Abstract
Breast cancer is a complex disease displaying different profiles involving genetic as well as epigenetic factors. MicroRNAs (miRNAs) are small, non-coding RNAs that regulate gene expression. Recent studies demonstrated that miRNAs may display great potential for the development of novel biomarkers and therapeutic targets. In the present study, the levels of miR-21 and miR-145 were analyzed in the peripheral blood of 52 patients with luminal A breast cancer. miRNA expression was determined in serum samples from matched pre- and post-treatment patients with breast cancer by quantitative polymerase chain reaction. There were no statistically significant differences in miR-145 and miR-21 levels between pre- and post-treatment samples. In addition, the miRNA levels were not found to be associated with the clinical parameters.
Keywords: breast cancer, circulating microRNAs, serum samples
Introduction
Breast cancer is the most prevalent type of cancer among women, affecting approximately one million women worldwide (1). The development of breast cancer is a complex, multistep process. Breast cancer is considered to be a disease entity including different molecular subtypes with distinct biological behavior profiles, which affect treatment response and outcome. Accordingly, the biological classification of breast tumors uses hormone receptor expression and human epidermal growth factor receptor 2 (HER2) status with the aim to assess the potential response to treatment. Luminal A tumors are associated with the best prognosis among all breast cancer subtypes (2).
Over the last decade, the identification of small, non-coding RNA molecules, namely the microRNAs (miRNAs), has attracted considerable attention due to the accumulating evidence regarding their relevant regulatory functions on cancer initiation, proliferation and progression to a more aggressive phenotype. The interactions between miRNAs and their target mRNAs, usually within the 3′-untranslated region of the target genes, results in the degradation and/or translational inhibition of these genes. Approximately 1,000 miRNA genes are encoded in the human genome (3) and a total of 1,600 precursors and 2,042 mature miRNAs have been identified according to miRBase (http://www.mirbase.org/cgi-bin/browse.pl?org=hsa).
miRNAs have great potential as biomarkers for cancer detection due to their remarkable stability in the blood. Recent data indicated that, in addition to their regulatory function, miRNAs may play a predictive role in evaluating the response to therapy (4,5). Several studies reported the use of circulating miRNAs as biomarkers of disease and therapy response and as diagnostic and prognostic markers in breast cancer (6). Studies investigating the expression of miRNA molecules in breast cancer have revealed their significance as potential markers in tumor classification (7,8). It has even been suggested that circulating miRNAs may be specific biomarkers for breast cancer screening (9).
The development of sensitive, non-invasive markers in the blood circulation may facilitate early detection, monitoring of tumor progression and treatment response in patients with breast cancer. Free nucleic acids in the serum of cancer patients were first reported in 1977 by Leon et al (10). For this reason, free nucleic acids and oligonucleotides, such as miRNAs, have recently attracted attention in the field of cancer research.
Recent studies indicated that miR-34a is one of the key miRNA molecules with a tumor suppressor function, suggesting that it may play a role in DNA damage repair through inducing cell cycle arrest and apoptosis. The targets of miR-34 include cyclin D1, cyclin-dependent kinase 6, E2F transcription factor 3 and myc genes (11,12). miR-145 is another miRNA with tumor suppressor function, playing a role in suppressing cancer cell invasion and metastasis (13,14). miR-21 has been associated with an oncogenic potential and poor outcome in cancer (15,16). It has been suggested that miR-21 is directly involved in regulating apoptosis in breast cancer cells (17–19). miR-221 is another oncogenic miRNA, which is involved in the induction of angiogenesis and resistance to tamoxifen (5,20). In addition, upregulation of miR-10b has been reported in breast cancer and has been correlated with invasion, metastatic capacity and progression (21,22).
In this study, we analyzed the plasma levels of breast cancer-associated miRNAs (miR-21, −145, −34a, −10b and −221) during adjuvant chemotherapy in patients with luminal A breast cancer, in order to identify miRNA molecules that are differentially expressed in response to chemotherapy.
Patients and methods
Patients
A total of 52 patients with primary breast cancer who were treated at the Oncology Institute, University of Istanbul (Istanbul, Turkey), were included in the study. To ensure that all the tumors exhibited the same biological characteristics, only patients with luminal A tumors were included in the study. Patients with stage I–III cancers were primarily treated with conservative surgery or mastectomy. Tumor staging was performed according to the American Joint Committee on Cancer TNM classification and the pTNM was determined following pathological examination. Patients with metastatic disease and other previous tumors were excluded from the study. İmmunohistochemical screening for estrogen receptors, progesterone receptors and HER2 status was performed on formalin-fixed paraffin-embedded tissue samples. The serum samples were obtained prior to the initiation and following completion of the adjuvant chemotherapy. Data regarding patient demographics, histology, receptor status, type of surgical treatment and adjuvant therapy were obtained by reviewing the patients' medical records. This study was approved by the local Ethics Committee of Istanbul University.
RNA isolation
All sera were immediately stored at −80°C until RNA isolation. Circulating RNA molecules were isolated from the sera using the TriPure isolation reagent (Roche Diagnostics GmbH, Mannheim, Germany). Briefly, a 200-µl sample was mixed with 800 µl of TriPure reagent and the mixture was incubated at room temperature for complete dissociation of the nucleoprotein complexes. Following incubation, 0.2 ml chloroform was added to each sample and incubated at room temperature for 15 min. Following centrifugation at 12,000 × g for 15 min, the RNA phase was transferred into Eppendorf tubes and 0.5 ml isopropanol was added. The mixture was incubated at room temperature for 10 min and centrifuged at 12,000 × g for 10 min. The supernatant was discarded, the RNA-containing pellet was washed with 75% ethanol and centrifuged at 7,500 × g for 5 min. The supernatant was removed, air-dried and resuspended in RNAse-free water. The RNA-containing sample was incubated at 55–60°C for 15 min and stored in −80°C until usage.
Conversion of total RNA into complementary DNA (cDNA)
cDNA was synthesized using the miScript Reverse Transcription kit (Qiagen, Valencia, CA, USA) according to the instructions of the manufacturer. cDNA synthesis was performed at 37°C for 60 min and the reaction was inactivated by incubation at 95°C for 5 min.
Quantitative polymerase chain reaction (qPCR)
To quantitate the miRNA molecules, the miScript Primer Assay (Qiagen) was used, which includes a universal primer specific to the poly-A tail and a miRNA-specific primer. Selected miRNA primers were obtained from the miScript Primer Assay, as presented in Table I. SYBR®-Green (Qiagen) was used as the fluorescent molecule. The amplified PCR product had a size of ~80 bp. The miR-16 molecule was used as a reference for normalization of the expression levels of the miR panel. qPCR was performed using the LightCycler 480 (Roche Diagnostics GmbH). The PCR program included a fast start step of 15 min at 95°C, followed by 45 cycles of amplification, with each cycle consisting of denaturation at 94°C for 15 sec, annealing at 55°C for 30 sec and elongation at 70°C for 30 sec. A relative quantification determined the ratio between the amount of target miRNA and reference amplicon. Melting curve analyses were performed to verify the specifity and identity of the PCR products.
Table I.
List of microRNAs used in the experiment.
| Assay name | Qiagen cat. no. | miRBase accession no. |
|---|---|---|
| Hs_miR-145_1_1 | MS00003528 | MIMAT0000437 |
| Hs_miR-21_2 | MS00009079 | MIMAT0000076 |
miR-21 was detected in paired samples from 32 patients, while miR-145 was analyzed in paired samples from 49 patients. Under identical experimental conditions, the remaining miRNAs exhibited very low levels and were detected only in a very small number of samples; therefore, they were not included in the statistical analyses.
Statistical analysis
We compared the plasma levels of miR-21 and miR-145 between the pre- and post-treatment samples of each patient. The statistical analysis was performed using the Mann-Whitney U test. P<0.05 was considered to indicate a statistically significant difference. The association of miR-21 and miR-145 expression with clinical stage was evaluated by the Wilcoxon signed-ranks test.
Results
Clinicopathological characteristics
The main clinicopathological characteristics of the patients in our series were as follows: The mean age was 47.7 years. Stage I was reported in 3 (5.8%), stage II in 33 (63.4%) and stage III in 16 patients (30.8%). All the patients underwent surgery: Conservative surgery was performed in 28 (53.8%) and mastectomy was performed in 24 patients (46.1%). The hormone receptor status was positive and the HER2 status was negative in all the patients. Radiotherapy was delivered to 44 patients (84.6%). Adjuvant chemotherapy was administered to all patients: A total of 15 patients (28.9%) received anthracycline-based therapy, 1 patient (1.9%) received taxane-based therapy and 36 patients (69.2%) received anthracycline and taxane-based regimens. Endocrine therapy was administered to 51 of the 52 patients (98.0%) (Table II).
Table II.
Characteristics of breast cancer patients.
| Characteristics | Patient no. (%) (n=52) |
|---|---|
| Age, years | |
| Mean | 47.7 |
| Median | 46 |
| Stage | |
| I | 3 (5.8) |
| II | 33 (63.4) |
| III | 16 (30.8) |
| Surgery | |
| Conservative surgery | 28 (53.8) |
| Mastectomy | 24 (46.2) |
| Radiotherapy treatment | 44 (84.6) |
| Chemotherapy treatment regimens | |
| Anthracycline-based | 15 (28.9) |
| Taxane-based | 1 (1.9) |
| Anthracyclin and taxane-based | 36 (69.2) |
| Endocrine therapy | 51 (98.0) |
Pre- and post-treatment plasma levels of miR-21 and miR-145
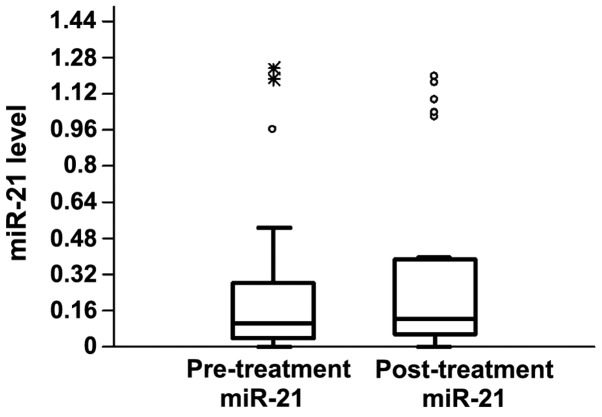
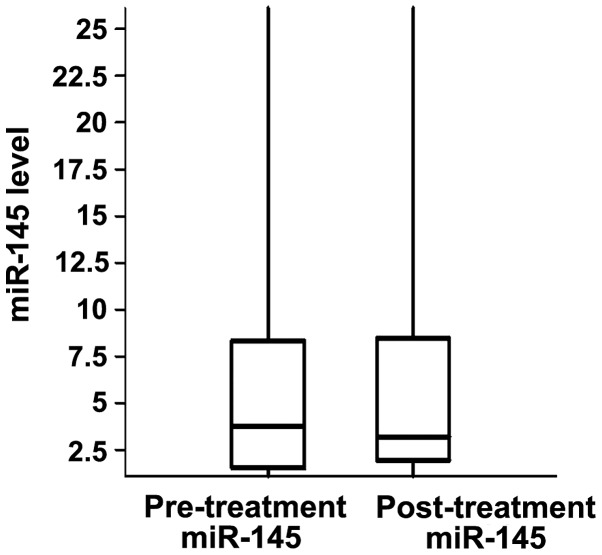
The plasma levels of miR-21 and miR-145 were measured in matched pre- and post-treatment samples of the patients. In the pre-treatment samples, the relative median levels of miR-21 and miR-145 were 0.1 and 3.84, respectively, while in the post-treatment samples the values were 0.12 and 3.08, respectively (Figs. 1 and 2). The differences between pre- and post-treatment samples were not statistically significant (P>0.05). Following chemotherapy, the plasma levels of miR-21 increased in 17 and decreased in 15 patients, when compared with pre-treatment levels. miR-145 expression was higher following chemotherapy in 20 patients, while it declined in the remaining 29 patients (Figs. 3 and 4).
Figure 1.
Levels of miR-21 in the pre-treatment and post-treatment sera samples in fluorescence units. The boxes indicate the upper and lower quartiles.
Figure 2.
Levels of miR-145 in the pre-treatment and post-treatment sera samples in fluorescence units. The boxes indicate the upper and lower quartiles.
Figure 3.
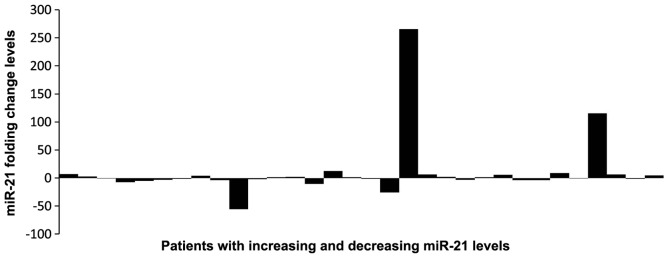
Changes in miR-21 levels in individiual patients. Increasing or decreasing miR-21 levels in pre- and post-treatment samples.
Figure 4.
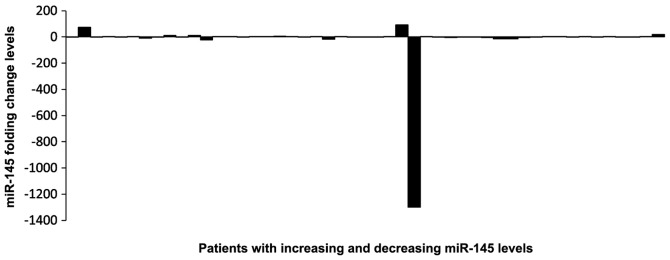
Changes in miR-145 levels in individiual patients. Increasing or decreasing miR-145 levels in pre- and post-treatment samples.
Plasma levels of miR-21 and miR-145 according to stage
We next compared miRNA levels according to clinical stage. Significantly higher levels of miR-21 were observed in post-treatment plasma samples from patients with stage III disease (P=0.04). In this subgroup, we also observed a decrease in miR-145 expression, although the difference was not statistically significant (P=0.06). In the evaluation of serum miRNA levels according to stage, the decline in stage III patients was significant compared with that in stage I+II patients (P=0.006) (Table III).
Table III.
Serum miR-21 and miR-145 levels according to stage.
| Stage I+II | Stage III | ||
|---|---|---|---|
| Variables | Median | Median | aP-value |
| miR-21 | 0.006 | ||
| Pre-treatment | 0.105 | 0.007 | |
| Post-treatment | 0.092 | 0.381 | |
| bP-value | >0.05 | 0.04 | |
| miR-145 | >0.05 | ||
| Pre-treatment | 3.5 | 6.52 | |
| Post-treatment | 2.73 | 3.08 | |
| bP-value | >0.05 | >0.05 |
Mann-Whitney U test
Wilcoxon signed-ranks test.
Discussion
miRNAs in the circulation may serve as biomarkers for cancer detection. It remains unclear how tumor-associated miRNAs enter the circulation. It has been suggested that tumor miRNAs may be present in circulation as a consequence of tumor cell death and lysis (23).
In this study, we evaluated pre- and post-treatment plasma levels of five miRNAs (miR-21, −145, −34a, −10b and −221) in patients with luminal A breast cancer undergoing adjuvant chemotherapy. The miR-10b, −34a and-221 could not be analyzed, as their serum levels were very low and several paired samples could not be matched.
In unselected patients, we observed no significant differences between pre- and post-treatment levels of miR-21 and −145. However, in patients with initial stage III tumors, the miR-21 plasma levels were found to be significantly increased following treatment. This finding indicates that chemotherapy-induced cancer cell apoptosis may contribute to increased circulating levels of miR-21. Aberrant expression of miR-21 in breast cancer has been previously reported (17–19,24). miR-21 has been found to be highly expressed in breast tumors compared with its expression in matched normal breast tissues, suggesting that miR-21 may play a significant role in tumorigenesis (25). A previous study on colorectal cancer, which indicated that increased miR-21 expression may represent a marker of poor prognosis, supports this finding (26).
miR-145 is considered to act as a tumor suppressor and has been shown to be downregulated in breast cancer. The expression levels of mature miR-21 and mature miR-145 were found to be higher in esophageal squamous cell carcinoma compared with those in the normal epithelium (27). Significantly reduced miR-145 expression was also reported in atypical and anaplastic tumors compared with benign meningiomas (28). In prostate cancer, miR-145, a direct target of p53, represses bone metastasis and is involved in regulating epithelial-to-mesenchymal transition (EMT) and cancer cell stemness. Recent data suggest that loss of wild-type p53 may promote bone metastasis of prostate cancer, at least partially through repressing miR-145, promote EMT and stemness of cancer cells (29). As stated above, we would have expected increased miR-145 levels following chemotherapy, but the miR-145 levels were in fact decreased.
To the best of our knowledge, this is the first study investigating the plasma levels of miR-21 and miR-145 in pre- and post-treatment breast cancer samples. Our results demonstrated that miR-21 expression is significantly higher in post-treatment plasma samples from patients with stage III tumors. These data support the fact that chemotherapy-induced apoptosis may contribute to increased circulating miRNA levels, including miR-21.
Acknowledgements
This study was supported by the Istanbul University Research Fund (project no. 17143).
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Chuthapisith S, Permsapaya W, Warnnissorn M, Akewanlop C, Sirivatanauksorn V, Prasarttong Osoth P. Breast cancer subtypes identified by the ER, PR and HER-2 status in Thai women. Asian Pac J Cancer Prev. 2012;13:459–462. doi: 10.7314/APJCP.2012.13.2.459. [DOI] [PubMed] [Google Scholar]
- 3.Fu SW, Chen L, Man YG. miRNA biomarkers in breast cancer detection and management. J Cancer. 2011;2:116–122. doi: 10.7150/jca.2.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salter KH, Acharya CR, Walters KS, et al. An intergrated approach to the prediction of chemotherapeutic response in patients with breast cancer. PLoS One. 2008;3:e1908. doi: 10.1371/journal.pone.0001908. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortez MA, Welsh JW, Calin GA. Circulating microRNAs as noninvasive biomarkers in breast cancer. Recent Results Cancer Res. 2012;195:151–161. doi: 10.1007/978-3-642-28160-0_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Wang BC, Tang JH. Clinical significance of microRNA-155 expression in human breast cancer. J Surg Oncol. 2012;106:260–266. doi: 10.1002/jso.22153. [DOI] [PubMed] [Google Scholar]
- 9.Ng EK, Li R, Shin VY, et al. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One. 2013;8:e53141. doi: 10.1371/journal.pone.0053141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 11.Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 12.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 13.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei P, Xie J, Wang L, Yang X, Dai Z, Hu Y. microRNA-145 inhibits osteosarcoma cell proliferation and invasion by targeting ROCK1. Mol Med Rep. 2014;10:155–160. doi: 10.3892/mmr.2014.2195. [DOI] [PubMed] [Google Scholar]
- 15.Song B, Wang C, Liu J, Wang X, Lv L, Wei L, Xie L, Zheng Y, Song X. MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J Exp Clin Cancer Res. 2010;29:29. doi: 10.1186/1756-9966-29-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair VS, Maeda LS, Ioannidis JP. Clinical outcome prediction by microRNAs in human cancer: A systematic review. J Natl Cancer Inst. 2012;104:528–540. doi: 10.1093/jnci/djs027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 18.Qi L, Bart J, Tan LP, Platteel I, Sluis T, Huitema S, Harms G, Fu L, Hollema H, Berg A. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer. 2009;9:163. doi: 10.1186/1471-2407-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 20.Le Bot N. MicroRNAs in angiogenesis. Nat Cell Biol. 2012;14:342. doi: 10.1038/ncb2484. [DOI] [Google Scholar]
- 21.Han X, Yan S, Weijie Z, Feng W, Liuxing W, Mengquan L, Qingxia F. Critical role of miR-10b in transforming growth factor-β1-induced epithelial-mesenchymal transition in breast cancer. Cancer Gene Ther. 2014;21:60–67. doi: 10.1038/cgt.2013.82. [DOI] [PubMed] [Google Scholar]
- 22.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 23.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 24.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 25.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 26.Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79:313–320. doi: 10.1159/000323283. [DOI] [PubMed] [Google Scholar]
- 27.Akagi I, Miyashita M, Ishibashi O, Mishima T, Kikuchi K, Makino H, Nomura T, Hagiwara N, Uchida E, Takizawa T. Relationship between altered expression levels of MIR21, MIR143, MIR145 and MIR205 and clinicopathologic features of esophageal squamous cell carcinoma. Dis Esophagus. 2011;24:523–530. doi: 10.1111/j.1442-2050.2011.01177.x. [DOI] [PubMed] [Google Scholar]
- 28.Kliese N, Gobrecht P, Pachow D, et al. miRNA-145 is downregulated in atypical and anaplastic meningiomas and negatively regulates motility and proliferation of meningioma cells. Oncogene. 2013;32:4712–4720. doi: 10.1038/onc.2012.468. [DOI] [PubMed] [Google Scholar]
- 29.Ren D, Wang M, Guo W, Zhao X, Tu X, Huang S, Zou X, Peng X. Wild-type p53 suppresses the epithelial-mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR-145. Int J Oncol. 2013;42:1473–1481. doi: 10.3892/ijo.2013.1825. [DOI] [PubMed] [Google Scholar]