Abstract
microRNAs (miRNAs) are involved in the regulation of almost all physiological and pathological processes, including osteoblast proliferation and differentiation. miR-375 has been reported to be aberrantly expressed in various tumor types and to inhibit cell proliferation and invasion by targeting a number of key genes. However, the effects of miR-375 on osteogenic differentiation have not been investigated. Thus, the present study aimed to elucidate the function served by miR-375 in osteogenic differentiation. The expression levels of miR-375 were observed to decrease in a C2C12 cell model of osteogenic differentiation. Overexpression of miR-375 inhibited the activity of key osteoblast markers, including osteocalcin (OC), alkaline phosphatase (ALP) and collagen, type I, α 1 (COL1A1). By contrast, inhibition of miR-375 expression resulted in an increase in the osteogenic potential, as indicated by the enhanced expression levels of OC, ALP and COL1A1. In addition, a dual-luciferase reporter assay indicated that runt-related transcription factor 2 (RUNX2) was a target of miR-375. Western blot analysis revealed that the inhibition of miR-375 led to a significant increase in the protein expression levels of RUNX2. In addition, overexpression of RUNX2 was observed to attenuate the miR-375-mediated suppression of osteogenic differentiation. Therefore, the results demonstrated that miR-375 was able to inhibit osteogenic differentiation via the regulation of RUNX2 expression.
Keywords: osteoblast, osteogenic differentiation, microRNA-375, runt-related transcription factor 2
Introduction
Skeletal development and metabolic homeostasis depend primarily on the activity of osteoblast cells, which are derived from mesenchymal stem cells (1). Three osteoblast-specific transcription factors, activating transcription factor 4, Sp7 (also known as Osterix) and runt-related transcription factor 2 (RUNX2), coregulate the differentiation of osteochondral progenitor cells into fully differentiated osteoblasts (2,3). Furthermore, transcription factors, such as cAMP-response element binding protein, forkhead box protein O1 and members of the activator protein 1 family, also contribute to osteoblast differentiation and function (4,5). Increasing evidence has indicated that osteogenic differentiation is regulated by post-transcriptional mechanisms, most significantly by temporally expressed microRNAs (miRNAs) (6,7).
miRNAs are short (17–25 nucleotides), non-coding RNAs that regulate gene expression at the post-transcriptional level (8). The biological function of the majority of miRNAs are not known; however, they are hypothesized to play critical roles in the regulation of almost all physiological and pathological processes, including cell proliferation, apoptosis, skeletal muscle development and tumorigenesis (9–11). Previous studies have indicated that certain miRNAs are involved in the regulation of osteoblast differentiation (12). For example, miRNA-204/211 targets RUNX2 in bone marrow-derived mesenchymal stem cells and inhibits osteoblastic differentiation (13). An additional study observed that miR-27a was able to regulate osteoblast differentiation by specifically targeting the pro-osteoblastic transcription factor, stabilin 2 (14). The aim of the present study was to characterize the expression of miR-375 and investigate its effects on osteoblast differentiation.
Materials and methods
Reagents, antibodies and plasmids
Recombinant bone morphogenetic protein 2 (BMP2) was purchased from Invitrogen Life Technologies (Carlsbad, CA, USA). Anti-RUNX2 (sc-390351) and anti-GAPDH (sc-365062) mouse monoclonal antibodies were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). miR-375 mimic/inhibitor, non-specific control and RUNX2-pcDNA3.1 were obtained from Santa Cruz Biotechnology, Inc.
Cell culture, differentiation and transfection
A C2C12 cell line was obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in Dulbeccos modified Eagles medium (DMEM; Gibco Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum, 100 µg/ml streptomycin and 100 U/ml penicillin (Sigma-Aldrich, St. Louis, MO, USA). Cultured cells were incubated in a humidity chamber (Thermo Fisher Scientific, Waltham, MA, USA) containing 5% CO2 at 37°C. To induce osteogenic differentiation, cells were treated with 2 nM BMP2 (Invitrogen Life Technologies) for 24 h. For transfection, Lipofectamine 2000 (Invitrogen Life Technologies) was mixed with the aforementioned small interfering (si)RNA or DNA, according to the manufacturers instructions. The solutions were subsequently combined with the C2C12 cells in 24-well culture plates at a density of 3.0×104 cells/well.
Quantitative polymerase chain reaction (qPCR)
Total RNA was isolated using TRIzol reagent (Invitrogen Life Technologies), and cDNA was synthesized from 1 µg total RNA using a Reverse Transcription kit (Invitrogen Life Technologies), according to the manufacturers instructions. qPCR was performed using an ABI 7500 Real-Time PCR System (Applied Biosystems Life Technologies, Foster City, CA, USA), using the following protocol: 95°C for 3 min, 40 cycles of 95°C for 15 sec, 60°C for 15 sec and 72°C for 30 sec. The primers used were as follows: Osteocalcin (OC) forward, 5-TGC TTG TGA CGA GCT ATC AG-3 and reverse, 5-GAG GAC AGG GAG GAT CAA GT-3; collagen, type I, α 1 (COL1A1) forward, 5-GAG AGC ATG ACC GAT GGA TT-3 and reverse, 5-ATG TAG GCC ACG CTG TTC TT-3; alkaline phosphatase (ALP) forward, 5-GAC AAG AAG CCC TTC ACT GC-3 and reverse, 5-AGA CTG CGC CTG GTA GTT GT-3; GAPDH forward, 5-ACC ACA GTC CAT GCC ATC AC-3 and reverse, 5-TCC ACC CTG TTG CTG TA-3. For the quantification of miRNA expression, specific primers for miR-375 and U6 (Applied Biosystems Life Technologies) were used.
ALP staining and measurement
C2C12 cells transfected with the indicated siRNA were fixed with 10% formalin for 20 min, followed by fixation in an ice-cold solution of ethanol and acetone. Subsequent to washing with phosphate-buffered saline (PBS), the cells were stained with an ALP staining solution (Sigma-Aldrich) at 37°C for 20 min. For the measurement of ALP activity, a p-Nitrophenyl Phosphate Liquid Substrate System (Sigma-Aldrich) was added and the absorbance was measured at 405 nm using a microplate reader (BD Biosciences, Franklin Lakes, NJ, USA).
Western blot analysis
Cells were washed twice with PBS and lysed in a buffer containing 50 mM Tris (pH 7.6), 150 mM NaCl, 1 mM EDTA, 10% glycerol and 0.5% NP-40 and protease inhibitor cocktail (Sigma-Aldrich). Protein samples were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. The membranes were incubated with the primary antibodies at 4°C overnight, followed by incubation with a horseradish peroxide-conjugated mouse IgG secondary antibody (sc-2025; Santa Cruz Biotechnology, Inc.). Proteins were visualized using an enhanced chemiluminescence method kit (GE Healthcare Life Sciences, Piscataway, NJ, USA), and the protein band intensity was quantified via densitometric analysis using Quantity One software (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Luciferase Reporter Assay
miRanda (http://www.microrna.org/microrna/getDownloads.do), TargetScan (http://www.targetscan.org) and PicTar database (http://pictar.mdc-berlin.de) software was used in the study to identify miR-375 targets. Then, cells were seeded in 24-well plates at a density of 5×104 cells per well, one day prior to transfection. miRNA-375 mimic or inhibitor (50 pmol), 500 ng luciferase reporter and 40 ng pRL-TK were added to each well. Cells were collected at 48 h after transfection and analyzed using a Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA).
Statistical analysis
Each experiment was performed in triplicate and repeated a minimum of three times, with all the data presented as the mean ± standard deviation. Statistical analyses were performed using SPSS software, version 16.0 (SPSS, Inc., Chicago, IL, USA). Comparisons between groups were conducted using the Students t-test or analysis of variance, where P<0.05 was considered to indicate a statistically significant difference.
Results
miR-375 is downregulated during osteogenic differentiation
Expression levels of miR-375 during osteogenic differentiation were analyzed using qPCR. C2C12 cells were cultured in DMEM without serum and treated with 2 nM BMP2 for 24 h to induce osteogenic differentiation. A number of osteogenic factors, including OC, ALP and COL1A1, were used as phenotypic markers of osteogenic differentiation. The results from the qPCR revealed that the mRNA expression levels of OC, ALP and COL1A1 were markedly increased in the cells treated with BMP2, indicating that osteogenic differentiation had been successfully induced (Fig. 1A–C). In addition, a significant reduction in miR-375 expression was observed in the BMP2-treated cells, as compared with the non-BMP2-induced cells during osteogenic differentiation (Fig. 1D). Collectively, these results demonstrated that miR-375 was involved in the progression of osteogenic differentiation.
Figure 1.
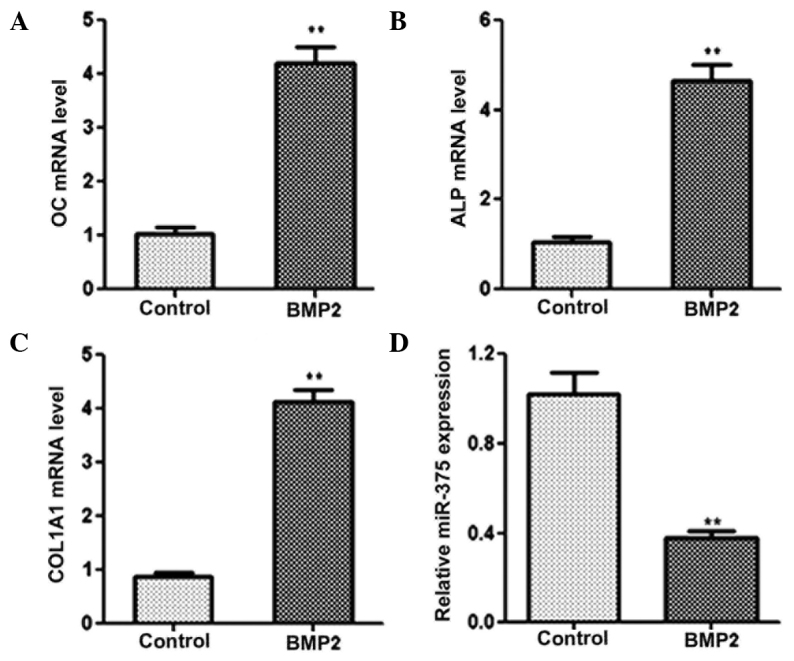
miR-375 expression is downregulated during osteogenic differentiation. Quantitative polymerase chain reaction was performed to determine the expression levels of (A) OC, (B) ALP, (C) COL1A1 and (D) miR-375 in control and BMP2-induced C2C12 cells. GAPDH was used as a control for the mRNA expression levels, while U6 was used as an internal control for miRNA expression. **P<0.01, vs. control group. BMP2, bone morphogenetic protein 2; OC, osteocalcin; ALP, alkaline phosphatase; COL1A1, collagen, type I, α 1; miRNA, microRNA.
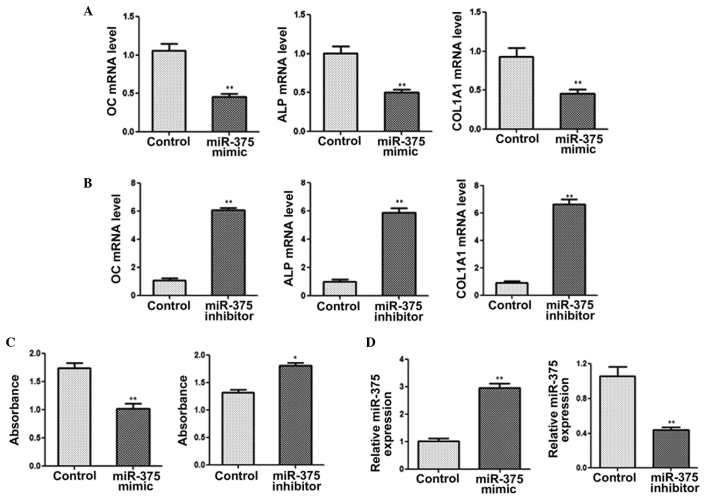
miR-375 inhibits osteogenic differentiation
C2C12 cells were transfected with miR-375 mimic or inhibitor in order to study the physiological role of miR-375 on osteogenic differentiation. The qPCR results revealed the mRNA expression levels of OC, ALP and COL1A1 to be markedly reduced in the cells transfected with the miR-375 mimic for 48 h (Fig. 2A). By contrast, C2C12 cells transfected with the miR-375 inhibitor exhibited increased mRNA expression levels of OC, ALP and COL1A1 (Fig. 2B). Furthermore, cells that overexpressed miR-375 exhibited a significant reduction in ALP activity, while the inhibition of miR-375 expression resulted in a notable increase in ALP activity (Fig. 2C). qPCR was also performed to determine the efficiency of transfection with miR-375 mimic/inhibitor (Fig. 2D). Collectively, the results indicated that miR-375 was able to inhibit osteogenic differentiation.
Figure 2.
miR-375 inhibits osteogenic differentiation. Quantitative polymerase chain reaction (qPCR) was performed to determine the mRNA expression levels of OC, ALP and COL1A1 in C2C12 cells transfected with (A) miR-375 mimic or (B) miR-375 inhibitor. (C) ALP activity was measured following transfection with miR-375 mimic or inhibitor. (D) Expression levels of miR-375 were determined using qPCR in C2C12 cells transfected with miR-145 mimic, miR-145 inhibitor or with no treatment (control) for 48 h. GAPDH was used as a control for the mRNA expression levels, while U6 was used as an internal control for miRNA expression. *P<0.05 and **P<0.01, vs. control group. OC, osteocalcin; ALP, alkaline phosphatase; COL1A1, collagen, type I, α 1; miRNA, microRNA.
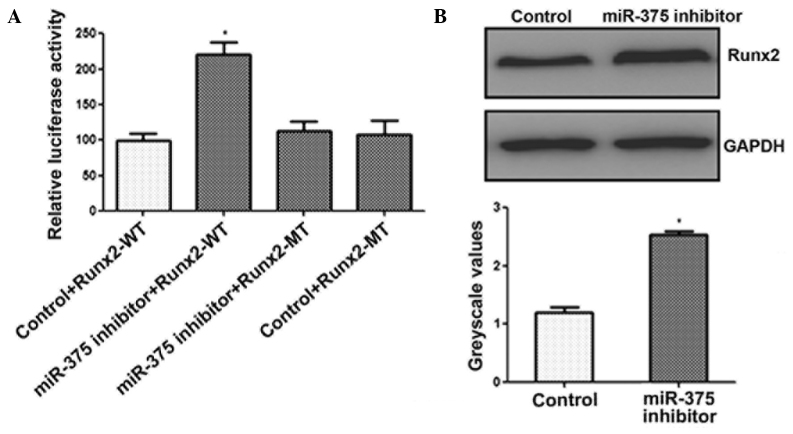
miR-375 reduces the expression levels of RUNX2
To elucidate the molecular mechanism by which miR-375 suppressed osteogenic differentiation, miRanda, TargetScan and PicTar databases were used to search for putative miR-375 targets. The search identified a 3′ untranslated region of RUNX2 that contained the conserved putative miR-375 binding site. A dual-luciferase activity assay was performed to confirm that RUNX2 was a target of miR-375. As shown in Fig. 3A, luciferase activity was observed to increase significantly in the C2C12 cells following transfection with the miR-375 inhibitor, whereas no increase was observed in the activity of the mutant luciferase (Fig. 3A). In addition, the results of the western blot analysis demonstrated that inhibition of miR-375 expression in C2C12 cells significantly enhanced the protein expression levels of RUNX2 (Fig. 3B). These results demonstrated that miR-375 downregulated the expression of RUNX2.
Figure 3.
miR-375 reduces the expression levels of RUNX2. (A) C2C12 cells were cotransfected with the miR-375 inhibitor and a reporter construct containing the WT or MT 3′ untranslated region of RUNX2 for 48 h. A luciferase activity assay was subsequently performed. (B) C2C12 cells were transfected with the miR-375 inhibitor for 48 h, and western blot analysis was used to quantify the protein expression levels of RUNX2. *P<0.05, vs. control group. RUNX2, runt-related transcription factor 2; WT, wild type, MT, mutant; miR, microRNA.
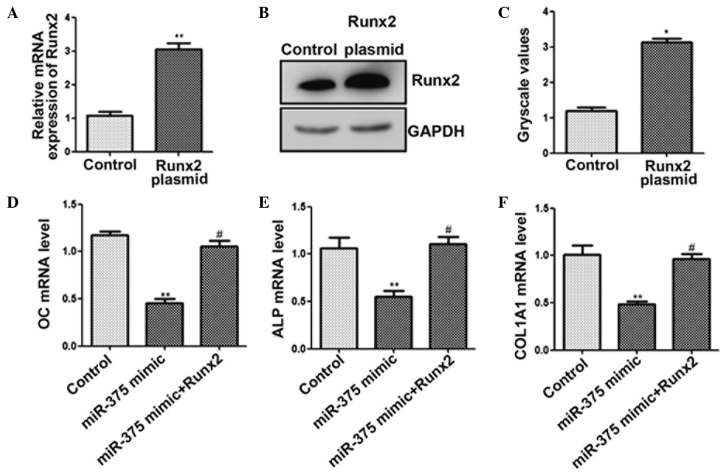
miR-375 inhibits osteogenic differentiation by targeting RUNX2
Finally, whether overexpression of RUNX2 attenuated the inhibitory effect of miR-375 on osteogenic differentiation was investigated. The results of the qPCR (Fig. 4A) and western blot analysis (Fig. 4B and C) indicated that RUNX2 expression was upregulated. Furthermore, cotransfection of miR-375 mimic with a RUNX2 overexpression plasmid significantly increased the mRNA expression levels of OC, ALP and COL1A1 (Fig. 4D–F). Thus, overexpression of RUNX2 appeared to attenuate the miR-375-mediated suppression of osteogenic differentiation. These results indicated that miR-375 inhibited osteogenic differentiation, in part via the downregulation of RUNX2 expression.
Figure 4.
miR-375 inhibits osteogenic differentiation by targeting RUNX2. C2C12 cells were transfected with a RUNX2 plasmid for 48 h. (A) Quantitative polymerase chain reaction (qPCR) and (B and C) western blot analysis was used to determine the expression levels of RUNX2. C2C12 cells were cotransfected with miR-375 mimic and RUNX2-pcDNA3.1 for 48 h. The mRNA expression levels of (D) OC, (E) ALP and (F) COL1A1 (F) were determined using qPCR. *P<0.05 and **P<0.01, vs. control group; #P<0.05, vs. miR-375 mimic transfection group. RUNX2, runt-related transcription factor 2; OC, osteocalcin; ALP, alkaline phosphatase; COL1A1, collagen, type I, α 1; miR, microRNA.
Discussion
miRNAs have been reported to function as regulators in skeletal muscle development, myeloblast differentiation and osteoblastogenesis (15). The results of the present study indicated that miR-375 functions as an inhibitor of osteogenic differentiation by modulating RUNX2 expression.
The C2C12 cell line is a typical pluripotent mesenchymal precursor cell line that possesses the potential to differentiate into myoblasts, chondroblasts and osteoblasts, which are commonly used in cellular models of osteogenic differentiation (16,17). The results of the present study indicated that the expression of miR-375 was downregulated in the C2C12 cells during osteogenic differentiation.
Furthermore, OC, ALP and COL1A1 are typical osteoblast differentiation markers (18). In the present study, C2C12 cells were used as a model of osteoblast differentiation and the successful establishment of the model was indicated by a significant elevation in the expression levels of OC, ALP and COL1A1. Overexpression of miR-375 was shown to suppress osteogenic differentiation, as indicated by reductions in the mRNA expression levels of OC, ALP and COL1A1. By contrast, the inhibition of miR-375 expression resulted in an increase in the expression levels of these markers. In addition, ALP activity was reduced in miR-375 mimic-transfected cells; however, this effect was reversed by the inhibition of miR-375 expression. Collectively, these results suggested that miR-375 was able to suppress osteogenic differentiation.
Previous studies have reported aberrant expression of miR-375 in a number of tumor types, in addition to the inhibition of cell proliferation and invasion by miR-375 through targeting numerous key genes (19,20). Furthermore, a number of additional studies have indicated that miR-375 is involved in the regulation of insulin secretion, the maintenance of blood homeostasis and the inhibition of neurite differentiation (21,22). The results of the present study revealed that miR-375 was capable of suppressing the osteogenic differentiation of C2C12 cells. Further investigation into the mechanism underlying this suppressive effect, by searching for target genes using bioinformatic analysis, identified RUNX2 as a potential target of miR-375.
RUNX2 is a member of the RUNX family of transcription factors and has a runt DNA-binding domain. RUNX2 has been identified as a key transcription factor in the regulation of osteogenesis and chondrogenesis (23). miRNAs, as small molecular regulators of gene expression, have critical roles in osteoblast function by regulating vital proteins involved in differentiation (24). For example, miR-204 and miR-211 are known to function as crucial endogenous negative regulators of RUNX2, promoting adipogenesis and inhibiting the osteogenesis of mesenchymal progenitor cells (13). Furthermore, the results of a previous study indicated that miR-3960 and miR-2861 affected osteoblast differentiation via a novel RUNX2/miR-3960/miR-2861 regulatory feedback loop (25). In the present study, miR-375 overexpression was demonstrated to result in the suppression of RUNX2 protein expression, indicating that RUNX2 is regulated by miR-375. A dual-luciferase reporter assay further identified RUNX2 as a direct target of miR-375. In addition, cotransfection of miR-375 mimic with a RUNX2 overexpression plasmid was demonstrated to attenuate the miR-375-mediated inhibition of osteogenic differentiation. Therefore, these results indicated that miR-375 negatively targets RUNX2 and subsequently inhibits osteogenic differentiation.
In conclusion, the results of the present study indicated that miR-375 inhibits osteoblast differentiation via the regulation of RUNX2 expression. These observations further the understanding into the biological roles played by miRNAs in osteoblast differentiation. These observations further understanding of the biological functions miRNAs in osteoblast differentiation, and may provide a novel therapeutic target for treatment of skeletal dysfunction.
References
- 1.Sabbieti MG, Agas D, Marchetti L, et al. BMP-2 differentially modulates FGF-2 isoform effects in osteoblasts from newborn transgenic mice. Endocrinology. 2013;154:2723–2733. doi: 10.1210/en.2013-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu S, Zhu K, Lai Y, et al. atf4 promotes β-catenin expression and osteoblastic differentiation of bone marrow mesenchymal stem cells. Int J Biol Sci. 2013;9:256–266. doi: 10.7150/ijbs.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeong HM, Choi YH, Jeong HG, et al. Bromopropane compounds inhibit osteogenesis by ERK-dependent Runx2 inhibition in C2C12 cells. Arch Pharm Res. 2014;37:276–283. doi: 10.1007/s12272-013-0178-3. [DOI] [PubMed] [Google Scholar]
- 4.Zhang R, Edwards JR, Ko SY, et al. Transcriptional regulation of BMP2 expression by the PTH-CREB signaling pathway in osteoblasts. PLoS One. 2011;6:e20780. doi: 10.1371/journal.pone.0020780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2011;13:27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- 6.Li Z, Hassan MQ, Volinia S, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci USA. 2008;105:13906–13911. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, Hassan MQ, Jafferji M, et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 9.Roy S, Banerjee J, Gnyawali SC, et al. Suppression of induced microRNA-15b prevents rapid loss of cardiac function in a Dicer depleted model of cardiac dysfunction. PLoS One. 2013;8:e66789. doi: 10.1371/journal.pone.0066789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rissland OS, Hong SJ, Bartel DP. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol Cell. 2011;43:993–1004. doi: 10.1016/j.molcel.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasheed SA, Teo CR, Beillard EJ, et al. MicroRNA-182 and microRNA-200a control G-protein subunit α-13 (GNA13) expression and cell invasion synergistically in prostate cancer cells. J Biol Chem. 2013;288:7986–7995. doi: 10.1074/jbc.M112.437749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei J, Shi Y, Zheng L, et al. miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J Cell Biol. 2012;197:509–521. doi: 10.1083/jcb.201201057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo D, Li Q, Lv Q, et al. MiR-27a targets sFRP1 in hFOB cells to regulate proliferation, apoptosis and differentiation. PLoS One. 2014;9:e91354. doi: 10.1371/journal.pone.0091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamouille S, Subramanyam D, Blelloch R, Derynck R. Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr Opin Cell Biol. 2013;25:200–207. doi: 10.1016/j.ceb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katagiri T, Yamaguchi A, Komaki M, et al. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol. 1994;127:1755–1766. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin CS, Lecanda F, Sheikh S, et al. Relative abundance of different cadherins defines differentiation of mesenchymal precursors into osteogenic, myogenic, or adipogenic pathways. J Cell Biochem. 2000;78:566–577. doi: 10.1002/1097-4644(20000915)78:4<566::AID-JCB6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 18.Gámez B, Rodríguez-Carballo E, Bartrons R, et al. MicroRNA-322 (miR-322) and its target protein Tob2 modulate Osterix (Osx) mRNA stability. J Biol Chem. 2013;288:14264–14275. doi: 10.1074/jbc.M112.432104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding L, Xu Y, Zhang W, et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 20.Wang F, Li Y, Zhou J, et al. miR-375 is down-regulated in squamous cervical cancer and inhibits cell migration and invasion via targeting transcription factor SP1. Am J Pathol. 2011;179:2580–2588. doi: 10.1016/j.ajpath.2011.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdelmohsen K, Hutchison ER, Lee EK, et al. miR-375 inhibits differentiation of neurites by lowering HuD levels. Mol Cell Biol. 2010;30:4197–4210. doi: 10.1128/MCB.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krützfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006;4:9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Takarada T, Hinoi E, Nakazato R, et al. An analysis of skeletal development in osteoblast-specific and chondrocyte-specific runt-related transcription factor-2 (Runx2) knockout mice. J Bone Miner Res. 2013;28:2064–2069. doi: 10.1002/jbmr.1945. [DOI] [PubMed] [Google Scholar]
- 24.Inose H, Ochi H, Kimura A, et al. A microRNA regulatory mechanism of osteoblast differentiation. Proc Natl Acad Sci USA. 2009;106:20794–20799. doi: 10.1073/pnas.0909311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu R, Liu W, Li H, et al. A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation. J Biol Chem. 2011;286:12328–12339. doi: 10.1074/jbc.M110.176099. [DOI] [PMC free article] [PubMed] [Google Scholar]