three strains were observed for RuBP, a metabolite specific to the CBB cycle as a substrate for Rubisco. RuBP was detected not in H16G∆cbbR but in H16G and H16G∆∆cbbLS. The abundance of [13C1]-RuBP in H16G harbouring intact cbb operons increased during incubation with [1-13C1]-glucose from 2 to 12 h, whereas the abundance decreased in H16G∆∆cbbLS lacking Rubiscos. Ribulose-5-phosphate (Ru5P), ribose-5-phosphate (R5P) and sedoheptulose-7-phosphate (S7P) were detected in all strains. The [13C1]-isotopomers of these pentose phosphates and heptose phosphate had already constituted 40–60% of the total metabolites after incubation with [1-13C1]-glucose for 2 h. The abundances of the [13C1]-isotopomers then decreased in H16G∆∆cbbLS but increased in H16G during further incubation, as seen for RuBP in the respective strains. 13C accumulation into these sugar phosphates was relatively stable in H16G∆cbbR after rapid increase until 2 h.
Several carboxylic acids in the tricarboxylic acid (TCA) cycle showed similar changes in 13C accumulation among the three strains. The abundances of the [13C1]-isotopomers slightly increased during incubation with [1-13C1]-glucose from 2 to 12 h, with the rates of increase in H16G higher than those in the other two mutants. The analysis also detected several amino acids in the extracts, as shown in supplementary Table S2, and the changes in 13C accumulation were similar to those of the related carboxylic acids in the respective strains. For example, the profiles observed for serine and glutamate were very similar to those for 3PGA and 2-oxoglutarate, respectively. The abundances of the 13C-labelled isotopomers of the most detectable amino acids were higher in H16G than in H16G∆cbbR and H16G∆∆cbbLS.
13C incorporation into P(3HB) synthesized from [1-13C1]-glucose
The abundance of the [1-13C1] glucose-derived 13C atom in P(3HB) was determined by gas chromatography-mass spectrometry (GC-MS) analysis of methyl 3-hydroxybutyrate formed by methanolysis of the polymer. The 13C abundances in P(3HB) synthesized by H16G∆cbbR and H16G∆∆cbbLS (2.3% and 1.9%, respectively) were slightly higher than the natural abundance (1.1%), whereas that in P(3HB) synthesized by H16G was intriguingly higher (5.6%) (Fig. 2 and Table 1). These observations indicated that the CBB cycle in H16G was responsible for the incorporation of 13C derived from [1-13C1] glucose into P(3HB).
Table 1. Production of and 13C abundance in P(3HB) on [1-13C1]-glucose, and carbon yield of P(3HB) on glucose with R. eutropha H16G and CBB cycle-inactivated strains.
| R. eutropha strain | Time (h) | P(3HB)a (g/L) | 13C abundance in P(3HB)b (%) | P(3HB) yieldc (g/g-glucose) |
|---|---|---|---|---|
| H16G | 2 | 0.21 ± 0.21 | 2.09 ± 0.00 | 0.345 |
| 12 | 1.61 ± 0.06 | 5.64 ± 0.00 | ||
| H16G∆cbbR | 2 | 0.20 ± 0.00 | 1.79 ± 0.00 | 0.294 |
| 12 | 1.38 ± 0.03 | 2.34 ± 0.00 | ||
| H16G∆∆cbbLS | 2 | 0.22 ± 0.04 | 1.68 ± 0.08 | 0.286 |
| 12 | 1.00 ± 0.02 | 1.91 ± 0.02 |
aP(3HB) produced from [1-13C1]-glucose by 2-step cultivation.
bMeans of 13C/12C ratio calculated from isotopomer abundances of the two fragments (m/z 45 and 87) derived from 3HB methyl ester.
cDetermined from relationship between P(3HB) production and glucose consumption obtained by 6 ~ 8 independent 2-step cultivations for various incubation periods.
Carbon yields P(3HB) on glucose by R. eutropha strains
The carbon yields of the conversion of P(3HB) to glucose were determined for the three R. eutropha strains by two-step cultivation. Because small amounts of P(3HB) (approximately 7–10 wt%) had been accumulated within the cells after the first step of cultivation in the rich medium, the yield during the second step of cultivation was calculated from the linear relationship between P(3HB) production and glucose consumption observed during several independent cultivations for various incubation periods. As shown in Table 1, the P(3HB) yields of H16G∆cbbR and H16G∆∆cbbLS were approximately half of the theoretical yield of 0.53 g-P(3HB)/g-glucose, and that of H16G was determined to be 1.2-fold higher than those of the two mutant strains lacking cbbR or cbbLSs.
Discussion
The 13C abundance in P(3HB) synthesized by H16G∆∆cbbLS, in which the CBB cycle cannot function owing to the lack of Rubiscos, was slightly increased from the natural abundance of 1.1% to 1.9% after incubation with [1-13C1]-glucose for 12 h. Considering that R. eutropha H16 does not possess phosphofructokinase and 6-phosphogluconate dehydrogenase in the Embden–Meyerhof (EM) and pentose phosphate (PP) pathways, respectively, the ED pathway is a unique sugar degradation pathway, so that [1-13C1]-pyruvate and unlabeled glyceraldehyde 3-phosphate (GAP) were produced from [1-13C1]-glucose. When [1-13C1]-pyruvate was converted to acetyl-CoA by decarboxylation or anaplerosis and the successive TCA cycle, 13CO2 was emitted and unlabeled acetyl-CoA was produced, even in the case of fixation of H13CO3− by anaplerotic reactions. Thus, the 13C atom derived from [1-13C1]-glucose was not incorporated into P(3HB) through the first round of the ED pathway (supplementary Fig. S1). Besides decarboxylation, [1-13C1]-pyruvate could be converted via the EM pathway, resulting in distribution of the 13C atom at the 1-position of triose phosphates, followed by formation of [3-13C1]-, [4-13C1]- or [3,4-13C2]-fructose-6-phosphate (F6P). While, [1-13C1]-F6P was formed by 6-phosphorylation and isomerization of [1-13C1]-glucose. These 13C atoms were distributed at the 1-, 2-, 3- and/or 4-positions of F6P through interconversion of C4-C7 sugar phosphates by a non-oxidative branch of the PP pathway [supplementary Fig. S5(A) and (C)] or RuBP regeneration steps in CBB cycle [supplementary Fig. S5(B) and (D)]. The 13C-labelled F6P molecules were converted to acetyl-CoA containing 13C at the 1 and/or 2 positions by the ED pathway and oxidative decarboxylation of pyruvate. Thus, the 13C atoms could be incorporated into P(3HB) without Rubisco-mediated CO2 fixation (supplementary Fig. S2). This event explained the slight increase of 13C abundance in P(3HB) to 1.9% in the H16G∆∆cbbLS strain.
When the CBB cycle in the H16G strain operated during P(3HB) biosynthesis from glucose, the CO2 molecules emitted by oxidative decarboxylation of pyruvate had a chance to couple with RuBP by the Rubisco-mediated reaction. As shown in supplementary Fig. S3, the complete CBB cycle likely established a new pathway for conversion of the 13C-labelled RuBP derived from [1-13C1]-glucose to 13C-labelled acetyl-CoA, even when 12CO2 was fixed. It was expected that the fixation of 13CO2, generated from [1-13C1]-pyruvate, by Rubisco enriched 13C atoms in the sugar phosphates via interconversion of sugar phosphates, leading to incorporation of more 13C into P(3HB) via 13C-enriched acetyl-CoA (supplementary Fig. S4). Indeed, the 13C abundance in P(3HB) synthesized from [1-13C1]-glucose by H16G increased to 5.6%. This observation demonstrated the actual fixation of CO2 emitted by decarboxylation during glucose degradation by the CBB cycle, and incorporation of the fixed carbon into P(3HB) in R. eutropha.
Metabolomic analysis revealed the increase of the [13C1]-isotopomers of most sugar phosphates, including RuBP and 3PGA, in the H16G strain during incubation with [1-13C1]-glucose (Fig. 2). This observation provided strong evidence for the actual flux of the CBB cycle with fixation of the [1-13C1]-glucose-derived 13CO2. In contrast, the [13C1]-isotopomers decreased in H16G∆∆cbbLS. The presence of intracellular RuBP without increase of 13C was reasonable, because RuBP could be generated by phosphoribulokinase (CbbP), but not be converted owing to the lack of Rubiscos in this strain. The absence of RuBP in H16G∆cbbR was consistent with the too low expression of cbbP and other cbb genes caused by the deletion of the transcriptional activator CbbR. However, the abundances of the [13C1]-isotopomers for many metabolites in H16G∆cbbR tended to show changes between those in H16G and H16G∆∆cbbLS. These results suggested that the subtly expressed cbb genes even in the absence of the activator CbbR, as shown by qRT-PCR, which resulted in the slightly higher 13C-abundance in P(3HB) synthesized from [1-13C1]-glucose by H16G∆cbbR (2.3%) than by H16G∆∆cbbLS (1.9%).
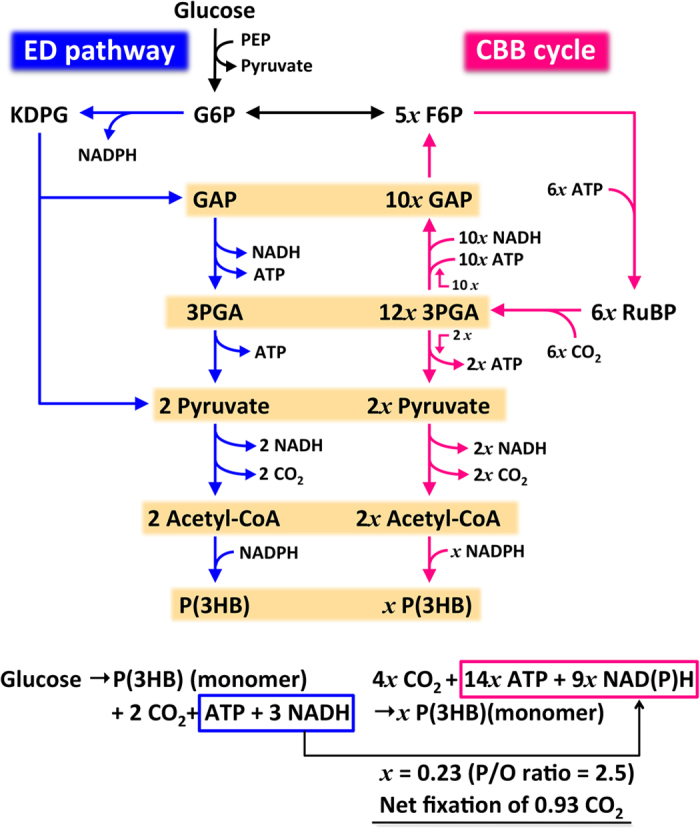
By R. eutropha H16G, one molecule of glucose is converted to P(3HB) monomer [(R)-3HB-CoA] along with two molecules of CO2 and surplus energy and reducing equivalents via the ED pathway, as shown as the left-hand equation in Fig. 2. The conversion of CO2 to P(3HB) monomer by the combination of the CBB cycle and P(3HB) biosynthesis is shown as the right-hand equation in Fig. 2. On the assumption that NADH is equivalent to NADPH and alternatively acted as an electron donor for generation of 2.5 ATP through aerobic respiration (P/O ratio = 2.527), it was estimated that the energy and reducing equivalents released during conventional P(3HB) synthesis from glucose correspond to those essential for fixation of 0.93 CO2 into P(3HB) (Fig. 3). Thus, when the CBB cycle is functional during heterotrophic P(3HB) biosynthesis not associated with cell growth, 0.23 molecules of 3HB monomer are expected to be additionally obtained from the fixed 0.93 molecules of CO2 (46.5% recovery from 2 molecules of CO2 emitted from 1 molecule of glucose), leading to increase of the P(3HB) yield to 123%. This calculation agreed well with the finding that R. eutropha H16G produced P(3HB) in 117–122% yield relative to H16G∆cbbR and H16G∆∆cbbLS (Table 1). Apparently, the active CBB cycle under heterotrophic conditions was an advantage in P(3HB) production for R. eutropha. However, the net increase of 13C abundance in P(3HB) synthesized from [1-13C1]-glucose by H16G was 4.5% of the natural abundance (1.1%), which was lower than the 7.8% simply estimated from the fixation of 0.93 CO2 in the two CO2 molecules derived from [1-13C1]-glucose. We initially supposed that this discrepancy was due to stable carbon isotope discrimination by Rubisco, but this reason is unlikely because the ratio of reaction rate toward 12CO2 to that toward 13CO2 (ε-values) for Rubisco from R. eutropha was only 1.9% by in vitro assay28. Considering that the yields of P(3HB) by the R. eutropha strains were lower than the theoretical maximum yield, some acetyl-CoA molecules were inferred to have been completely degraded to CO2 by the TCA cycle to obtain energy under aerobic conditions for maintaining various cellular functions. This process would reduce the abundance of 13CO2 within cells, reflecting the lower-than-expected 13C abundance in P(3HB). The turnover of the TCA cycle in the P(3HB) accumulation phase not associated with cell growth was supported by the increase of the 13C-labelled isotopomers of intermediate acids in the TCA cycle in H16G (Fig. 2).
Figure 3. P(3HB) biosynthesis from glucose in R. eutropha H16G through an integrated pathway with the ED pathway and CBB cycle.
Recently, Guadalupe-Medina et al. have reported that functional expression of type II Rubisco and phosphoribulokinase in Saccharomyces cerevisiae established a bypass for glucose degradation not producing excess NADH and resulting in reduced glycerol formation during bioethanol production under anaerobic conditions29. In the present study, we found that the CBB cycle in R. eutropha plays a role in fixation of CO2 emitted by oxidative decarboxylation during sugar degradation and reutilized the fixed CO2 as a source of P(3HB). It should be further noted that this novel function of CBB cycle was expressed under aerobic heterotrophic conditions, differently from those of the CBB cycle in purple non-sulphur bacteria under anaerobic photoheterotrophic conditions and chemoautotrophic bacteria under mixotrophic conditions. There are expected to be three important factors for this: heterotrophic derepression of the CBB cycle by an unique intercellular PEP sensor CbbR8 at a probable low intracellular concentration of PEP attributed to P(3HB) formation, a high ratio of carboxylase activity to oxygenase activity (τ value) of 75 for the red-type Rubisco from R. eutropha30 and simple modulation of Rubisco activity by a AAA+ protein CbbX specific for red-type Rubiscos31. The more accumulation of the storage compound may be beneficial for survival in natural habitats. Moreover, the present results suggested that the CBB cycle, under heterotrophic conditions, could raise base yields of useful compounds by reutilization of the carbon atom emitted from carbon sources, when the reducing equivalents obtained by heterotrophic metabolisms were greater than those required for biosynthesis of the end products. Although the generation and consumption of reducing equivalents were equally balanced in typical anaerobic fermentation such as production of ethanol and 1-butanol, surplus reducing equivalents are available in biosynthesis of some bioproducts; for example, P(3HB), optically active 3-hydroxybutyrate and 2-propanol. The functional integration of carbon-fixing enzyme/pathways into the metabolic networks of industrial microorganisms may be a useful strategy for avoiding the loss of biomass-derived carbons in such cases.
Materials and Methods
Construction of disruption plasmids and recombinant strains
General cultivation of bacterial strains and construction plasmids pK18ms∆cbbLSc and pK18ms∆cbbLSp for deletion of cbbLSc and cbbLSp, respectively, have been reported in previously17. A plasmid pK18ms∆cbbR for deletion of cbbR from chromosome 2 of R. eutropha H16 was constructed as follows. First, upstream and downstream regions (1 kbp) of cbbR were individually amplified by PCR with genomic DNA of R. eutropha H16 as a template and primer sets of cbbR-up5’ (GCTCTAGAAGCCATTTGGCAATCACGCGGA)/cbbR-up3’ (TCCTGGAACCGGGCGGTTGGGGGCGGCTTTGGAT) and cbbR-down5’ (CCAACCGCCCGGTTCCAGGAGGGTTGGCTGGGATT)/cbbR-down3’ (GGAATTCTGGTAGCGGCGTTGTCATACACAT), respectively. The underlined sequence showed the restriction sites for XbaI and EcoRI, respectively. The second PCR with the amplified fragments using cbbR-up5’/cbbR-down3’ primers gave a fused fragments of upstream and downstream regions of cbbR. The resulting fragment was digested by EcoRI and XbaI and then ligated with pK18mobsacB32 at the corresponding sites to obtain pK18ms∆cbbR.
Transconjugation of the mobilizable plasmids from E. coli S17-1 to R. eutropha H16G and isolation of strains generated by pop in-pop out recombination using pK18mobsacB-based suicide plasmids were preformed as described previously12,14. The strains H16G∆∆cbbLS and H16G∆cbbR were obtained by double deletion of cbbLSc and cbbLSp, and single deletion of cbbR in R. eutropha H16G, respectively.
P(3HB) production by two-step cultivation and determination of the carbon yield
R. eutropha H16G, H16G∆cbbR, and H16G∆∆cbbLS strains were firstly cultivated in 100 ml of a nutrient rich medium in 500 ml flask, with reciprocal shaking (117 strokes/min) for 14 ~ 15 h at 30 °C. The grown cells in 40 ml of the culture broth were harvested by centrifugation (5,000 g, 4 °C for 3 min), and washed with a mineral salt solution (9 g/l Na2HPO4 • 12 H2O, 1.5 g/l KH2PO4 in deionized water). The cell pellet was resuspended with 40 ml of a nitrogen-free MB medium composed by 9 g/l Na2HPO4 • 12 H2O, 1.5 g/l KH2PO4, 0.2 g/l MgSO4 and 40 μl of trace element solution33 and 5 g/l naturally-labeled glucose in a 200 ml flask. The cell suspension was further incubated with reciprocal shaking (128 strokes/min) at 30 °C as the second step cultivation for P(3HB) biosynthesis.
After the incubation for various times (2 h ~ 30 h), the cells and supernatant were separated by centrifugation (5,000 g, 10 min, 4 °C). Glucose concentration in the supernatant was measured by glucose oxidase method using a glucose kit GLU-NEO (SHINO-TEST, Tokyo, Japan). The recovered cells were washed with cold deionized water, and then lyophilized. The cellular P(3HB) content was determined by gas chromatography after methanolysis of the dried cells in the presence of 15% (v/v) sulfuric acid in methanol as described previously33. The carbon yield of P(3HB) was calculated from the amount of the produced P(3HB) and consumption of glucose during the cultivation.
Quantitative real-time PCR (qRT-PCR)
The two-step cultivation of the R. eutropha strains was performed as mentioned above, and total RNA was isolated from the cells incubated with glucose under a nitrogen-depleted condition (the second stage) for 2 h by using RNeasy Midi Kit (Qiagen, Valencia, CA, USA). cDNA was synthesized by using ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO, Osaka, Japan) according to the manufacturer’s instruction. Real-time PCR was performed by using Thunderbird SYBR qPCR Mix (TOYOBO, Osaka, Japan) with Thermal Cycler Dice Real Time System (Takara Bio, Otsu, Japan). The reaction conditions were: 1 min at 95 oC, 40 cycles of 10 s at 95 oC and 30 s at 60 oC). bfr2 (h16_A0328) was used as an inner control gene17, and primer sequences are listed in supplementary Table S3. The primers for amplification of a region in cbbL, cbbP and cbbF were designed to bind to both the copies in cbbc and cbbp operons.
13C-labeling of metabolites and P(3HB) using [1-13C1]-glucose
R. eutropha strains were cultivated by the two-step cultivation, where the second step was done with 5 g/l [1-13C1]-glucose (1-13C 98-99%, Cambridge Isotope Laboratories, Andover, MA, USA). 5-ml portion of the culture broth were taken at 2 h or 12 h for metabolomics analysis as follows. The remaining culture broth was used to determine the cellular content of P(3HB) as described above, and the 13C-abundance in the resulting P(3HB) was measured by GC-MS according to the procedure described previously17.
Metabolite extraction and sample preparation
The 5-ml portion of the culture broth was put into 30 ml of 60% (v/v) methanol pre-cooled in ethanol/dry ice bath in a 50-ml centrifuge tube. The quenched cells were immediately recovered by centrifugation (8,000 g, 5 min, –8 °C), and dried in vacuo. The extraction of the intracellular metabolites from the dried cells was performed according to the procedure previously reported34 with slight modifications. Three milliliters of methanol/water/chloroform (2.5:1:1) was added into the dried cells in the 50-ml tube, and vigorously shaken by using vortex mixer for 30 s. The cell suspension was kept in –80 °C for 30 min, and then allowed to thaw at –30 °C and sonicate for 1 min. This cycle of freezing, thawing, and sonicating of the cells at the low temperatures was repeated three times for efficient extraction of the intercellular metabolites without decomposition. The precipitated proteins were removed by centrifugation at 16,000 g, 4 °C for 30 min. The two milliliters of the resulting supernatant was divided to 1 ml each in two micro tubes, and mixed with 200 μl of ultrapure water. The mixture was centrifuged, and the 1 ml aliquots of the polar phase were combined and concentrated using a concentrator VC-36S (Taitech Co., Tokyo, Japan) to a final volume of approximately 50 μl. The 20 μl portions of the extracts were taken and stored at –80 °C as metabolite extracts to be analyzed.
Mass spectrometry
The metabolite extracts were analyzed byRP-IP-LC/QqQ-MS by using a Nexera UHPLC system equipped with LCMS 8030 Plus (Shimadzu, Kyoto, Japan). A PE capped CERI L-column 2 ODS (150 mm × 2.1 mm I. D., particle size 3 μm, Chemicals Evaluation and Research Institute, Tokyo, Japan) was used for RP-IP-LC. The conditions were as follows: mobile phase, 10 mM tributylamine and 15 mM acetic acid in water (A) and methanol (B); flow rate, 0.3 ml/min; gradient curve, 0% B at 0–1 min, 0–15% B at 1–2 min, 15-50% B at 4–9 min, 50–55% B at 9–11.5 min, 55–100% B at 11.5–12 min and 100-0% B at 13–13.5 min and 0% B at 13.5–18 min; injection volume, 3 μl; and column oven temperature, 45 °C. The mode of mass analysis was negative ion mode. The probe position was +1.5 mm, the desolvation line temperature was 250 °C, the nebulizer gas flow was 2 l/min, the drying gas flow was 15 l/min, and the heat block temperature was 400 °C. The other MS parameters were determined by auto-tuning. The scheduled multiple reaction monitoring (MRM) mode was applied throughout the analysis. The peaks of isotopomers of each target metabolite were identified by comparison of the shapes and retention times with that of the authentic compounds, and those areas were determined by LabSolutions version 5.60 (Shimadzu). The LC/QqQ–MS parameters of metabolites and details of MRM method are shown in supplementary Table S4. The data was obtained from three independent cultivations.
Additional Information
How to cite this article: Shimizu, R. et al. New Insight into the Role of the Calvin Cycle: Reutilization of CO2 Emitted through Sugar Degradation. Sci. Rep. 5, 11617; doi: 10.1038/srep11617 (2015).
Supplementary Material
Acknowledgments
We thank Asssoc. Prof. T. Tsuge and Dr. A. Hiroe (Department of Innovative and engineered Materials, Interdisciplinary Graduate School of Science and Engineering, Tokyo Institute of Technology, Japan) for GC-MS analysis. This work was supported by JSPS KAKENHI Grant Numbers 25292058 and 26011292, and Solutions Research Laboratory of Tokyo Institute of Technology.
The authors declare no competing financial interests.
Author Contributions R.S. and T.F. designed the experiments and wrote the main manuscript. R.S. performed the experiments and analyzed the data. Y.D., Y.N., T.B. and E.F. carried out LC-MS analysis and data processing, and suggested the experimental design. S.N. participated in the design and coordination of the study. All authors read and approved the manuscript.
06/20/2016
A correction has been published and is appended to both the HTML and PDF versions of this paper. The error has not been fixed in the paper.
References
- Woodrow I. E. & Berry J. A. Enzymatic regulation of photosynthetic CO2, fixation in C3 plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 533–594 (1988). [Google Scholar]
- Parry M. A. J. et al. Regulation of Rubisco by inhibitors in the light. Plant. Cell Environ. 20, 528–534 (1997). [Google Scholar]
- Schürmann P. & Jacquot J.-P. Plant thioredoxin systems revisited. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 371–400 (2000). [DOI] [PubMed] [Google Scholar]
- Matsumura H. et al. Structure basis for the regulation of glyceraldehyde-3-phosphate dehydrogenase activity via the intrinsically disordered protein CP12. Structure 19, 1846–1854 (2011). [DOI] [PubMed] [Google Scholar]
- Tamoi M., Miyazaki T., Fukamizo T. & Shigeoka S. The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J. 42, 504–513 (2005). [DOI] [PubMed] [Google Scholar]
- Kusian B. & Bowien B. Organization and regulation of cbb CO2 assimilation genes in autotrophic bacteria. FEMS Microbiol. Rev. 21, 135–155 (1997). [DOI] [PubMed] [Google Scholar]
- Bowien B. & Kusian B. Genetics and control of CO2 assimilation in the chemoautotroph Ralstonia eutropha. Arch. Microbiol. 178, 85–93 (2002). [DOI] [PubMed] [Google Scholar]
- Grzeszik C., Jeffke T., Schäferjohann J., Kusian B. & Bowien B. Phosphoenolpyruvate is a signal metabolite in transcriptional control of the cbb CO2 fixation operons in Ralstonia eutropha. J. Mol. Microbiol. Biotechnol. 2, 311–320 (2000). [PubMed] [Google Scholar]
- Friedrich C. G., Friedrich B. & Bowien B. Formation of enzymes of autotrophic metabolism during heterotrophic growth of Alcaligenes eutrophus. J. Gen. Microbiol. 122, 69–78 (1981). [Google Scholar]
- Bowien B., Windhövel U., Yoo J.-G., Bednarski R. & Kusian B. Genetics of CO2 fixation in the chemoautotroph Alcaligenes eutrophus. FEMS Microbiol. Lett. 87, 445–450 (1990). [Google Scholar]
- Budde C. F., Riedel S. L., Willis L. B., Rha C. & Sinskey A. J. Production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from plant oil by engineered Ralstonia eutropha strains. Appl. Environ. Microbiol. 77, 2847–2854 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima Y. et al. Characterization and functional analyses of R-specific enoyl coenzyme A hydratases in polyhydroxyalkanoate-producing Ralstonia eutropha. Appl. Environ. Microbiol. 78, 493–502 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifune J., Nakamura S. & Fukui T. Engineering of pha operon on Cupriavidus necator chromosome for efficient biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from vegetable oil. Polym. Degrad. Stab. 95, 1305–1312 (2010). [Google Scholar]
- Insomphun C. et al. Modification of β-oxidation pathway in Ralstonia eutropha for production of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from soybean oil. J. Biosci. Bioeng. 117, 184–190 (2014). [DOI] [PubMed] [Google Scholar]
- Insomphun C. et al. Improved artificial pathway for biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) with high C6-monomer composition from fructose in Ralstonia eutropha. Metab. Eng. 27, 38–45 (2015). [DOI] [PubMed] [Google Scholar]
- Brigham C. J., Speth D. R., Rha C. & Sinskey A. J. Whole-genome microarray and gene deletion studies reveal regulation of the polyhydroxyalkanoate production cycle by the stringent response in Ralstonia eutropha H16. Appl. Environ. Microbiol. 78, 8033–8044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu R. et al. Detection of phase-dependent transcriptomic changes and Rubisco-mediated CO2 fixation into poly (3-hydroxybutyrate) under heterotrophic condition in Ralstonia eutropha H16 based on RNA-seq and gene deletion analyses. BMC Microbiol. 13, 169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peplinski K. et al. Genome-wide transcriptome analyses of the “Knallgas” bacterium Ralstonia eutropha H16 with regard to polyhydroxyalkanoate metabolism. Microbiology 156, 2136–2152 (2010). [DOI] [PubMed] [Google Scholar]
- Fukui T. et al. Metabolite profiles of polyhydroxyalkanoate-producing Ralstonia eutropha H16. Metabolomics 10, 190–202 (2014). [Google Scholar]
- Chinen A., Kozlov Y. I., Hara Y., Izui H. & Yasueda H. Innovative metabolic pathway design for efficient L-glutamate production by suppressing CO2 emission. J. Biosci. Bioeng. 103, 262–269 (2007). [DOI] [PubMed] [Google Scholar]
- Clasquin M. F. et al. Riboneogenesis in yeast. Cell 145, 969–980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putri S. P. et al. Current metabolomics: Practical applications. J. Biosci. Bioeng. 115, 579–589 (2013). [DOI] [PubMed] [Google Scholar]
- Nakayama Y., Putri S. P., Bamba T. & Fukusaki E. Metabolic distance estimation based on principle component analysis of metabolic turnover. J. Biosci. Bioeng. 118, 350–355 (2014). [DOI] [PubMed] [Google Scholar]
- Hasunuma T. et al. Metabolic turnover analysis by a combination of in vivo13C-labelling from 13CO2 and metabolic profiling with CE-MS/MS reveals rate-limiting steps of the C3 photosynthetic pathway in Nicotiana tabacum leaves. J. Exp. Bot. 61, 1041–1051 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita I., Iwazawa R., Nakamura S. & Fukui T. Identification of mutation points in Cupriavidus necator NCIMB 11599 and genetic reconstitution of glucose-utilization ability in wild strain H16 for polyhydroxyalkanoate production. J. Biosci. Bioeng. 113, 63–69 (2012). [DOI] [PubMed] [Google Scholar]
- Jeffke T. et al. Mutational analysis of the cbb operon (CO2 assimilation) promoter of Ralstonia eutropha. J. Bacteriol. 181, 4374–4380 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkle P. C., Kumar M. A., Resetar A. & Harris D. L. Mechanistic stoichiometry of mitochondrial oxidative phosphorylation. Biochemistry 30, 3576–3582 (1991). [DOI] [PubMed] [Google Scholar]
- Boller A. J. Stable carbon isotope discrimination by rubisco enzymes relevant to the global carbon cycle. Ph.D. dissertation, University of South Florida (2012). [Google Scholar]
- Guadalupe-Medina V. et al. Carbon dioxide fixation by Calvin-Cycle enzymes improves ethanol yield in yeast. Biotechnol. Biofuels 6, 125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horken K. M. & Tabita F. R. Closely related form I ribulose bisphosphate carboxylase/oxygenase molecules that possess different CO2/O2 substrate specificities. Arch. Biochem. Biophys. 361, 183–194 (1999). [DOI] [PubMed] [Google Scholar]
- Mueller-Cajar O. et al. Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature 479, 194–199 (2011). [DOI] [PubMed] [Google Scholar]
- Schäfer A. et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73 (1994). [DOI] [PubMed] [Google Scholar]
- Kato M., Bao H. J., Kang C.-K., Fukui T. & Doi Y. Production of a novel copolyester of 3-hydroxybutyric acid and medium-chain-length 3-hydroxyalkanoic acids by Pseudomonas sp. 61-3 from sugars. Appl. Microbiol. Biotechnol. 45, 363–370 (1996). [Google Scholar]
- Dempo Y., Ohta E., Nakayama Y., Bamba T. & Fukusaki E. Molar-based targeted metabolic profiling of cyanobacterial strains with potential for biological production. Metabolites 4, 499–516 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.