Abstract
Dental pulp cells (DPCs) are a promising source of transplantable cells in regenerative medicine. However, DPCs have not been fully characterized at the molecular level. The aim of the present study was to distinguish DPCs from various source-derived mesenchymal stem cells (MSCs), fibroblasts (FBs) and other cells by the expression of several DPC-characteristic genes. DPCs were isolated from human pulp tissues by the explant method or the enzyme digestion method, and maintained with media containing 10% serum or 7.5% platelet-rich plasma. RNA was isolated from the cells and from dental pulp tissue specimens. The mRNA levels were determined by DNA microarray and quantitative polymerase chain reaction analyses. The msh homeobox 1, msh homeobox 2, T-box 2 and ectonucleoside triphosphate diphosphohydrolase 1 mRNA levels in DPCs were higher than that of the levels identified in the following cell types: MSCs derived from bone marrow, synovium and adipose tissue; and in cells such as FBs, osteoblasts, adipocytes and chondrocytes. The enhanced expression in DPCs was consistently observed irrespective of donor age, tooth type and culture medium. In addition, these genes were expressed at high levels in dental pulp tissue in vivo. In conclusion, this gene set may be useful in the identification and characterization of DPCs in basic studies and pulp cell-based regeneration therapy.
Keywords: dental pulp cells, gene expression, mesenchymal stem cells, fibroblasts, regenerative medicine
Introduction
Dental pulp cells (DPCs) show odontoblast-like differentiation, with increased expression of alkaline phosphatase and osteocalcin and matrix calcification in vitro following exposure to osteogenesis induction medium (1–3). In addition, transplantation of DPCs enhances the regeneration of various tissues, including dental, skeletal and nerve tissues (4,5). However, DPCs have not been fully characterized at the molecular level.
DPCs derived from pulp tissue explants by direct outgrowth have been widely used in dental biology. In addition, dental pulp stem cells (DPSCs), stem cells from human exfoliated deciduous teeth and cluster of differentiation 105 (CD105)-positive DPCs were isolated from dental pulp by enzymatic digestion, clonal expansion and/or fluorescence-activated cell sorting (4,6,7). These cells may be more multipotent than DPCs (8). In the present study, DPCs and DPSCs obtained by outgrowth and enzymatic disaggregation were used, respectively, to characterize dental pulp-derived stromal cells.
DPCs and DPSCs were maintained in Dulbecco's modified Eagle's medium (DMEM) or α-MEM. The media were supplemented with 10% serum or 7.5% platelet-rich plasma (PRP), as PRP enhanced the proliferation and calcification of DPCs (9).
DPCs and DPSCs may be similar to mesenchymal stem cells (MSCs), as MSCs also have osteogenic potential and MSC-like cells can be isolated from various tissues such as bone marrow, adipose tissue and synovium. In addition, certain fibroblasts (FBs) have osteogenic potential at low levels (10) and gingival FBs have an MSC-like activity (11), suggesting a similarity among DPCs/DPSCs, MSCs derived from other tissues and FBs. Therefore, it is noteworthy to examine whether certain genes are expressed at high levels only in DPCs/DPSCs among numerous types of MSCs and FBs.
In the present study, four genes were expressed selectively in DPCs and DPSCs but not in various MSCs, FBs, osteoblasts (OBs), chondrocytes (CHs) and adipocytes (ADs). These genes may be useful in characterization of dental pulp stromal cells in basic studies and regenerative medicine.
Materials and methods
Isolation of DPCs by the explant outgrowth method
Healthy teeth were obtained with informed consent from donors (Table I), following protocols approved by the Hiroshima University Ethics Authorities (permit no. D88-2; Hiroshima, Japan). DPCs were grown out of dental pulp tissue explants in the presence of DMEM (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS; Biowest LLC, Miami, FL, USA or GE Healthcare Life Sciences, HyClone Laboratories, Logan, UT, USA) and a 1% antibiotic-antimycotic solution (Invitrogen Life Technologies, Carlsbad, CA, USA) in 95% air (5% CO2) at 37°C (1). Migrating cells were briefly incubated with a 0.05% trypsin-EDTA solution (Sigma-Aldrich) and harvested cells were plated at 4×104 cells/cm2 in 10-cm tissue culture dishes (BD Biosciences, San Jose, CA, USA) and incubated with DMEM supplemented with 10% FBS, 1% antibiotic-antimycotic solution and 1 ng/ml fibroblast growth factor-2 (FGF-2) (medium A). These DPCs were confirmed to induce matrix calcification following exposure to osteogenesis induction medium (data not shown).
Table I.
Information on human cells and tissues used in the present study.
| Donor information | |||
|---|---|---|---|
| Cells/tissues | Age, years | Gender | Origin |
| DPCs-1 | 18 | Male | Permanent molar |
| DPCs-2 | 24 | Female | Permanent molar |
| DPCs-3 | 25 | Female | Permanent molar |
| DPCs-4 | 25 | Female | Permanent molar |
| DPCs-5 | 27 | Female | Permanent molar |
| DPSCs-1 | 18 | Male | Permanent molar |
| DPSCs-2 | 6 | Male | Primary incisor |
| DPSCs-3 | 7 | Female | Primary incisor |
| DPSCs-4 | 6 | Male | Primary incisor |
| DPSCs-5 | 7 | Female | Primary incisor |
| FBs-1 | 29 | Female | Skin |
| FBs-2 | 33 | Female | Skin |
| FBs-3 | 18 | Female | Gum |
| FBs-4 | 41 | Female | Gum |
| FBs-5 | 29 | Female | Skin |
| FBs-6 | 47 | Male | Skin |
| BM-MSCs-1 | 18 | Male | Bone marrow |
| BM-MSCs-2 | 24 | Male | Bone marrow |
| BM-MSCs-3 | 26 | Male | Bone marrow |
| BM-MSCs-4 | 29 | Male | Bone marrow |
| BM-MSCs-5 | 56 | Female | Bone marrow |
| BM-MSCs-6 | 22 | Female | Bone marrow |
| BM-MSCs-7 | 24 | Male | Bone marrow |
| A-MSCs-1 | 32 | Female | Adipose tissue |
| A-MSCs-2 | 37 | Female | Adipose tissue |
| A-MSCs-3 | 41 | Female | Adipose tissue |
| A-MSCs-4 | 42 | Male | Adipose tissue |
| A-MSCs-5 | 62 | Female | Adipose tissue |
| OA-MSCs-1 | 54 | Male | Synovial tissue |
| OA-MSCs-2 | 54 | Female | Synovial tissue |
| OA-MSCs-3 | 61 | Male | Synovial tissue |
| RA-MSCs-1 | 30 | Female | Synovial tissue |
| RA-MSCs-2 | 40 | Female | Synovial tissue |
| RA-MSCs-3 | 72 | Female | Synovial tissue |
| Pulp-1 | 18 | Male | Permanent molar |
| Pulp-2 | 8 | Male | Primary incisor |
| Pulp-3 | 8 | Male | Primary incisor |
| Pulp-4 | 57 | Female | Permanent premolar |
| Gingiva-1 | 18 | Male | Gum |
DPCs, dental pulp cells; FBs, fibroblasts; BM-MSCs, bone marrow-derived mesenchymal stem cells; A-MSCs, adipose tissue-derived MSCs; OA-MSCs, osteoarthritis arthritis synovium-derived MSCs; RA-MSCs, rheumatoid arthritis synovium-derived MSCs.
Isolation of DPSCs by the enzymatic digestion method
DPSCs were isolated at Hiroshima University with informed consent from donors (Table I), using the enzyme digestion method (6,7). Pulp tissues were digested with 3 mg/ml collagenase type 1 (Life Technologies, Carlsbad, CA, USA) and 4 mg/ml dispase (Life Technologies), in the presence of DMEM, in 95% air (5% CO2) at 37°C for 1 h. Dispersed cells were filtered through a 70-µm mesh (BD Biosciences) and seeded at a low density of 1×104 cells/10-cm tissue culture dishes in DMEM supplemented with 20% FBS and a 1% antibiotic-antimycotic solution. Colony-forming cells were incubated briefly with a 0.05% trypsin-EDTA solution (Sigma-Aldrich) and harvested cells were plated at 4×105 cells/cm2 in 10-cm tissue culture dishes and incubated with medium A. These DPSCs also induced calcification following exposure to osteogenesis-induction medium (data not shown). In certain studies, cells were incubated with α-MEM supplemented with 10% FBS or 7.5% PRP. PRP was prepared using a kit from AdiStem Ltd. (Hong Kong, China).
Preparation of FBs and MSCs
Human skin FBs were obtained from Kurabo Industries Ltd. (Osaka, Japan). Human gingival FBs were obtained with informed consent from donors and were isolated as described by Kawahara and Shimazu (12). Bone marrow-derived MSCs (BM-MSCs) were isolated with informed consent from donors (13), or were obtained from Cambrex Bio Science Walkersville Inc. (Walkersville, MD, USA) and PromoCell GmbH (Heidelberg, Germany). Osteoarthritis and rheumatoid arthritis synovium-derived MSCs (OA-MSCs and RA-MSCs) and adipose tissue-derived MSCs (A-MSCs), were obtained from Cell Applications, Inc., (San Diego, CA, USA) and Zen-Bio, Inc., (Research Triangle Park, NC, USA) (Table I).
DNA microarray analysis
Different cell types (donor information in Table I) were cultured by incubating the cells with medium A at specific passage numbers, as follows: DPCs (DPCs-2, −3 and −4), BM-MSCs (BM-MSCs-1, −2 and −3), A-MSCs (A-MSCs-1, −2 and −3), OA-MSCs (OA-MSCs-1, −2 and −3) and RA-MSCs (RA-MSCs-1, −2 and −3) at passages 5–9; and skin FBs (FBs-1 and −2) and gingival FBs (FBs-3) at passages 7–14. Cells were cultured under similar conditions using the same batch of FBS. These cells were expanded with FGF-2 to maintain their multipotent nature throughout several mitotic divisions (14). FGF-2 was removed from the culture medium 72 h before the isolation of RNA to decrease a direct effect of the growth factor on gene expression. The total RNA was isolated, 24 h after the cultures reached confluency, using TRIzol (Life Technologies) and an RNeasy Mini kit (Qiagen, Chatsworth, CA, USA). In addition, total RNA was isolated from OBs, ADs and CHs, which were derived from BM-MSC (BM-MSCs-1, −2 and −3) cultures exposed to appropriate differentiation-inducing media for 28 days (13). DNA microarray analyses were carried out by a Kurabo GeneChip Custom Analysis Service with the Human Genome U133 Plus 2.0 chips (Affymetrix Inc., Santa Clara, CA, USA). Raw data were standardized by the global median normalization method using GeneSpring (Silicon Genetics, Redwood City, CA, USA). Normalization was limited by flag values and the median was calculated using genes that exceeded the present or marginal flag restrictions. The raw data have been deposited at the Gene Expression Omnibus database (accession nos. GSE9451 and GSE66084) (15).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
First-strand cDNA was synthesized from total RNA using ReverTra Ace reverse transcriptase (Toyobo, Osaka, Japan) and oligo(dT) primer. RT-qPCR analyses were carried out using an ABI PRISM® 7900HT Sequence Detection System instrument and software (Applied Biosystems Inc., Foster City, CA, USA), based on the comparative Ct method. The cDNA were amplified using the Universal PCR Master mix (Applied Biosystems Inc.) with a forward and reverse primer (Table II). The PCR cycling conditions included an incubation of 50°C for 2 min, denaturation of 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. TaqMan probes were purchased from Roche Diagnostics (Basel, Switzerland). Data were normalized against 18S rRNA levels.
Table II.
Primers and probes used for the quantitative polymerase chain reaction.
| Gene | Primer sequence (5′→3′) | Roche Universal probe no. |
|---|---|---|
| MSX1 | F: ctcgtcaaagccgagagc | 7 |
| R: cggttcgtcttgtgtttgc | ||
| MSX2 | F: tcggaaaattcagaagatgga | 70 |
| R: caggtggtagggctcatatgtc | ||
| TBX2 | F: ctgacaagcacggcttca | 28 |
| R: gttggctcgcactatgtgg | ||
| ENTPD1 | F: caggaaaaggtgactgagatga | 52 |
| R: cctttactccagcgtaagatgttt |
MSX1, msh homeobox 1; MSX2 msh homeobox 2; TBX2, T-box 2; ENTPD1, ectonucleoside triphosphate diphosphohydrolase 1; F, forward; R, reverse.
Statistical analysis
Data were analyzed by a two-way analysis of variance (ANOVA) and are expressed as mean ± standard deviation. The statistical differences between two groups were evaluated using Bonferroni's test when the ANOVA indicated a significant difference among the groups. In all analyses, P<0.05 was considered to indicate a statistically significant difference.
Results
Enhanced expression of msh homeobox 1 (MSX1), msh homeobox 2 (MSX2), T-box 2 (TBX2) and ectonucleoside triphosphate diphosphohydrolase 1 (ENTPD1) in DPCs
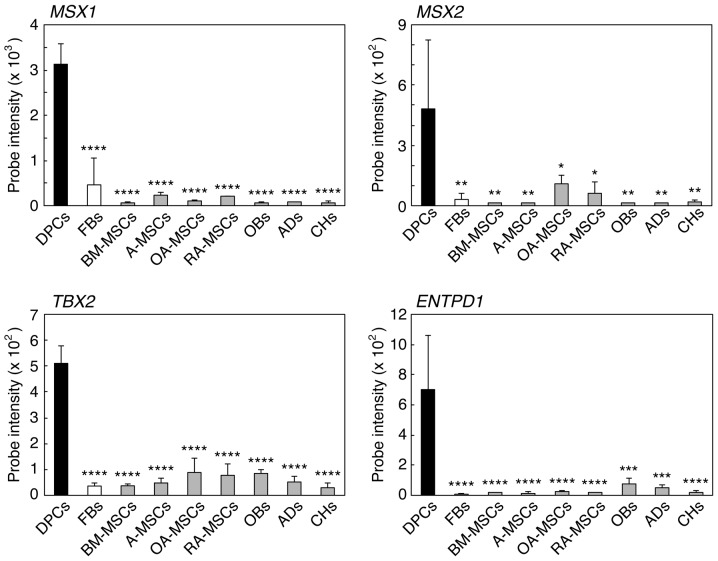
In DNA microarray analyses, MSX1, MSX2, TBX2 and ENTPD1, which are involved in tooth development and calcification (16–19), were expressed at significantly higher levels in DPCs (>4-fold, P<0,05) compared to FBs, BM-MSCs, A-MSCs, OA-MSCs, RA-MSCs, OBs, ADs and CHs (Fig. 1).
Figure 1.
Comparison of the expression levels of the dental pulp cell (DPC)-characteristic genes in various types of cells by DNA microarray analyses. Total RNA was extracted from DPCs-(cultures 2–4), fibroblasts (FBs)-(1–3), bone marrow-derived mesenchymal stem cells (BM-MSCs)-(1–3), adipose tissue-derived MSCs (A-MSCs)-(1–3), osteoarthritis arthritis synovium-derived MSCs (OA-MSCs)-(1–3), rheumatoid arthritis synovium-derived MSCs (RA-MSCs)-(1–3), osteoblasts (OBs), adipocytes (ADs) and chondrocytes (CHs). Gene expression levels of the four genes [msh homeobox 1 (MSX1), msh homeobox 2 (MSX2), T-box 2 (TBX2) and ectonucleoside triphosphate diphosphohydrolase 1 (ENTPD1)] were estimated by measuring the probe intensity in the microarray analyses. The values expressed are mean ± standard deviation for three cultures. *P<0.05, **P<0.01, ***P<0.005, ****P<0.001 vs. DPCs.
RT-qPCR analyses of the expression of the DPC-characteristic genes in DPCs, DPSCs, FBs and BM-MSCs
Subsequently, the mRNA levels of the DPC-characteristic gene candidates were quantitated by qPCR in DPCs, DPSCs, FBs and BM-MSCs (Fig. 2). DPCs were isolated from permanent molar teeth by the explant method and DPSCs were isolated from primary incisor and permanent molar teeth by the enzyme digestion method (Table I). These cells consistently showed the enhanced expression of MSX1, MSX2, TBX2 and ENTPD1 mRNA when compared to FBs and BM-MSCs, irrespective of the different isolation procedures, donor age, donor gender and tooth types. Furthermore, the selective expression of the four genes in DPCs/DPSCs was observed with cells from different donors by DNA microarray and qPCR analyses (Fig. 2).
Figure 2.
Quantitative polymerase chain reaction analyses of the expression of the dental pulp cell (DPC)-characteristic genes in various cells. The DPCs-(cultures 1 and 5), fibroblasts (FBs)-(4–6) and bone marrow-derived mesenchymal stem cells (BM-MSCs)-(4–7) were different from the cells used in the microarray analyses (Table I). The mRNA levels in confluent cultures of these cells, in addition to DPCs-(2–4) and dental pulp stem cells (DPSCs)-(1–3), were compared. The values are mean ± standard deviation for three or four cultures and are expressed as relative levels compared to the averages of the mRNA levels in FBs. *P< 0.05, **P<0.01 vs. DPCs or DPSCs. MSX1, msh homeobox 1; MSX2, msh homeobox 2; TBX2, T-box 2; ENTPD1, ectonucleoside triphosphate diphosphohydrolase 1.
Expression of the DPC-characteristic genes in DPSCs under different culture conditions
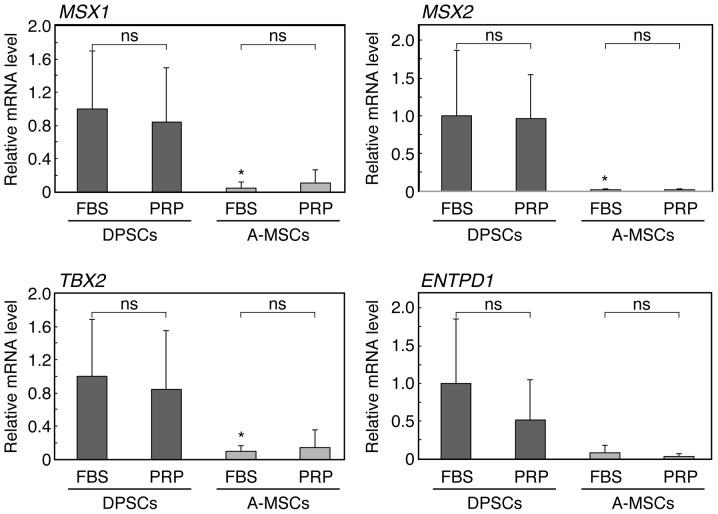
DPSCs were incubated in α-MEM with either 10% FBS or 7.5% PRP, as culture media may affect the expression of the DPC-characteristic genes. In these media, DPSCs showed a higher expression of MSX1, MSX2, TBX2 and ENTPD1 compared to A-MSCs, irrespective of whether FBS or PRP was used (Fig. 3). Therefore, the enhanced expression of these genes in DPSCs could also be observed with α-MEM (Fig. 3) and DMEM (Fig. 2). In the presence of PRP, the average expression levels of these genes were also higher in DPSCs compared to A-MSCs, although the observed differences were not statistically significant.
Figure 3.
Quantitative polymerase chain reaction analyses of the expression of the dental pulp cell (DPC)-characteristic genes in dental pulp stem cells (DPSCs) and adipose tissue-derived mesenchymal stem cells (A-MSCs) maintained under various culture conditions. The DPSCs-(cultures 4 and 5) and A-MSCs-(4 and 5) cells were isolated and expanded with α-modified Eagle's medium (α-MEM) with 10% fetal bovine serum (FBS). These cells were incubated with α-MEM in the presence of 10% FBS or 7.5% platelet-rich plasma (PRP) until cultures became confluent. The values are mean ± standard deviation for 3–11 cultures. *P<0.05 vs. DPSCs; NS, not significant; MSX1, msh homeobox 1; MSX2, msh homeobox 2; TBX2, T-box 2; ENTPD1, ectonucleoside triphosphate diphosphohydrolase 1.
In a pilot study, whether the passage number affects the DPC-characteristic gene expression was evaluated in DPSCs. The MSX1, MSX2, TBX2 and ENTPD1 genes were expressed at high levels in DPSCs, irrespective of the passage number; at least until the 11th passage (data not shown).
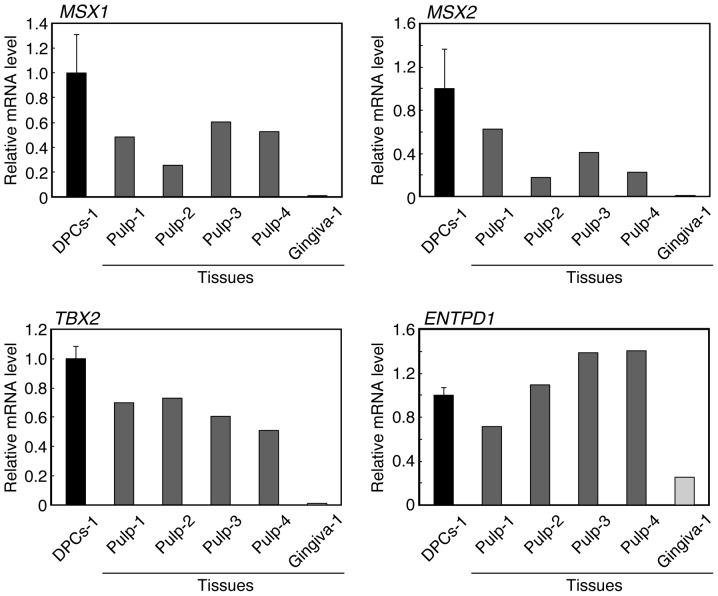
Expression of the DPC-characteristic genes in vivo
Certain DPC-characteristic genes may be expressed in dental pulp tissue in vivo. MSX1, MSX2, TBX2 and ENTPD1 were expressed at comparable levels in cultured DPCs-1 and in the dental pulp specimens from 4 donors (Fig. 4). The expression levels of these genes were comparable between Pulp-1 and DPCs-1, as these were isolated from the same tooth. However, although Pulp-1 and Gingiva-1 were obtained from the same donor (Table I), the expression levels of these genes were higher in Pulp-1 compared to Gingiva-1.
Figure 4.
Quantitative polymerase chain reaction analyses of the expression of the dental pulp cell (DPC)-characteristic genes in dental pulp and gingival tissues. Tissues (Pulp-1, Pulp-2, Pulp-3, Pulp-4 and Gingiva-1) were obtained from several donors (Table I). The pulp cells DPCs-1 were cultured with Dulbecco's modified Eagle's medium in the presence of 10% fetal bovine serum. Of note, DPCs-1, Pulp-1 and Gingiva-1 were obtained from the same donor. The values for tissues are averages of duplicate determinations, and those for DPCs are mean ± standard deviation of four cultures. MSX1, msh homeobox 1; MSX2, msh homeobox 2; TBX2, T-box 2; ENTPD1, ectonucleoside triphosphate diphosphohydrolase 1.
Discussion
Cells with mesenchymal stem characteristics can be obtained from virtually all postnatal organs and tissues (20). Therefore, DPCs may be a subtype of MSCs and could be either a heterogeneous cell population consisting of MSCs and FBs, or unique cells that are different from the others. The enhanced expression of the four genes, MSX1, MSX2, TBX and ENTPD1, in DPCs and DPSCs indicates that dental pulp-derived stromal cells differ from other stromal cells, such as MSCs and FBs, at the molecular level. The uniqueness of DPCs may be associated with dentinogenesis. DPCs show a greater dentinogenic potential compared to BM-MSCs in the transplantation sites of various animals (4,6,21). However, A-MSCs were also reported to induce dentinogenesis following transplantation with bone morphogenetic protein-2 at a site of tooth extraction in rabbits (22).
Certain genes may be artificially upregulated by the in vitro microenvironment depending upon culture conditions. However, the expression levels of MSX1, MSX2, TBX2 and ENTPD1 in DPCs in culture were comparable with those in dental pulp tissue in vivo. Furthermore, the enhanced expression of MSX1, MSX2, TBX2 and ENTPD1 was consistently observed in DPCs, irrespective of donor age, tooth type and culture medium. Therefore, these genes have proven to be useful in the characterization of DPCs.
The expression of MSX1 and MSX2 is essential for the development of teeth, craniofacial structures and/or limb structures in embryos (15,18,23), but their role in postnatal teeth is unknown. In our preliminary studies, knockdown of the protein MSX1, using small interfering RNA, suppressed the mineralization of DPCs (data not shown). TBX2 is involved in the development of tooth, heart and bone (17,24,25) and regulates Sonic hedgehog, bone morphogenetic protein and FGF signaling (26). Previous microarray analyses also showed greater TBX2 expression in DPCs compared to BM-MSCs (27). The ENTPD1/CD39 protein is an adenosine triphosphate (ATP) diphosphohydrolase involved in extracellular ATP metabolism, purinergic signaling, immunological reaction, angiogenesis and matrix calcification (16,28–30). High expression of ENTPD1 in DPCs/DPSCs may act to promote reparative dentin formation by scavenging extracellular ATP, as ATP released from OBs, stromal cells and osteocytes stimulates osteoclast formation and inhibits bone mineralization (31). In future studies, the roles of the four molecules in dental pulp and regenerating tissues should be investigated.
Molecular characterization of transplantable cells is indispensable in cell-based regeneration therapy. Currently, cell surface antigens/CD antigens are used in cell-based therapy, as well as in routine tests for stem cells, including MSCs and DPCs. However, CD antigens may not be specific for a particular cell type. For example, CD105 and STRO-1 have been used as markers for MSCs and DPCs. Therefore, a unique set of DPC-characteristic genes identified in the present study may be useful in pulp cell-based regeneration therapy.
In conclusion, MSX1, MSX2, TBX2 and ENTPD1 proved to be useful in the identification and characterization of DPCs.
Acknowledgements
The present study was supported by Grant-in-Aid for Challenging Exploratory Research (no. 24659876) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to Dr Yukio Kato. The authors would like to thank Dr Eiso Hiyama at the Natural Science Center for Basic Research and Development and Hiroshima University for the use of the equipment.
Glossary
Abbreviations
- DPCs
dental pulp cells
- DPSCs
dental pulp stem cells
- MSCs
mesenchymal stem cells
- FBs
fibroblasts
- BM-MSCs
bone marrow-derived mesenchymal stem cells
- OA-MSCs
osteoarthritis arthritis synovium-derived mesenchymal stem cells
- RA-MSCs
rheumatoid arthritis synovium-derived mesenchymal stem cells
- A-MSCs
adipose tissue-derived mesenchymal stem cells
- MSX1
msh homeobox 1
- MSX2
msh homeobox 2
- TBX2
T-box 2
- ENTPD1
ectonucleoside triphosphate diphosphohydrolase 1
References
- 1.Couble ML, Farges JC, Bleicher F, Perrat-Mabillon B, Boudeulle M, Magloire H. Odontoblast differentiation of human dental pulp cells in explant cultures. Calcif Tissue Int. 2000;66:129–138. doi: 10.1007/PL00005833. [DOI] [PubMed] [Google Scholar]
- 2.Min JH, Ko SY, Cho YB, Ryu CJ, Jang YJ. Dentinogenic potential of human adult dental pulp cells during the extended primary culture. Hum Cell. 2011;24:43–50. doi: 10.1007/s13577-011-0010-7. [DOI] [PubMed] [Google Scholar]
- 3.Spath L, Rotilio V, Alessandrini M, Gambara G, De Angelis L, Mancini M, Mitsiadis TA, Vivarelli E, Naro F, Filippini A, et al. Explant-derived human dental pulp stem cells enhance differentiation and proliferation potentials. J Cell Mol Med. 2010;14:1635–1644. doi: 10.1111/j.1582-4934.2009.00848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iohara K, Imabayashi K, Ishizaka R, Watanabe A, Nabekura J, Ito M, Matsushita K, Nakamura H, Nakashima M. Complete pulp regeneration after pulpectomy by transplantation of CD105+ stem cells with stromal cell-derived factor-1. Tissue Eng Part A. 2011;17:1911–1920. doi: 10.1089/ten.tea.2010.0615. [DOI] [PubMed] [Google Scholar]
- 5.Sakai K, Yamamoto A, Matsubara K, Nakamura S, Naruse M, Yamagata M, Sakamoto K, Tauchi R, Wakao N, Imagama S, et al. Human dental pulp-derived stem cells promote locomotor recovery after complete transection of the rat spinal cord by multiple neuro-regenerative mechanisms. J Clin Invest. 2012;122:80–90. doi: 10.1172/JCI59251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci USA. 2000;97:13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, Shi S. SHED: Stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA. 2003;100:5807–5812. doi: 10.1073/pnas.0937635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeon M, Song JS, Choi BJ, Choi HJ, Shin DM, Jung HS, Kim SO. In vitro and in vivo characteristics of stem cells from human exfoliated deciduous teeth obtained by enzymatic disaggregation and outgrowth. Arch Oral Biol. 2014;59:1013–1023. doi: 10.1016/j.archoralbio.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Lee UL, Jeon SH, Park JY, Choung PH. Effect of platelet-rich plasma on dental stem cells derived from human impacted third molars. Regen Med. 2011;6:67–79. doi: 10.2217/rme.10.96. [DOI] [PubMed] [Google Scholar]
- 10.Sudo K, Kanno M, Miharada K, Ogawa S, Hiroyama T, Saijo K, Nakamura Y. Mesenchymal progenitors able to differentiate into osteogenic, chondrogenic and/or adipogenic cells in vitro are present in most primary fibroblast-like cell populations. Stem Cells. 2007;25:1610–1617. doi: 10.1634/stemcells.2006-0504. [DOI] [PubMed] [Google Scholar]
- 11.Fournier BP, Ferre FC, Couty L, Lataillade JJ, Gourven M, Naveau A, Coulomb B, Lafont A, Gogly B. Multipotent progenitor cells in gingival connective tissue. Tissue Eng Part A. 2010;16:2891–2899. doi: 10.1089/ten.tea.2009.0796. [DOI] [PubMed] [Google Scholar]
- 12.Kawahara K, Shimazu A. Expression and intracellular localization of progesterone receptors in cultured human gingival fibroblasts. J Periodontal Res. 2003;38:242–246. doi: 10.1034/j.1600-0765.2003.00654.x. [DOI] [PubMed] [Google Scholar]
- 13.Kubo H, Shimizu M, Taya Y, et al. Identification of mesenchymal stem cell (MSC)-transcription factors by microarray and knockdown analyses, and signature molecule marked MSC in bone marrow by immunohistochemistry. Genes to cells. 2009;14:407–424. doi: 10.1111/j.1365-2443.2009.01281.x. [DOI] [PubMed] [Google Scholar]
- 14.Tsutsumi S, Shimazu A, Miyazaki K, Pan H, Koike C, Yoshida E, Takagishi K, Kato Y. Retention of multilineage differentiation potential of mesenchymal cells during proliferation in response to FGF. Biochem Biophys Res Commun. 2001;288:413–419. doi: 10.1006/bbrc.2001.5777. [DOI] [PubMed] [Google Scholar]
- 15.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Bei M, Woo I, Satokata I, Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- 17.Demenis MA, Furriel RP, Leone FA. Characterization of an ectonucleoside triphosphate diphosphohydrolase 1 activity in alkaline phosphatase-depleted rat osseous plate membranes: Possible functional involvement in the calcification process. Biochim Biophys Acta. 2003;1646:216–225. doi: 10.1016/S1570-9639(03)00021-9. [DOI] [PubMed] [Google Scholar]
- 18.Saadi I, Das P, Zhao M, Raj L, Ruspita I, Xia Y, Papaioannou VE, Bei M. Msx1 and Tbx2 antagonistically regulate Bmp4 expression during the bud-to-cap stage transition in tooth development. Development. 2013;140:2697–2702. doi: 10.1242/dev.088393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshizawa T, Takizawa F, Iizawa F, Ishibashi O, Kawashima H, Matsuda A, Endo N, Kawashima H. Homeobox protein MSX2 acts as a molecular defense mechanism for preventing ossification in ligament fibroblasts. Mol Cell Biol. 2004;24:3460–3472. doi: 10.1128/MCB.24.8.3460-3472.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.da Silva Meirelles L, Chagastelles PC, Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci. 2006;119:2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 21.Shi S, Robey PG, Gronthos S. Comparison of human dental pulp and bone marrow stromal stem cells by cDNA microarray analysis. Bone. 2001;29:532–539. doi: 10.1016/S8756-3282(01)00612-3. [DOI] [PubMed] [Google Scholar]
- 22.Hung CN, Mar K, Chang HC, Chiang YL, Hu HY, Lai CC, Chu RM, Ma CM. A comparison between adipose tissue and dental pulp as sources of MSCs for tooth regeneration. Biomaterials. 2011;32:6995–7005. doi: 10.1016/j.biomaterials.2011.05.086. [DOI] [PubMed] [Google Scholar]
- 23.Bensoussan-Trigano V, Lallemand Y, Saint Cloment C, Robert B. Msx1 and Msx2 in limb mesenchyme modulate digit number and identity. Dev Dyn. 2011;240:1190–1202. doi: 10.1002/dvdy.22619. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Zhong Q, Wang J, Cameron RS, Borke JL, Isales CM, Bollag RJ. Microarray analysis of Tbx2-directed gene expression: A possible role in osteogenesis. Mol Cell Endocrinol. 2001;177:43–54. doi: 10.1016/S0303-7207(01)00456-7. [DOI] [PubMed] [Google Scholar]
- 25.Verhoeven MC, Haase C, Christoffels VM, Weidinger G, Bakkers J. Wnt signaling regulates atrioventricular canal formation upstream of BMP and Tbx2. Birth Defects Res A Clin Mol Teratol. 2011;91:435–440. doi: 10.1002/bdra.20804. [DOI] [PubMed] [Google Scholar]
- 26.Farin HF, Lüdtke TH, Schmidt MK, Placzko S, Schuster-Gossler K, Petry M, Christoffels VM, Kispert A. Tbx2 terminates shh/fgf signaling in the developing mouse limb bud by direct repression of gremlin1. PLoS Genet. 2013;9:e1003467. doi: 10.1371/journal.pgen.1003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamada Y, Fujimoto A, Ito A, Yoshimi R, Ueda M. Cluster analysis and gene expression profiles: A cDNA microarray system-based comparison between human dental pulp stem cells (hDPSCs) and human mesenchymal stem cells (hMSCs) for tissue engineering cell therapy. Biomaterials. 2006;27:3766–3781. doi: 10.1016/j.biomaterials.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Drosopoulos JH, Broekman MJ, Islam N, Maliszewski CR, Gayle RB, III, Marcus AJ. Site-directed mutagenesis of human endothelial cell ecto-ADPase/soluble CD39: Requirement of glutamate 174 and serine 218 for enzyme activity and inhibition of platelet recruitment. Biochemistry. 2000;39:6936–6943. doi: 10.1021/bi992581e. [DOI] [PubMed] [Google Scholar]
- 29.Pulte ED, Broekman MJ, Olson KE, Drosopoulos JH, Kizer JR, Islam N, Marcus AJ. CD39/NTPDase-1 activity and expression in normal leukocytes. Thromb Res. 2007;121:309–317. doi: 10.1016/j.thromres.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Q, Yan J, Putheti P, et al. Isolated CD39 expression on CD4 T cells denotes both regulatory and memory populations. Am J Transplant. 2009;9:2303–2311. doi: 10.1111/j.1600-6143.2009.02777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orriss IR, Burnstock G, Arnett TR. Purinergic signalling and bone remodelling. Curr Opin Pharmacol. 2010;10:322–330. doi: 10.1016/j.coph.2010.01.003. [DOI] [PubMed] [Google Scholar]