Abstract
The highest risk areas of gastric cancer are currently Japan, Korea and China; Qinghai, a high-altitude area, has one of the highest gastric cancer rates in China. The incidence of gastric cancer is higher in the Tibetan ethnic group compared to that in the Han ethnic group in Qinghai. This study was conducted to determine the clinical characteristics of mitochondrial DNA (mtDNA) mutations and copy numbers among Tibetans with gastric cancer residing at high altitudes and investigate the association between adaptations to hypoxic conditions and oncogenesis. A total of 23 Tibetan gastric cancer patients and 40 matched controls were recruited in this study. Leukocyte mtDNA genes and copy numbers were analyzed. The haplogroups were classified based on mitochondrial gene sequences. A total of 56.5% of the study participants had used alcohol at some point in their lives and 73.9% were positive for Helicobacter pylori (H. pylori). Eight mutations in 8 mitochondrial genes were identified in 43.4% of the Tibetan cancer patient group. There were no significant differences in leukocyte mtDNA copy number levels based on smoking status, alchohol consumption, obesity or H. pylori infection between the control and cancer groups. Statistical differences were also not found between gastric cancer patients with and those without mtDNA mutations. The majority of Tibetan patients with gastric cancer belonged to the mitochondrial haplogroup M9. In conclusion, Tibetans with gastric cancer residing at high altitudes exhibited a wide spectrum of mtDNA mutations. However, leukocyte mtDNA copy numbers in stage II gastric cancer were not statistically different compared to those in healthy Tibetans.
Keywords: gastric cancer, mtDNA copy number, Tibetan, hypoxia
Introduction
Despite a sharp decline in the incidence and mortality rates of gastric cancer, this condition remains the second leading cause of cancer-related mortality worldwide, following lung cancer. Gastric cancer is frequently diagnosed at an advanced stage. The 5-year survival rate is ~20% in developing countries (1). There is a wide variation in the incidence of gastric cancer in different geographical regions. The highest risk areas with an age-standardized incidence rate are Japan, Korea and China (2). The distribution of gastric cancer is not uniform across China. In particular, gastric cancer is the leading type of cancer in Qinghai, a high-altitude area in Northwest China. Moreover, the Chinese National Cancer Center in 2012 reported that Qinghai has one of the highest rates of gastric cancer in China. The incidence of gastric cancer is higher in the Tibetan compared to that in the Han ethnic group in Qinghai, a finding that has attracted significant attention over the last few years.
The marked variation of gastric cancer risk in different geographical areas and the striking differences in potential environmental risk factors between regions prompted the investigation of gastric cancer etiology. Mitochondria are semi-autonomous organelles within cells that play an essential role in cellular energy metabolism, generation of reactive oxygen species (ROS) and apoptosis. The mitochondrial DNA (mtDNA) codes for 37 genes essential to the oxidative phosphorylation system and is present in thousands of copies per cell. In humans, severe mtDNA mutations result in multisystem diseases, whereas certain functional population-specific polymorphisms appear to have enabled humans to adapt to new environments. The copy number and mutations of mtDNA have been found to contribute to the initiation and progression of gastric cancer (3). Hypoxia is one of the most prominent environmental factors at high altitudes. Tibetan individuals inhabiting high-altitude regions exhibit adaptations that enable them to acclimatize to a hypoxic and cold environment; they also exhibit a higher incidence of gastric cancer. The association of gastric cancer with adaptations to a hypoxic and cold environment remains unclear. This study aimed to determine the mtDNA characteristics of Tibetan patients with gastric cancer and investigate the role of mitochondria in gastric cancer under hypoxic conditions.
Materials and methods
Patients
A total of 23 Tibetan patients with gastric cancer were recruited in this study. All the patients were admitted to the Affiliated Hospital of Qinghai University and diagnosed according to standard clinical and pathological criteria within the 30 months preceding the initiation of the study. The patients were diagnosed with stage II gastric cancer according to the approved international standards described by the American Joint Committee on Cancer Cancer Staging Manual (4). A total of 40 Tibetans were randomly selected among healthy volunteers who were free of cancer and donated a blood sample. The control subjects were individually matched to the gastric cancer patients with respect to age, body mass index, educational level and Helicobacter pylori (H. pylori) infection.
This study was approved by the Ethics Committee of the Affiliated Hospital of Qinghai University and conformed to the tenets of the Helsinki Declaration. All the patients provided written informed consent prior to their inclusion in this study.
Mitochondrial gene sequence analysis and haplogroup classification
Mitochondrial gene sequence analysis was performed on all the participants. Total DNA was extracted from blood samples by conventional salting-out protocols. The entire mitochondrial genome was amplified by polymerase chain reaction (PCR) into 8 overlapping fragments using sets of light- and heavy-strand oligonucleotide primers. Each fragment was purified and analyzed by direct sequencing. The sequence results were then compared with the revised Cambridge reference sequence (http://www.mitomap.org), GenBank ID: NC_012920.1. PCR was performed in a 25-µl reaction containing 50 ng DNA, PCR buffer, 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.3 µM of each primer and 1.25 U Taq DNA polymerase. The reaction was performed with an initial denaturation step at 95°C for 5 min, followed by 30 cycles at 95°C for 30 sec, at 55°C for 30 sec and at 72°C for 45 sec, with a final elongation step at 72°C for 4 min.
The haplogroups of mtDNA were determined using phylogenetic analysis based on the mitochondrial phylogenetic tree as updated by van Oven and Kayser (5). The classification details were previously described by Fan and Yao (6).
mtDNA copy number assay
The mtDNA copy number assay was previously described in detail (7). The ratio between the multi-copy mtDNA ND1 gene and the double-copy nuclear gene hemoglobin β was determined using an Applied Biosystems 7900HT Sequence Detection system (Applied Biosystems, Foster City, CA, USA). The mitochondrial ND1 and the nuclear hemoglobin β genes were individually amplified, using the ND1-forward (F)/ND1-reverse (R) and globin-F/globin-R primers, respectively. The primer pair for the mitochondrial ND1 gene was: ND1-F, 5′-ATTCGATGTTGAAGCCTGAGACT-3′; and ND1-R, 5′-TGACCCTTGGCCATAATATGATT-3′. The primer pair for the globin gene was: globin-F, 5′-ACCTCAAGGGCACCTTTGC-3′; and globin-R, 5′-AAAACATCAAGCGTCCCATAGAC-3′.
The PCR reaction mixture (15 µl) contained 0.5 ng DNA, 1X SYBR-Green master mix (Applied Biosystems) and 300 nmol of each primer. The PCR amplification consisted of a single denaturation-enzyme activation step for 8 min at 95°C, followed by 50 amplification cycles of 10 sec at 95°C, 10 sec at 60°C and 6 sec at 72°C. The data were analyzed using LightCycler software, version 3.5.3 (Roche Diagnostics, Mannheim, Germany). The ratio of mtDNA copy number to nuclear DNA was used as a measurement of mtDNA content in each specimen. All the samples were examined in triplicate and the average value was used for the ratio calculations. The R2 for each standard curve was ≥0.99. Standard deviations for the cycle of threshold (Ct) value were accepted at 0.25. The relative mtDNA copy number was calculated by a comparative Ct method, using the following equation: relative copy number (mtDNA/nDNA) = 2ΔCt (ΔCt = Ctβ-globin - CtND1). High reproducibility was observed with this assay, with a coefficient of variance of 8% (8).
Statistical analysis
Statistical analyses were performed by Statistical Product and Service Solutions software, version 15.0 (SPSS, Inc., Chicago, IL, USA). The Student's t-test and the Chi-square test were used to compare the distributions of demographics and clinical characteristics between the gastric cancer and control groups. Wilcoxon's non-parametric and ANOVA tests were used to evaluate the differences among controls in relation to selected characteristics. For samples with normal distribution, the Student's t-test was used; for samples with non-normal distribution, comparisons were performed with Wilcoxon's non-parametric tests.
Results
Patient characteristics
The main clinical characteristics of Tibetan patients with gastric cancer and control subjects are summarized in Table I. There were no significant differences between the two groups in any of the investigated characteristics. The age at diagnosis of gastric cancer was 56.3±9.1 years. Two prominent clinical characteristics were observed. A total of 56.5% of the study participants had used alcohol at some point in their lives and 73.9% were positive for H. pylori.
Table I.
Characteristics of gastric cancer patients and controls in a Tibetan population.
| Characteristics | Cases, no. (%) (n=23) | Controls, no. (%) (n=40) | P-value |
|---|---|---|---|
| Age at diagnosis, years median ± SD | 56.3±9.1 | 55.8±9.2 | 0.8180 |
| BMI, kg/m2 | 0.9280 | ||
| <24 | 6 (26.1) | 12 (30.0) | |
| >24 and <30 | 10 (43.5) | 15 (37.5) | |
| >30 | 7 (30.4) | 13 (32.5) | |
| Smoking | 13 (56.5) | 13 (32.5) | 0.0622 |
| Alcohol | 13 (56.5) | 19 (47.5) | 0.4904 |
| Education | 0.1219 | ||
| Lower than high school | 15 (65.2) | 18 (55.0) | |
| High school or higher | 8 (34.8) | 22 (45.0) | |
| Family history of gastric cancer | 2 (8.7) | 2 (5.0) | 0.7817 |
| H. pylori-positive | 17 (73.9) | 28 (70.0) | 0.7406 |
| Copy no. | 1.277±0.472 | 1.165±0.567 | 0.4218 |
BMI, body mass index; H. pylori, Helicobacter pylori.
mtDNA mutations
Eight mutations in 8 mitochondrial genes were identified in 10 gastric cancer patients (43.4%). Missense mutations were common. Of these 8 mutations, 7 were previously reported in other diseases; the other was a novel mutation, 8686 T>C in the ATP6 gene (Table II).
Table II.
Mitochondrial DNA mutations detected in Tibetan patients with gastric cancer.
| Author | Mutation | Gene | Product | Case no. | (Refs.) |
|---|---|---|---|---|---|
| Munakata et al | 3644 T>C | ND1 | V-A | 1 | (9) |
| Jones et al | 15983 T>C | tRNA-pro | tRNA-pro | 1 | (10) |
| Porcelli et al | 15767 C>G | CYB | Q-E | 2 | (11) |
| Elstner et al | 960 insC | 12S rRNA | 12S rRNA | 1 | (12) |
| Okura et al | 15497 G>A | CYB | G-S | 1 | (13) |
| Present study | 8686 T>C | ATP6 | S-P | 1 | |
| Fauser et al | 11253 T>C | ND4 | I-T | 2 | (14) |
| Petros et al | 7080 T>C | COI | F-L | 1 | (15) |
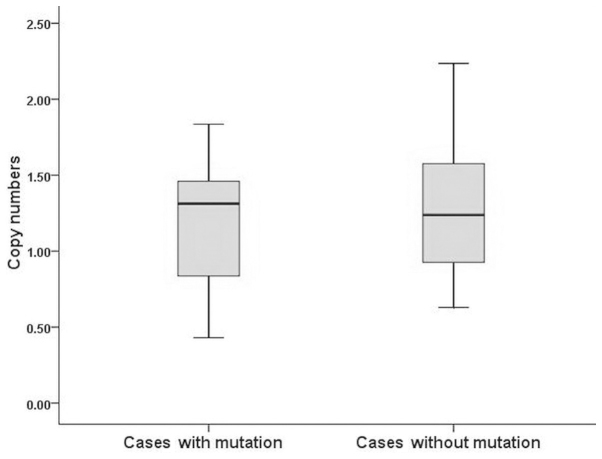
The leukocyte mtDNA copy number was 1.277±0.472 and 1.165±0.567 in Tibetan gastric cancer patients and controls, respectively. The difference between the two groups was not statistically significant (Table I). In addition, there was no significant difference in leukocyte mtDNA copy number levels by smoking status, alchohol consumption, obesity or H. pylori infection between the patient and control groups. The same findings were obtained when comparing gastric cancer patients with to those without mtDNA mutations (P>0.05; Fig. 1). Detailed information is provided in Table III.
Figure 1.
Distribution of mitochondrial DNA copy number between gastric cancer patients with (left) and those without (right) mtDNA mutation. There was no statistically significant difference between the two groups (P>0.05).
Table III.
Means of mtDNA copy numbers by selected variables among Tibetan gastric cancer patients and control subjects.
| Gastric cancer cases (n=23) | Controls (n=40) | |||||
|---|---|---|---|---|---|---|
| Characteristics | Cases | mtDNA copy no. | P-value | Cases | mtDNA copy no. | P-value |
| BMI, kg/m2 | 0.8971 | 0.2521 | ||||
| <24 | 6 | 1.445±0.429 | 12 | 1.135±0.493 | ||
| >24 and <30 | 10 | 1.3139±0.481 | 15 | 1.235±0.742 | ||
| >30 | 7 | 1.083±0.492 | 13 | 1.111±0.408 | ||
| Smoking status | 0.7426 | 0.7667 | ||||
| Never | 10 | 1.297±0.471 | 13 | 1.184±0.618 | ||
| Ever | 13 | 1.221±0.514 | 27 | 1.126±0.460 | ||
| Alchohol consumption | 0.2665 | 0.4026 | ||||
| Never | 10 | 1.404±0.462 | 21 | 1.237±0.635 | ||
| Ever | 13 | 1.180±0.472 | 19 | 1.084±0.484 | ||
| Family history of gastric cancer | 0.1483 | 0.4453 | ||||
| No | 21 | 1.233±0.453 | 38 | 1.181±0.573 | ||
| Yes | 2 | 1.743±0.559 | 2 | 0.862±0.373 | ||
| H. pylori infection | 0.5233 | 0.5257 | ||||
| Positive | 17 | 1.316±0.464 | 28 | 1.203±0.613 | ||
| Negattive | 6 | 1.169±0.520 | 12 | 1.076±0.452 | ||
mtDNA, mitochondrial DNA; BMI, body mass index; H. pylori, Helicobacter pylori.
The mtDNA D-loop sequences were analyzed based on the Revised Cambridge Reference Sequence criteria. The 23 Tibetan gastric cancer patients were classified (GenBank ID: NC_012920.1) in 5 haplogroups as follows: haplogroup M9, 8 patients; M13, 6 patients; C, 4 patients; D, 3 patients; and M7, 2 patients.
Discussion
Tibetans have lived in a significantly cold climate combined with hypoxia for several generations. A higher incidence of gastric cancer among Tibetans was observed in high-altitude areas; therefore, a potential mechanism linking the adaptations enabling Tibetans to survive at high altitudes and gastric cancer was suspected. mtDNA is essential for the biogenesis, maintenance and bioenergetic metabolism of mitochondria. Oxidative phosphorylation damage has been proposed to play an important role in the altered energy metabolism in a number of cancer types. mtDNA is extremely susceptible to mutations due to the high levels of ROS generated in the mitochondrion. Previous studies revealed a number of tumor-specific mtDNA mutations. Wu et al (16) reported that 48% of the gastric cancer cases investigated harbored mtDNA control region tumor-specific mtDNA mutations. Mutations in the 12S rRNA gene and the tRNAPhe gene have been identified in gastric cancer (17). It was previously reported that 51% of bladder, head and neck and lung cancers harbored tumor-specific mtDNA mutations (18). However, little is known regarding Tibetan gastric cancer patients residing in a high-altitude area. In the present study, the mtDNA of gastric cancer was analyzed in Qinghai Tibetans.
Eight point mutations in encoding regions of mtDNA were detected in 43.4% (10/23) of Tibetan patients. Missense mutations were common. The 15983 T>C mutation in the tRNA-pro gene, the 15767 C>G in the CYB gene and the 7080 T>C in the COI gene were previously reported in studies on pancreatic and head and neck cancer (10,11,15). Four additional mutations, namely the 3644 T>C in the ND1 gene, the 960 insC in the 12S rRNA gene, the 15497 G>A in the CYB gene and the 11253 T>C in the ND4 gene, were previously found in obesity, deafness and bipolar disorders (9,19). The 8686 T>C mutation in the ATP6 gene was detected as a novel mutation that changes a polar serine into a hydrophobic proline in a highly conserved region of the protein. The percentage of somatic point mutations in gastric cancer (43.4%) was lower compared to that previously reported (20). This discrepancy may result from differences in race or geographic area.
The somatic mtDNA mutations detected in this study exhibited a wide spectrum, which may provide some information on Tibetan gastric cancer patients. However, a limitation of this study was the small patient sample. In addition, several mtDNA mutations found in Tibetan subjects in this study were also found as sequence variants in a population database (21). The possible reason for this may be that gastric cancer cells are prone to acquire some of the same functional mtDNA mutations as they migrate into different environments. As reported by Brandon et al (21), the adaptive mtDNA mutations may enable tumor cells to thrive in different environments as they metastasize.
Since mtDNA is exclusively maternally inherited and lacks recombination capability, there is sequential accumulation of mtDNA mutations along radiating female lineages. The sequence variants linked to adaptive mutations are also enriched. As a result, mtDNA is used in population genetics. Haplogroup association studies have been used to assess the role of mtDNA variants in various complex diseases. Therefore, the mitochondrial haplogroups of Tibetan gastric cancer patients were analyzed. A total of 23 Tibetans with gastric cancer were classified in 5 different haplogroups, namely M9, M13, C, D and M7. A total of 34.7% (8/23) patients were classified into haplogroup M9. The high frequency of haplogroup M9 in Tibetan patients is in agreement with the findings reported by Gu et al (22). It is well known that the origin and development of races and nationalities are particularly complicated. The findings from the present study may provide information on Tibetan migration patterns and cancer etiology.
The leukocyte mtDNA copy number has been investigated in several types of cancer (7,23,24). Several studies observed that a higher cancer risk is likely accompanied by an increase in the mtDNA copy number (21). The association between a lower mtDNA content and renal cell cancer risk was previously reported (7). The present study demonstrated that leukocyte mtDNA copy numbers were not statistically different between Tibetans with stage II gastric cancer and matched controls, a finding which is in agreement with the findings of a previous retrospective study (8). Considering the lack of association between mtDNA mutations and mtDNA copy number, we performed further copy number comparisons between Tibetan patients harboring mtDNA mutations and those without mutations. There were no significant differences between the two groups, which may be explained as follows: First, the comparisons may have been limited by the small sample. Second, the Tibetan population residing at high altitudes is well adapted to the hypoxic environment; however, the genetic mechanisms of long-term survival in this environmental extreme remain unclear. Better mitochondrial compensatory or adaptive functions may contribute to the indistinctive alteration of mtDNA copy numbers, particularly in early-stage cancer. The mtDNA mutations and copy numbers may be selective influences on Tibetans and are active on cells undergoing neoplastic transformation (21). However, mtDNA mutations possibly occur prior to mitochondrial copy number alterations in Tibetans with stage II gastric cancer. In conclusion, 43.4% of the gastric cancers cases in this study among Tibetans residing at high altitudes carried mtDNA mutations, although the leukocyte mtDNA copy number may not change in early-stage cancer. A larger sample size is required in order to elucidate the mechanisms underlying the development of gastric cancer in Tibetans.
Acknowledgements
This study was supported by a grant from the Qinghai University Science Foundation (no. 2011-QYY-4) and the National Nature Sciense Foundation of China (no. 31160219).
References
- 1.Ahn HS, Yook JH, Park CH, et al. General perioperative management of gastric cancer patients at high-volume centers. Gastric Cancer. 2011;14:178–182. doi: 10.1007/s10120-011-0012-x. [DOI] [PubMed] [Google Scholar]
- 2.Malekzadeh R, Derakhshan MH, Malekzadeh Z. Gastric cancer in Iran: epidemiology and risk factors. Arch Iran Med. 2009;12:576–583. [PubMed] [Google Scholar]
- 3.Hung WY, Wu CW, Yin PH, et al. Somatic mutations in mitochondrial genome and their potential roles in the progression of human gastric cancer. Biochim Biophys Acta. 2010;1800:264–270. doi: 10.1016/j.bbagen.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC Cancer Staging Manual. 7th. Springer; New York, NY: 2010. [Google Scholar]
- 5.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 6.Fan L, Yao YG. An update to MitoTool: using a new scoring system for faster mtDNA haplogroup determination. Mitochondrion. 2013;13:360–363. doi: 10.1016/j.mito.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Xing J, Chen M, Wood CG, et al. Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst. 2008;100:1104–1112. doi: 10.1093/jnci/djn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao LM, Baccarelli A, Shu XO, et al. Mitochondrial DNA copy number and risk of gastric cancer: a report from the Shanghai Women's Health Study. Cancer Epidemiol Biomarkers Prev. 2011;20:1944–1949. doi: 10.1158/1055-9965.EPI-11-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munakata K, Tanaka M, Mori K, Washizuka S, et al. Mitochondrial DNA 3644T→C mutation associated with bipolar disorder. Genomics. 2004;84:1041–1050. doi: 10.1016/j.ygeno.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Jones JB, Song JJ, Hempen PM, Parmigiani G, Hruban RH, Kern SE. Detection of mitochondrial DNA mutations in pancreatic cancer offers a ‘mass’-ive advantage over detection of nuclear DNA mutations. Cancer Res. 2001;61:1299–1304. [PubMed] [Google Scholar]
- 11.Porcelli AM, Ghelli A, Ceccarelli C, et al. The genetic and metabolic signature of oncocytic transformation implicates HIF1alpha destabilization. Hum Mol Genet. 2010;19:1019–1032. doi: 10.1093/hmg/ddp566. [DOI] [PubMed] [Google Scholar]
- 12.Elstner M, Schmidt C, Zingler VC, et al. Mitochondrial 12S rRNA susceptibility mutations in aminoglycoside-associated and idiopathic bilateral vestibulopathy. Biochem Biophys Res Commun. 2008;377:379–383. doi: 10.1016/j.bbrc.2008.09.134. [DOI] [PubMed] [Google Scholar]
- 13.Okura T, Koda M, Ando F, Niino N, Tanaka M, Shimokata H. Association of the mitochondrial DNA 15497G/A polymorphism with obesity in a middle-aged and elderly Japanese population. Hum Genet. 2003;113:432–436. doi: 10.1007/s00439-003-0983-8. [DOI] [PubMed] [Google Scholar]
- 14.Fauser S, Luberichs J, Besch D, Leo-Kottler B. Sequence analysis of the complete mitochondrial genome in patients with Leber's hereditary optic neuropathy lacking the three most common pathogenic DNA mutations. Biochem Biophys Res Commun. 2002;295:342–347. doi: 10.1016/S0006-291X(02)00672-1. [DOI] [PubMed] [Google Scholar]
- 15.Petros JA, Baumann AK, Ruiz-Pesini E, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci USA. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu CW, Yin PH, Hung WY, et al. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosomes Cancer. 2005;44:19–28. doi: 10.1002/gcc.20213. [DOI] [PubMed] [Google Scholar]
- 17.Han CB, Li F, Zhao YJ, et al. Variations of mitochondrial D-loop region plus downstream gene 1 2S rRNA-tRNAPHE and gastric carcinomas. World J Gastroenterol. 2003;9:1925–1929. doi: 10.3748/wjg.v9.i9.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fliss MS, Usadel H, Caballero OL, et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 19.Pereira L, Soares P, Radivojac P, Li B, Samuels DC. Comparing phylogeny and the predicted pathogenicity of protein variations reveals equal purifying selection across the global human mtDNA diversity. Am J Hum Genet. 2011;88:433–439. doi: 10.1016/j.ajhg.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maximo V, Soares P, Seruca R, Rocha AS, Castro P, Sobrinho-Simoes M. Microsatellite instability, mitochondrial DNA large deletions, and mitochondrial DNA mutations in gastric carcinoma. Genes Chromosomes Cancer. 2001;32:136–143. doi: 10.1002/gcc.1175. [DOI] [PubMed] [Google Scholar]
- 21.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 22.Gu M, Dong X, Shi L, et al. Differences in mtDNA whole sequence between Tibetan and Han populations suggesting adaptive selection to high altitude. Gene. 2012;496:37–44. doi: 10.1016/j.gene.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 23.Shen J, Platek M, Mahasneh A, Ambrosone CB, Zhao H. Mitochondrial copy number and risk of breast cancer: a pilot study. Mitochondrion. 2010;10:62–68. doi: 10.1016/j.mito.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thyagarajan B, Wang R, Nelson H, Barcelo H, Koh WP, Yuan JM. Mitochondrial DNA copy number is associated with breast cancer risk. PLoS One. 2013;8:e65968. doi: 10.1371/journal.pone.0065968. [DOI] [PMC free article] [PubMed] [Google Scholar]