Abstract
Background
Species of Bryopsidales form ecologically important components of seaweed communities worldwide. These siphonous macroalgae are composed of a single giant tubular cell containing millions of nuclei and chloroplasts, and harbor diverse bacterial communities. Little is known about the diversity of chloroplast genomes (cpDNAs) in this group, and about the possible consequences of intracellular bacteria on genome composition of the host. We present the complete cpDNAs of Bryopsis plumosa and Tydemania expeditiones, as well as a re-annotated cpDNA of B. hypnoides, which was shown to contain a higher number of genes than originally published. Chloroplast genomic data were also used to evaluate phylogenetic hypotheses in the Chlorophyta, such as monophyly of the Ulvophyceae (the class in which the order Bryopsidales is currently classified).
Results
Both DNAs are circular and lack a large inverted repeat. The cpDNA of B. plumosa is 106,859 bp long and contains 115 unique genes. A 13 kb region was identified with several freestanding open reading frames (ORFs) of putative bacterial origin, including a large ORF (>8 kb) closely related to bacterial rhs-family genes. The cpDNA of T. expeditiones is 105,200 bp long and contains 125 unique genes. As in B. plumosa, several regions were identified with ORFs of possible bacterial origin, including genes involved in mobile functions (transposases, integrases, phage/plasmid DNA primases), and ORFs showing close similarity with bacterial DNA methyltransferases. The cpDNA of B. hypnoides differs from that of B. plumosa mainly in the presence of long intergenic spacers, and a large tRNA region. Chloroplast phylogenomic analyses were largely inconclusive with respect to monophyly of the Ulvophyceae, and the relationship of the Bryopsidales within the Chlorophyta.
Conclusions
The cpDNAs of B. plumosa and T. expeditiones are amongst the smallest and most gene dense chloroplast genomes in the core Chlorophyta. The presence of bacterial genes, including genes typically found in mobile elements, suggest that these have been acquired through horizontal gene transfer, which may have been facilitated by the occurrence of obligate intracellular bacteria in these siphonous algae.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1418-3) contains supplementary material, which is available to authorized users.
Keywords: Horizontal gene transfer, Introns, Mobile elements, Phylogenomics, Plastid genome, Viridiplantae
Background
The circa 70 chloroplast genomes (cpDNAs) of green algae sequenced to date have revealed a remarkable array of genomic architectures, including a wide range of genome sizes, gene repertoires and arrangements, and nucleotide compositions [1-3]. Many green algal cpDNAs have a quadripartite structure also found in land plants, characterized by the presence of two copies of a large inverted repeat sequence separating a small and a large single-copy region. Although this architecture is believed to be ancestral in the green algae, many species (including several Chlorophyta) do not have a quadripartite structure [4-10].
Complete cpDNA sequences are only available for three species of Ulvophyceae (Figure 1), including the marine flagellate Oltmannsiellopsis viridis (Oltmannsiellopsidales), the freshwater microfilamentous Pseudendoclonium akinetum (Ulotrichales), and the marine siphonous species Bryopsis hypnoides (Bryopsidales) [11-13]. The cpDNAs of Oltmannsiellopsis (151.9 kb) and Pseudendoclonium (195.8 kb) share the quadripartite architecture, and a similar large complement of genes (104 and 105 genes, respectively), but differ in size due to different intron numbers (five in Oltmannsiellopsis vs 27 in Pseudendoclonium). The 153.4 kb cpDNA of B. hypnoides differs in several aspects from the two other species of Ulvophyceae. It does not feature a quadripartite architecture, and it includes 10 tRNA genes that are not present in any other green algal cpDNA. Another peculiarity of the B. hypnoides cpDNA is the presence of multimeric forms of the cpDNA, including monomers, dimers, trimers, tetramers, and higher-order multimers, which were detected by pulsed-field gel electrophoresis and Southern blot methods [13]. 111 genes were reported in the cpDNA of B. hypnoides but our preliminary analysis indicated a higher number of genes, as also suggested by others [1]. Apart from completely sequenced cpDNAs, partial chloroplast genome data in the Bryopsidales has long been available for Codium fragile [14] and Caulerpa sertularoides [15] through Southern hybridization analysis of restriction fragments. In addition partial DNA sequence data is available for Caulerpa filiformis [16].
Figure 1.
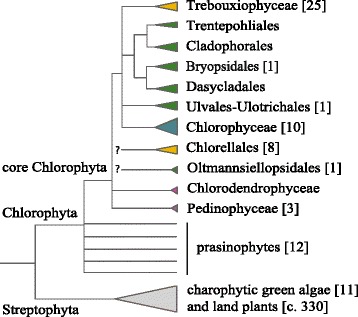
Phylogenetic relationships among the main clades of green algae, focusing on the core Chlorophyta. The tree is a composite of accepted relationships based on molecular phylogenetic evidence from different studies [17-19]. Uncertain or conflicting relationships are indicated by polytomies or question marks. Numbers in square brackets indicate the number of (nearly) complete cpDNAs available to date. Clades currently classified as Ulvophyceae are in green, and Trebouxiophyceae are in yellow.
The relationships within the core Chlorophyta have recently been evaluated based on multigene datasets [17-19], which is relevant in the light of comparative genomics. Although monophyly of the Ulvophyceae was supported in a 10-gene phylogeny (eight nuclear and two chloroplast encoded genes) [17], chloroplast (cp) multigene phylogenetic analyses generally failed to recover the Ulvophyceae as a clade. For example, phylogenetic analysis of 23 cp genes recovered Caulerpa (Bryopsidales) as more closely related to Chlorella (Trebouxiophyceae) than to the other two ulvophycean taxa in the phylogeny (Oltmannsiellopsis and Pseudendoclonium) [16], and a phylogeny inferred from 42 cp genes indicated a relationship between Bryopsis and Chlorophyceae [13]. Phylogenomic analyses with increased taxon sampling (53 taxa, 7 cp genes + 18S; and 38 taxa, 53 cp genes) suggested a relationship between Oltmannsiellopsis and Tetraselmis (Chlorodendrophyceae), and like in the two previous studies did not provide support for a monophyletic Ulvophyceae [18]. In addition, cp multigene phylogenies have rejected monophyly of the Trebouxiophyceae, which was shown to fall apart in at least two main clades [18,19]. In general, relationships among the major lineages of core Chlorophyta remain poorly supported. As illustrated in Figure 1, complete data on chloroplast genomes is scarce in the Ulvophyceae compared to other green algal lineages, with data lacking for several important clades, including the Cladophorales, Dasycladales, and Trentepohliales.
This paper focusses on the siphonous marine green algae Bryopsis and Tydemania, two members of the order Bryopsidales [20]. Species in this order form thalli that are composed of a single, giant cell, containing millions of nuclei, chloroplasts and mitochondria that move about freely by cytoplasmic streaming. Bryopsis includes more than 20 species, all characterized by morphologically simple feather-like thalli, mostly only a few cm tall. The genus is distributed worldwide in tropical to temperate seas. Tydemania forms more complex and larger thalli composed of whorls of siphons arising from a branched main axis. The genus includes a single species, T. expeditiones, that is found in the marine tropical Indo-West Pacific. The order Bryopsidales includes two main clades, Bryopsidineae and Halimedineae. Bryopsis is a member of the Bryopsidineae, which also includes Derbesia and Codium, and some other, less known genera. Tydemania is a member of the Halimedineae, which also includes some of the better known siphonous green seaweeds such as Caulerpa and Halimeda. The placement of the Bryopsidales in the class Ulvophyceae has been implied by 18S data and nuclear multi-gene phylogenetic analysis [17,21], but as mentioned above, analyses based on cp genes have casted doubt on the affinity of siphonous green algae with other clades of Ulvophyceae [13,16,18].
In this paper we report on the complete cpDNAs of Bryopsis plumosa and Tydemania expeditiones, and a re-annotated cpDNA of B. hypnoides. We compared these genomes with previously published cpDNAs in the core Chlorophyta to gain a better understanding of the evolution of chloroplast genomes in this group, and to assess relationships among the main clades of core Chlorophyta.
Results and discussion
cpDNA assembly, size and organization
For both Bryopsis plumosa and Tydemania expeditiones, assemblies yielded a single cpDNA sequence, which could be closed into a circle by an overlap of more than 300 base pairs. Both assemblies showed high overall read coverage, and as a result did not contain any ambiguous regions (Additional file 1). For B. plumosa, 467,353 reads of a total of 7.2 million mapped to the cpDNA with a mean coverage of 430× (min. 4×, max. 871×). The 4× coverage was 3 bp long and situated in the 3’-end of the cysT gene, and was spanned by paired-end reads. For T. expeditiones, 7.1 million reads of a total of 14.7 million mapped to the cpDNA with a mean coverage of 6,667× (min. 749×, max. 12,274×).
The circular cpDNAs of B. plumosa (Figure 2) and T. expeditiones (Figure 3) consist of 106,859 bp and 105,200 bp, respectively. This is smaller than most published cpDNAs of free-living species of core Chlorophyta, but similar to those found in Pedinomonas minor (98 kb), Marvania geminata (108 kb), Pseudochloris wilhelmii (110 kb), and Planctonema lauterbornii (114 kb) [9,19,22]. Most prasinophytes have even smaller cpDNAs, ranging between 64-86 kb, with the exception of Nephroselmis, which has larger chloroplast genomes (125 and 201 kb) [3]. Unexpectedly, the cpDNA of B. plumosa is much smaller than the 153,429 bp cpDNA of B. hypnoides. This size difference is mainly a result of large intergenic spacers (up to 20,875 bp in length), totaling 59,842 bp or 38.9% of the cpDNA in B. hypnoides (Additional file 2). Intergenic spacers in B. plumosa and T. expeditiones account for only 20.3% and 16.3% of the total cpDNA. Nevertheless, some long intergenic spacer regions are present in both genomes. In B. plumosa, the largest intergenic spacer was 3,307 bp long. This region contains a few ORFs (>300 bp), none of which showed significant homology to known proteins (blastp E-values > 0.1). In T. expeditiones, a large spacer region (6,583 bp) contains ten ORFs (>300 bp), none with significant homology to known proteins (blastp E-values > 1). GC content of the two genomes falls within the limits of other ulvophycean and green algal cpDNAs (Table 1) [1,2].
Figure 2.

Gene map of the chloroplast genome of Bryopsis plumosa . The 106,859 bp genome contains 115 unique genes, including three ribosomal RNA genes, 26 transfer RNA genes, and 86 protein coding genes. Genes shown on the outside of the circle are transcribed counterclockwise. Annotated genes are colored according to the functional categories shown in the legend bottom left. The red arcs indicate gene regions of putative bacterial origin.
Figure 3.
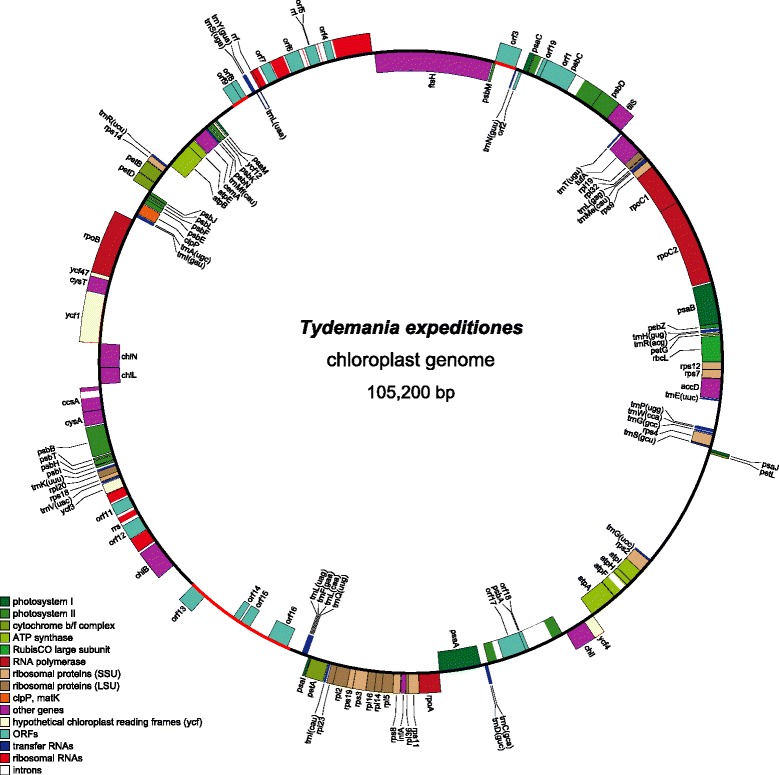
Gene map of the chloroplast genome of Tydemania expeditiones . The 105,200 bp genome contains 125 unique genes, including three ribosomal RNA genes, 28 transfer RNA genes, and 94 protein coding genes. Genes shown on the outside of the circle are transcribed counterclockwise. Annotated genes are colored according to the functional categories shown in the legend bottom left. The red arcs indicate gene regions of putative bacterial origin.
Table 1.
Summary of the Bryopsis plumosa and Tydemania expeditiones cpDNAs and comparison with other ulvophycean cpDNAs
| Species | cpDNA size (kb) | GC content (%) | Gene number (unique genes) | Protein coding genes/ORFs d | rRNA genes | tRNA genes | % Coding b | Intergenic space (%) | Intron (%) c | Intron (%) b | Intron number | Quadripartite structure | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B. plumosa | 106.9 | 30.8 | 115 | 79/7 | 3 | 26 | 74.4 | 20.1 | 5.5 | 8.3 | 13 | - | This study |
| T. expeditiones | 105.2 | 32.8 | 125 | 77/17 | 3 | 28 | 77.5 | 16.3 | 6.3 | 11.2 | 11 | - | This study |
| B. hypnoides a | 153.4 | 33.1 | 131 | 78/11 | 3 | 39 | 55.9 | 38.9 | 5.2 | 7.1 | 11 | - | [13]a |
| P. akinetum | 195.8 | 31.5 | 135 | 74/29 | 3 | 29 | 53.9 | 37.5 | 8.5 | 12.3 | 28 | + | [12] |
| O. viridis | 151.9 | 40.5 | 112 | 75/8 | 3 | 26 | 57.9 | 39.5 | 2.5 | 6.8 | 10 | + | [11] |
aRe-annotated in this study (see Additional file 2).
bIncluding intronic open reading frames (ORFs).
cExcluding intronic ORFs.
dOnly those ORFs are included, which were found to have a significant blastp result (E value < 1e-04).
The B. plumosa and T. expeditiones cpDNAs both lack a large inverted repeat, similar to the situation found in B. hypnoides [13]. The lack of a quadripartite architecture in Bryopsidales had earlier been implied based on Southern hybridization analysis of restriction fragments in Codium fragile and Caulerpa sertularoides [14,15]. Most plastid genomes have a quadripartite structure where a set of large inverted repeats (typically containing the rRNA genes and some other genes) divides the genome into two single-copy regions. Although the quadripartite structure is believed to be ancestral in Viridiplantae, loss of the inverted repeat has occurred multiple times in different green algal lineages [3,7-9,23,24].
Gene content
The B. plumosa cpDNA contains 115 unique genes, including 86 protein coding genes, 26 tRNA genes, and three rRNA genes (Tables 1, 2, Figure 4). In addition to expected, conserved plastid protein coding genes, several freestanding ORFs were identified that did not show any relationship with known plastid genes, but instead showed close similarity with bacterial genes (Table 3). The large orf3 (8,031 bp) showed close similarity to bacterial rhs-family proteins. A similar but slightly smaller ORF (7,173 bp, interrupted by a 267 bp intron) was also found in the re-annotated cpDNA of B. hypnoides (Additional file 2). Rhs genes are a family of composite genes that occur in several Gram-negative bacteria, including some Cyanobacteria. Their broad phylogenetic distribution has been explained by a complex evolutionary history of horizontal transfers and independent gene losses [25]. Despite their wide occurrence, the function of rhs genes remains elusive [26]. Other ORFs of putative bacterial origin include a gene encoding DNA polymerase with maturase-specific domain (orf7), and two ORFs (orf4 and orf5) showing close similarity to uncharacterized bacterial proteins. Orf3, orf4 and orf5 are grouped in a 13 kb region of the cpDNA, while orf7 is situated elsewhere (Figure 2, indicated in red).
Table 2.
Comparison of tRNA genes in the two Bryopsis species, Tydemania expeditiones and two other species of Ulvophyceae ( Oltmannsiellopsis viridis and Pseudendoclonium akinetum )
| B. hyp | B. plu | T. exp | P. aki | O. vir | |
|---|---|---|---|---|---|
| trnA(agc) | 1* | ||||
| trnA(cgc) | 1* | ||||
| trnA(ugc) | 1,1* | 1 | 1 | 2 | 2 |
| trnC(gca) | 1,1* | 1 | 1 | 1 | 1 |
| trnD(guc) | 1,1* | 1 | 1 | 1 | 1 |
| trnE(cuc) | 1* | ||||
| trnE(uuc) | 1 | 1 | 1 | 1 | 1 |
| trnF(gaa) | 1,1* | 1 | 1 | 1 | 1 |
| trnG(gcc) | 1,1* | 1 | 1 | 1 | 1 |
| trnG(ucc) | 1,1* | 2 | 1 | 1 | 1 |
| trnH(gug) | 1,1* | 1 | 1 | 1 | 1 |
| trnI(aau) | 1* | ||||
| trnI(cau) | 1 | 1 | 1 | 1 | |
| trnI(gau) | 1 | 1 | 1 | 2 | 2 |
| trnK(cuu) | 1* | ||||
| trnK(uuu) | 1 | 1 | 1 | 1 | 1 |
| trnL(caa) | 1* | 1 | 1 | ||
| trnL(uaa) | 1 | 1 | 1 | 1 | 1 |
| trnL(uag) | 1 | 1 | 1 | 1 | 1 |
| trnL(gag) | 1 | ||||
| trnM(cau)a | 1* | ||||
| trnMe(cau) | 1 | 1 | 1 | 1 | 1 |
| trnMf(cau) | 1 | 1 | 1 | 1 | 1 |
| trnN(guu) | 1,1* | 1 | 1 | 1 | 1 |
| trnP(agg) | 1* | ||||
| trnP(ugg) | 1,1* | 1 | 1 | 1 | 1 |
| trnQ(cug) | 1* | ||||
| trnQ(uug) | 1 | 1 | 1 | 1 | 1 |
| trnR(acg) | 1,1* | 1 | 1 | 1 | 1 |
| trnR(ucg) | 1* | ||||
| trnR(ccu) | 1* | 1 | 1 | ||
| trnR(ucu) | 1 | 1 | 1 | 1 | 1 |
| trnS(gcu) | 1,1* | 1 | 1 | 1 | 1 |
| trnS(uga) | 1 | 1 | 1 | 1 | 1 |
| trnT(agu) | 1* | ||||
| trnT(ugu) | 1 | 1 | 1 | 1 | 1 |
| trnV(acc) | 1* | ||||
| trnV(cac) | 1* | ||||
| trnV(uac) | 1 | 1 | 1 | 1 | 1 |
| trnW(cca) | 1,1* | 1 | 1 | 1 | 1 |
| trnY(gua) | 1,1* | 1 | 1 | 1 | 1 |
| Total | 53 | 27 | 28 | 27 | 27 |
| Unique | 40 | 26 | 28 | 27 | 27 |
aThe trnM(cau) gene situated in the large tRNA region in B. hypnoides showed similarity (blastn E value < 2e-10) with land plant nuclear encoded tRNA-Met initiator (Met-tRNA-i) genes.
Numbers indicate gene copy. An asterisks indicates that the gene or gene copy is situated in the large tRNA region in B. hypnoides (Additional file 2). B. hyp = Bryopsis hypnoides, B. plu = B. plumosa, T. exp = Tydemania expeditiones, P. aki = Pseudendoclonium akinetum, O. vir = Oltmannsiellopsis viridis.
Figure 4.
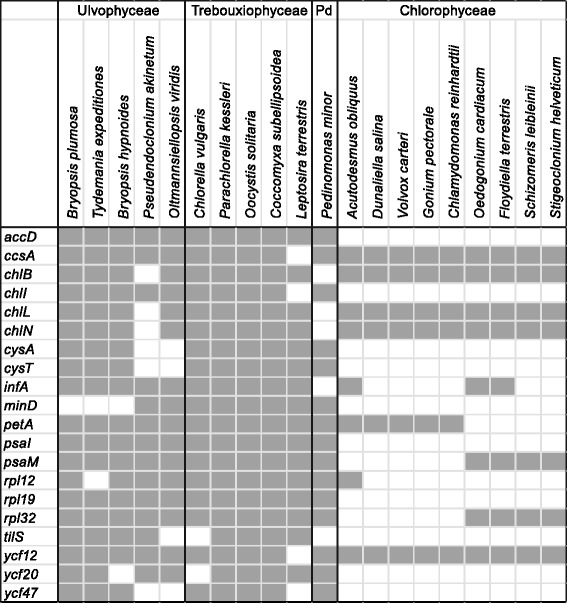
Comparison of protein coding genes content among core Chlorophyta. Pd = Pedinophyceae. 80 genes that are shared among the 20 cpDNAs are not included: atpA, B, E, F, H, cemA, clpP, ftsH, petB, D, G, L, psaA, B, C, J, psbA, B, C, D, E, F, H, I, J, K, L, M, N, T, Z, rbcL, rpl2, 5, 14, 16, 20, 23, 36, rps2, 3, 4, 7, 8, 9, 11, 12, 14, 18, 19, tufA, ycf1, ycf3, ycf4, rpoA, B, C1, C2, rrf, rrl, rrs, and 20 tRNA genes (trnA, C, D, E, F, G, H, I, K, L, M, N, P, Q, R, S, T, V, W, Y). ftsH and ycf1 are present in C. reinhardtii as ORF2971 and ORF1995, respectively, and ycf1 is present in C. vulgaris as ORF819 ([91,92] (supplementary Table II)). ycf47 is present in C. vulgaris as ORF70 (determined by blastx). Data sources: B. hypnoides [13], P. akinetum [12], O. viridis [11], C. vulgaris [6], P. kessleri, O. solitaria, P. minor [22], C. subellipsoidea [93], L. terrestris [8], A. obliquus [94], D. salina [95], V. carteri [96], G. pectorale [97], C. reinhardtii [98], O. cardiacum [99], F. terrestris [7], S. leibleinii [4], S. helveticum [5].
Table 3.
Freestanding ORFs in Bryopsis plumosa and Tydemania expeditiones cpDNAs
| Species orf number | Length (AA) | Closest BLAST hit(s) | blastp E-value | Note |
|---|---|---|---|---|
| B. plumosa orf3 | 2,676 | bacterial rhs-family proteins | <1e-39 | No similar green algal sequencesa (apart from B. hypnoides) |
| B. plumosa orf4 | 212 | hypothetical protein, Francisella spp. | 1e-26 | No similar green algal sequencesa |
| B. plumosa orf5 | 76 | hypothetical protein, Enterobacter sp. | 4e-19 | No similar green algal sequencesa (apart from B. hypnoides) |
| B. plumosa orf7 | 446 | Bacterial RNA-directed DNA polymerase | <1e-32 | Contains a group II intron, maturase-specific domain. Similarity to a RT group II intron protein of Caulerpa filiformis (also present in B. hypnoides). |
| T. expeditiones orf3 | 375 | Bacterial RNA-directed DNA polymerase | <1e-23 | Contains a Group II intron, maturase-specific domain. Similarity with RT in Caulerpa filiformis cpDNA |
| T. expeditiones orf8 | 96 | bacterial DNA methyltransferase | <1e-22 | Contains Cytosine-C5 specific DNA methylase domain. No similar green algal sequencesa |
| T. expeditiones orf9 | 202 | bacterial DNA methyltransferase | <1e-56 | Contains Cytosine-C5 specific DNA methylase domain. No similar green algal sequencesa |
| T. expeditiones orf13 | 287 | bacterial transposase | <1e-23 | Contains an integrase core domain protein. No similar green algal sequencesa |
| T. expeditiones orf14, orf15 | 117, 177 | bacterial DNA polymerase | < 1e-46 | Contains a DNA polymerase family A domain. No similar green algal sequencesa |
| T. expeditiones orf16 | 381 | bacterial DNA primase or phage/plasmid primase | < 1e-97 | No similar green algal sequencesa |
aTo verify whether homologous genes were present in green algae, blastp searcher were performed with organisms constrained to Viridiplantae (taxid:33090).
Most genes found in B. plumosa, with the exception of ycf20 and orf4, were also found in B. hypnoides [13] (Additional file 2). However, the cpDNA of B. hypnoides contains a region of about 6 kb including 27 tRNA genes, ten of which have not been found in any other green algal cpDNA [13]. This large tRNA region is not present in B. plumosa, nor is the set of unique tRNA genes (Table 2). As in B. plumosa, several ORFs in B. hypnoides showed high similarity to bacterial genes, including a putative rhs gene, and genes containing domains commonly found in mobile elements, such as reverse transcriptase, endonuclease and maturase (Additional file 2). Reverse transcriptase proteins catalyze the reverse transcription of RNA to complementary DNA, and have a key role in the mobility of retroviruses, retrotransposons, and organellar group II introns [27,28]. Endonuclease-like proteins catalyze cleavage of retrotransposons before transcription [29], and are also involved in mobility and homing of introns [30]. Maturase-like proteins are best known from bacterial and organellar introns, where they aid in intron splicing [30].
The T. expeditiones cpDNA contains 125 unique genes, including 94 protein coding genes, three rRNA genes, and 28 tRNA genes (Tables 1, 2, Figure 4). In addition to the conserved set of chloroplast genes, several freestanding ORFs were found coding for bacterial or intron related genes (Table 3). Orf3 contains a maturase-specific domain, which are usually found within intronic ORFs. Two overlapping ORFs (orf8 and orf9) are related to bacterial C-5 cytosine-specific DNA methyltransferase genes, a family of genes coding for enzymes that catalyzes the methyl-transfer reaction in the process of DNA methylation, and in bacteria are often associated with restriction/modification systems, which function as a defense against infection of bacteria by bacteriophages [31]. DNA methyltransferases have up till now not been found in plastid genomes. However, the identification of DNA methyltransferase genes in organelles is not unprecedented as a gene of this nature has been identified in the mitochondrial genome of the streptophyte Klebsormidium, which was seen as a possible remnant of viral infection [32]. A region of about 7 kp contains four freestanding ORFs showing close similarity to bacterial genes involved in mobile functions. Orf13 showed high similarity to bacterial transposase and contains an integrase core domain. Transposases and integrases catalyze the movement and integration of DNA copies to new locations within and between genomes [33]. Orf14 and orf15 showed similarity to bacterial DNA polymerase, containing a DNA polymerase family A domain. Family A polymerases fill DNA gaps that arise during DNA repair, recombination and replication, and are found primarily in prokaryotes [34]. Orf16 showed high similarity to phage/plasmid DNA primases, which are known from phages and plasmids of Bacteria and Archaea [35]. The presence of these four ORFs indicates that the 7 kp region may be a mobile element of bacterial origin.
The presence of bacterial genes (including genes involved in mobile functions) in both the Bryopsis and Tydemania cpDNAs, and absent in other green algae, suggest that these genes have been acquired through horizontal gene transfer. Transfer of genes to plastid genomes is only rarely observed but some clear instances are known [36,37]. In the Oedogonium chloroplast genome, the unprecedented finding of int (a gene belonging to the family of tyrosine recombinases) and dpoB (a member of the B family of DNA-directed DNA polymerases) was seen as evidence of horizontal transfer, possibly from a mitochondrial genome donor [99]. In the cpDNA of Nephroselmis olivacea, two large regions (27 kb in total) are believed to have been acquired by lateral transfer from a bacterial donor based on a deviant base composition, the lack of genes typically found in cpDNAs, and the presence of an ORF showing similarity to phage associated DNA primases [9,38]. Horizontal gene transfer from bacteria to plastids has also been demonstrated in other groups of algae, including red algae [39], dinoflagellates [37,40], haptophytes and cryptophytes [41,42]. It is relevant to note that siphonous green algae (Bryopsidales) are known to harbor intracellular bacterial communities, with some bacteria showing close associations with the host (e.g., [43-45]). Based on fluorescence in situ hybridization, bacteria in Bryopsis were found in the vacuole or in the cytoplasmic layer of the siphonous cells, sometimes closely adhered to the chloroplast membrane [46]. These intracellular bacteria may facilitate gene transfer from the endobiontic bacteria to the host genome, possibly via vectors such as bacteriophages.
The two Bryopsis cpDNAs share nearly identical gene repertoires with T. expeditiones, except for rpl12, which is present in B. plumosa and B. hypnoides but was not found in T. expeditiones. In addition, several of the ORFs discussed above were uniquely found in B. plumosa or T. expeditiones (Table 3). A number of genes in Bryopsis and/or Tydemania, including psbM, rpl19, and rpl23, were found to be quite divergent from core chlorophytan orthologs based on visual inspection of amino acid alignments and branch lengths of resulting phylogenetic trees (Additional file 3). The petL gene is present in two identical copies in B. plumosa (see inverted repeats below). Also B. hypnoides contains two copies of petL, but here they differ in nucleotide composition as well as in length: a 96 bp copy identical to that of B. plumosa, and a 105 bp copy that is divergent from the bryopsidalean orthologs (Additional file 4). Tydemania expeditiones contains a single 96 bp copy of petL.
A comparison of gene repertoires between B. plumosa, T. expeditiones and 18 published core chlorophytan cpDNAs is shown in Figure 4. A total of 80 genes are shared among the 20 cpDNAs. A large proportion of genes are shared among species of Ulvophyceae and Trebouxiophyceae, while most Chlorophyceae have a smaller gene repertoire (Figure 4). Notably, several genes are shared between the Bryopsidales and Trebouxiophyceae, but are absent from Pseudendoclonium and/or Oltmannsiellopsis. These include three genes encoding subunits of protochlorophyllide reductase (chlB, L, N), tRNA Ile-lysidine synthetase (tilS) and hypothetical protein ycf47. Only one gene, the organelle division inhibitor factor, minD, was present in Pseudendoclonium, Oltmannsiellopsis, and Trebouxiophyceae but absent in Bryopsidales.
tRNA(Ile)-lysidine synthase (tilS), also known as ycf62, is present in all Ulvophyceae and Trebouxiophyceae, but absent from Chlorophyceae (Figure 4). In T. expeditiones, this gene seems to be pseudogenized by a stop codon at amino acid position 152 (492 bp) from the start methionine. A similar situation was found in Caulerpa filiformis (like Tydemania a member of the Halimedineae), which contains a tilS pseudogene with a frame shift at about the same position [16]. In B. plumosa and B. hypnoides the gene is interrupted by a 71 bp, AT-rich (80.3 and 81.7% AT) insertion at amino acid position 157.
Introns
In B. plumosa 13 introns are present in 12 genes, comprising 8.3% (including intronic ORFs) of the total cpDNA (Tables 1 and 4). Four introns were identified as group I introns, and five as group II introns, while for four introns the class could not be determined with certainty. In T. expeditiones 11 introns are present in 7 genes, comprising 11.2% (including intronic ORFs) of the total cpDNA. Seven introns were identified as group I introns, and two as group II introns, while the class of two introns could not be determined with certainty. Several of the introns in B. plumosa and T. expeditiones (psaA, psbB, rrl and rrs introns) are commonly found in orthologous genes of other green algae. Others are only rarely found in green algae or have not yet been observed in plastids in general. These are discussed below.
Table 4.
Distribution and characteristics of introns in Bryopsis plumosa and Tydemania expeditiones
| Gene | B. plu | T. exp | Length ( B. plu / T. exp ) (bp) | Insertion positions a | AT content (excl. intronic ORF) ( B.plu / T.exp ) | Intronic ORF | Class |
|---|---|---|---|---|---|---|---|
| atpA (intron 1) | + | - | 239 | 489 | 69% | - | group I intron |
| atpA (intron 2) | + | - | 728 | 753 | 70% | - | putative group II |
| atpF | + | + | 481/373 | 108/108 | 82/77% | - | undetermined |
| ccsA | + | + | 385/370 | 210/210 | 79/78% | - | undetermined |
| psaA | + | - | 2244 | 1797 | 69% | RT, IM | group II |
| psbA | - | + | 3227 | 576 | 67% | RT, IM | group II |
| psbB | + | - | 1103 | 600 | 73% | LHE | group I |
| psbC | - | + | 2491 | 1131 | 57% | RT, IM | group II |
| psbT | + | - | 427 | 27 | 82% | - | undetermined |
| rpl5 | + | - | 389 | 171 | 77% | - | putative group II |
| rpl23 | + | - | 351 | 24 | 85% | - | undetermined |
| rps19 | + | - | 848 | 141 | 64% | - | putative group II |
| rrl (intron 1) | - | + | 776 | 1917 | 73% | LHE | group I |
| rrl (intron 2) | - | + | 801 | 1923 | 69% | LHE | group I |
| rrl (intron 3) | - | + | 1049 | 1931 | 67% | LHE b | group I |
| rrl (intron 4) | + | + | 1037/730 | 2598/2598 | 63/71% | LHE | group I |
| rrs (intron 1) | - | + | 860 | 510 | 67% | LHE | group I |
| rrs (intron 2) | - | + | 988 | 794 | 71% | LHE | group I |
| trnL(uaa) | + | + | 205/152 | 35 | 73/65% | - | putative group I |
| ycf3 | + | - | 373 | 174 | 79% | - | putative group II |
aIntron insertion site positions correspond to the nucleotide immediately preceding the intron. Insertion sites in genes coding for proteins and the tRNA are given relative to the corresponding genes in Mesostigma viride cpDNA (GenBank NC_002186); insertion sites in the rRNA genes are given relative to the 16S and 23S rRNA genes of Escherichia coli (GenBank NC_004431).
bContains two LAGLIDADG homing endonuclease domains.
RT = reverse transcriptase, IM = intron maturase, LHE = LAGLIDADG homing endonuclease.
The atpA gene in B. plumosa contains two introns. The first was identified as a group I intron, which is commonly found in this gene in other green algae [4,12]. The second intron was identified as a putative group II intron (Additional file 5), which has until now only been found in the atpA gene of Volvox carteri.
The psbA gene in T. expeditiones has a large intron containing two overlapping ORFs: one encoding reverse transcriptase and intron maturase, the other encoding N-terminal domain of reverse transcriptase. The majority of psbA introns in green algae are group I introns. Thus far, only one other green algal psbA group II intron has been identified, in Floydiella terrestris [7]. The psbC intron in T. expeditiones was also identified as a group II intron based on the presence of an ORF encoding reverse transcriptase and intron maturase. Introns are common in green algal psbC genes, but are all group I introns. PsbC group II introns have thus far only been found in euglenoids [47,48].
The rpl5 and rps19 introns in B. plumosa are putative group II introns (RNA secondary structure showed 6 helices, helix V with conserved AGC, and helix VI with bulged A). Thus far, rpl5 introns have only been found in B. hypnoides. Plastid rps19 introns were until now only known in euglenoids (where they have been identified as group III introns) [48,49] and the red alga Porphyridium purpureum (unspecified intron class) [50]. These introns ranged between 95-216 bp in size, which is much smaller than the intron found in B. plumosa.
AT-rich introns (75-85% AT) of which the class could not be determined with certainty were found in several genes, including atpF, ccsA, psbT and rpl23. AtpF introns are also present in some charophytic green algae, where they have been identified as group II introns [23,51]. Introns in plastid rpl23 were until now only known in euglenoids (group III intron) and B. hypnoides. The length of the Bryopsis rpl23 intron falls within the range of those found in euglenoids (47-440 bp), and the high AT-content is similar to the euglenoid introns (76-91% AT). B. plumosa contains an AT-rich intron in the psbT gene. Introns in this gene have thus far only been found in euglenoids (all group II). Another AT-rich intron was found in the ccsA gene of B. plumosa and T. expeditiones, which is unexpected as this gene lacks introns in all other published plastids. Similar to the other AT-rich introns, the class could not be determined because we failed to identify conserved structural motives.
The trnL(uaa) gene in Bryopsis and Tydemania contains a group I intron. This intron has been assumed to have been present in the common ancestor of Cyanobacteria and chloroplasts because it is found across a wide diversity of Cyanobacteria and plastids where it is conserved in position, secondary structure and primary sequence [52]. In green algae, this intron is present in several charophytes, and core Chlorophyta, but is absent in the published cpDNAs of prasinophytes [3] and the ulvophytes Pseudendoclonium and Oltmannsiellopsis [11,12]. The absence from the early diverging prasinophytes implies multiple losses of the trnL intron.
Repeats
The B. plumosa cpDNA contains three relatively large inverted repeats (IRs) and one large tandem repeat (Figure 5). IR1 (237 bp) duplicates part of the ccsA intron in an intergenic region between rpoC1 and rpoB. IR9 (88 bp) duplicates trnG(ucc), and IR10 (300 bp) duplicates petL and part of psaJ. A tandem repeat of 2 × 265 bp duplicates part of psbK and part of ycf12.
Figure 5.

Sequence repeats (yellow) in Bryopsis plumosa and Tydemania expeditiones cpDNAs. Arrows indicate the direction of the repeats. Protein coding genes are indicated in green, tRNA genes in purple and rRNA genes in red.
In T. expeditiones several repeats are present within CDSs: rpoC2 contains two inverted repeats of 15 and 25 bp long, rpoC1 contains a tandem repeat of 2 × 38 bp, and rps11 contains a tandem repeat of 5 × 15 bp (Figure 5). The intergenic spacer region between orf16 and trnL(uag) contains a short inverted repeat and a tandem repeat of 5 × 9 bp.
In addition, B. plumosa and T. expeditiones contain numerous short inverted repeats near the 3’end of genes or overlapping with the 3’end of genes (Figure 5). Short IRs in the 3’untranslated regions (UTRs) of mRNAs are common in organellar genomes where they serve as RNA-processing signals [53-55]. In B. plumosa and T. expeditiones, several of these short IRs partially overlap with the 3’-end of the CDSs (e.g. psaM, chlN, clpP, rpl23, psbI in B. plumosa, and ycf1, chlN, rpoA in T. expeditiones), a condition also found in bacteria where hairpins preceding the stop codon are involved in termination of transcription by RNA polymerase at rho-independent sites, known as intrinsic termination [56].
Synteny
The Mauve alignments between the two Bryopsis cpDNAs and between B. plumosa and T. expeditiones (Figure 6) visualize locally collinear blocks (LCBs), representing homologous regions of sequences that do not contain any major rearrangements. The syntenic structure of the B. plumosa cpDNA is similar to that of B. hypnoides, separated by 25 rearrangements as calculated by the double cut and join (DCJ) analysis. When excluding all tRNA genes from the analysis, the DCJ distance is reduced to 7. This large difference is mainly due to numerous inversions of tRNA genes, and the absence of the large tRNA region in B. plumosa. The cpDNAs of the two Bryopsis species and Tydemania are more rearranged with respect to each other, with a DCJ distance of 36 (23 excluding all tRNA genes). This high dissimilarity in synteny is not surprising as high variability in cpDNA architecture has also been observed among species of Chlorophyceae and Trebouxiophyceae, including between congeneric taxa [4,22,57]. Even higher number of rearrangements separate the cpDNAs of Bryopsidales and Pseudendoclonium and Oltmannsiellopsis (DCJ distances up to 60).
Figure 6.
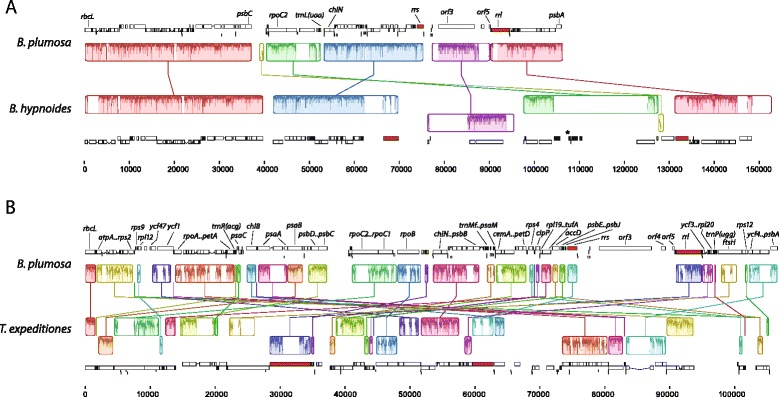
Alignments between the chloroplast genomes of Bryopsis plumosa , B. hypnoides , and Tydemania expeditiones . The Mauve algorithm [81] was used to align the cpDNAs between B. plumosa and B. hypnoides (A), and between B. plumosa and T. expeditiones (B). Corresponding colored boxes are locally collinear blocks (LCBs), which represent homologous regions of sequences that do not contain any major rearrangements. Inside each block a sequence identity similarity profile is shown. Inverted LCBs are presented as blocks below the center line. Annotations are shown above and below the LCBs: protein coding genes as white boxes, tRNA genes in green, rRNA genes in red and short repeats in pink. Lowered position of a box indicates inverted orientation. The asterisk indicates the large tRNA region in B. hypnoides.
Several, but not all, gene clusters conserved in the Ulvophyceae and Trebouxiophyceae [based on Turmel et al. [22]] are also conserved in Bryopsis and Tydemania. One notable exception is the petA-petL-petG cluster, which is absent in the Bryopsidales (Figure 7A). On the other hand, some gene clusters shared in the Bryopsidales are not present in any of the four analyzed Ulvophyceae and Trebouxiophyceae species (Figure 7B).
Figure 7.
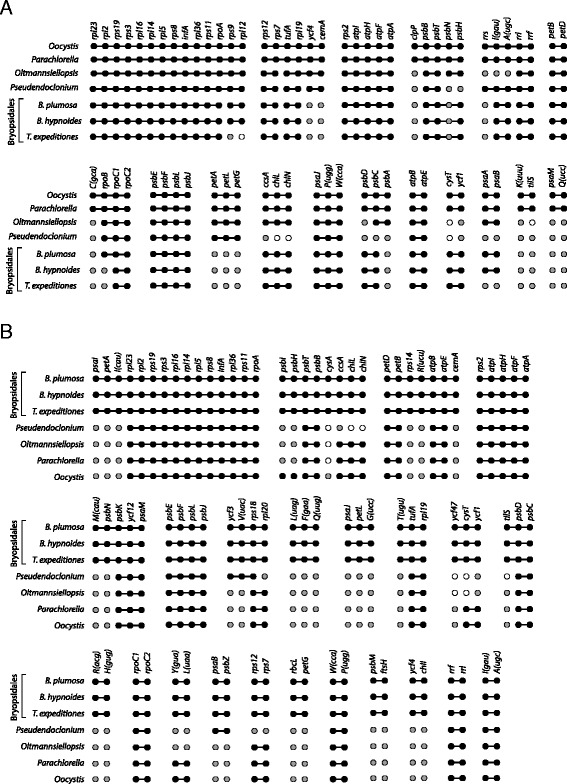
Comparison of conserved gene clusters. (A) Comparison of gene clusters conserved between at least two of the four depicted species, representing different ulvophycean and trebouxiophycean lineages [based on Turmel et al. [22]] with gene order found in the Bryopsidales (Bryopsis plumosa, B. hypnoides and Tydemania expeditiones). (B) Comparison of gene clusters conserved in Bryopsidales with gene order found in four species, representing different ulvophycean and trebouxiophycean lineages. Black connected circles indicate gene clusters. Grey circles indicate genes that are located elsewhere on the cpDNA. White circles indicated genes that are missing from the cpDNA.
Phylogenomic analyses
The phylogenetic trees resulting from the analyses of the 79-gene dataset are summarized in Figure 8. The topology of the trees are in general agreement with published chloroplast phylogenies of Chlorophyta [3,18,19]. Among prasinophytes, the position of Pycnococcus provasolii differs from the analyses of Lemieux et al. [3,19], but this may be attributable to a difference in tree rooting. The core Chlorophyta are comprised of several well supported main clades, including the Pedinophyceae, Chlorellales, the core Trebouxiophyceae, Chlorophyceae and two clades of Ulvophyceae. The relationships among these lineages are generally poorly supported, which contrasts with previous analyses that did not include members of the Bryopsidales [19]. Although the phylogenies recovered two separate Ulvophyceae clades, the relationship between the two is poorly supported and therefore our analyses are inconclusive with respect to monophyly of the class. The unstable phylogenetic position of the Bryopsidales is also apparent when comparing published chloroplast multi-gene phylogenies, which revealed different relationships depending on gene and taxon sampling: a sister relationship between Bryopsis and Chlorophyceae [13], between Caulerpa (Bryopsidales) and Chlorella [16], or between the core Trebouxiophyceae and a Bryopsidales + Dasycladales + Trentepohliales clade [18].
Figure 8.
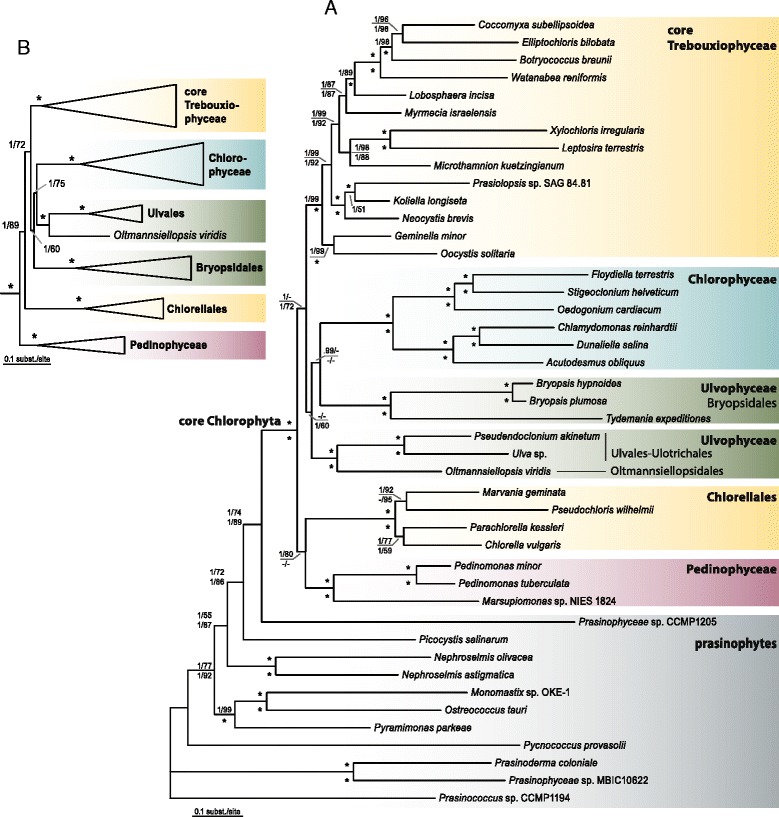
Phylogenetic trees of Chlorophyta inferred from a 79-gene dataset. (A) Bayesian majority rule tree showing all compatible partitions, inference from the protein alignment of 79 concatenated chloroplast genes (16,205 amino acid positions and 44 terminal taxa). Node support is given as Bayesian posterior probabilities and maximum-likelihood (ML) bootstrap values of the protein analyses (above branches), and the nucleotide analyses (below branches); values <0.9 and 50, respectively, are not shown; asterisks indicated full support in both the Bayesian and ML analyses. (B) Bayesian tree inference from the nucleotide alignment (first two codon positions) of 79 concatenated chloroplast genes (32,410 nucleotide positions and 44 terminal taxa). Node support is given as Bayesian posterior probabilities and maximum-likelihood (ML) bootstrap values, and asterisks indicate full support in both analyses.
The phylogenetic trees resulting from the analyses of the 50-gene dataset are summarized in Figure 9. The 50-gene dataset differs from the 79-dataset mainly in the inclusion of species of Trentepohliales and Dasycladales (both orders of Ulvophyceae) and Tetraselmis (Chlorodendrophyceae). Overall, the 50-gene phylogenies recovered the same main clades as the 79-gene phylogenies, in addition to two extra clades of Ulvophyceae (Trentepohliales and Dasycladales), and Tetraselmis. Similar to the 79-gene phylogenies, the relationships among the main clades of core Chlorophyta (including the different clades of Ulvophyceae) received little statistical support, and therefore the monophyly of the Ulvophyceae cannot be confirmed nor rejected by our data. The phylogenies inferred from the nucleotide alignment (Figure 9B) recovered the same main clades as the protein analyses, but the topology and branch support differs in several aspects, including a well-supported Trentepohliales + Dasycladales clade, and a different phylogenetic position of Tetraselmis.
Figure 9.
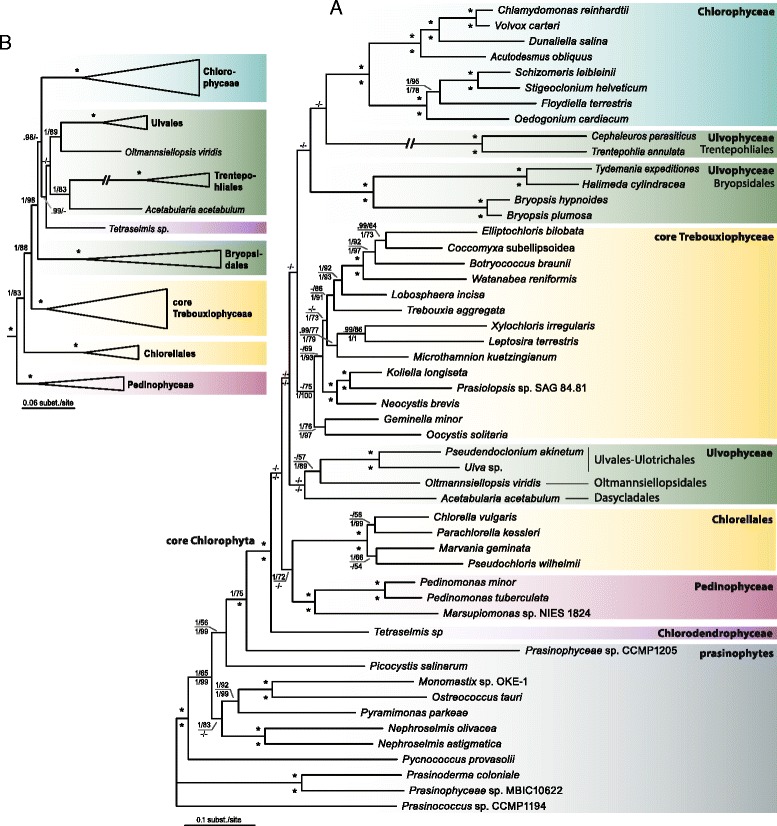
Phylogenetic trees of Chlorophyta inferred from a 50-gene dataset. (A) Bayesian majority rule tree showing all compatible partitions, inference from the protein alignment of 50 concatenated chloroplast genes (9,300 amino acid positions and 51 terminal taxa). Node support is given as Bayesian posterior probabilities and maximum-likelihood (ML) bootstrap values of the protein analyses (above branches), and the nucleotide analyses (below branches); values <0.9 and 50, respectively, are not shown; asterisks indicated full support in both the Bayesian and ML analyses. (B) Bayesian tree inference from the nucleotide alignment (first two codon positions) of 50 concatenated chloroplast genes (18,600 nucleotide positions and 51 terminal taxa). Node support is given as Bayesian posterior probabilities and maximum-likelihood (ML) bootstrap values, and asterisks indicate full support in both analyses.
The relationships within the Ulvophyceae and the monophyly of the class has been the subject of a longstanding debate [2,17,18,21]. Phylogenies based on nuclear ribosomal DNA data have remained ambiguous and often recovered two distinct clades: the Oltmannsiellopsidales + Ulvales-Ulotrichales clade, and a clade consisting of Trentepohliales, Cladophorales, Bryopsidales and Dasycladales (TCBD clade) [21,58-61]. Similarly, multigene chloroplast data have failed to provide support for monophyly of the Ulvophyceae [13,16,18]. Conversely, phylogenetic analyses based on 10 genes (eight nuclear- and two plastid-encoded) recovered the class as a monophyletic group with strong support [17,61]. These analyses also supported the division of the two above mentioned distinct ulvophycean clades. Within the TCBD clade, the Bryopsidales and Dasycladales were recovered as sister lineages with high support. The division of two distinct ulvophycean clades was also supported by independent molecular data, such as the distribution of elongation factor genes [60,62], and a non-canonical nuclear genetic code [63]. It is important to note that several important clades that were present in the analyses of Cocquyt et al. [17] and Škaloud et al. [61], such as the Cladophorales, Scotinosphaerales, Blastophysa, and Ignatius-clade, were not included in our study, which may have an important effect on phylogenetic reconstruction. A notable difference between our phylogenies and the ones of Fučíková et al. [18] is the position of Oltmannsiellopsis and Tetraselmis. Oltmannsiellopsis is currently classified as an ulvophyte based on morphological and molecular data [59,60,64]. Tetraselmis is a member of the Chlorodendrophyceae and has, in nuclear ribosomal-based phylogenies, been recovered as an early branching clade of the core Chlorophyta [2,65,66]. In contrast, chloroplast multi-gene analyses have recovered a clade uniting Tetraselmis and Oltmannsiellopsis, which was sister to the Ulvales-Ulotrichales [67] or branched early in the radiation of the core Chlorophyta [18]. Our analyses recovered Oltmannsiellopsis as sister to the Ulvales-Ulotrichales (Figures 8 and 9), similar to [13,16,19], while the position of Tetraselmis remained ambiguous, branching early in the core Chlorophyta in the protein tree (Figure 9A), or branching in the vicinity of ulvophycean lineages in the nucleotide-based phylogeny (Figure 9B). It should be noted that gene sampling in Tetraselmis was low, which may explain its unstable phylogenetic position.
An accurate phylogenetic reconstruction of the core Chlorophyta, and in particular the Ulvophyceae will require a richer sampling of taxa and genes. As has been shown in the Trebouxiophytes, expanding taxon sampling has greatly aided in resolving phylogenetic relationships [18,19]. Compared with other groups of core Chlorophyta, the Ulvophyceae are still underrepresented in chloroplast phylogenomic studies (Figure 1), and for several important clades chloroplast genomic data is lacking altogether (e.g. Scotinosphaerales, Cladophorales, Blastophysa, and Ignatius-clade). Although chloroplast genomes contain large amounts of genetic information, a combination of plastid and nuclear genomic data may be needed to test monophyly of the Ulvophyceae, resolve relationships within the Ulvophyceae, and among the main clades of core Chlorophyta.
Conclusions
The gene dense chloroplast genomes of Bryopsis plumosa and Tydemania expeditiones are amongst the smallest chloroplast genomes in the core Chlorophyta. Comparison of the cpDNA of Bryopsis plumosa with the previously published cpDNAs of Ulvophyceae highlighted some exceptional differences in genome size, gene content, and gene order, illustrating the remarkable plasticity of green algal chloroplast genomes, even between congeneric species.
The cpDNAs of B. plumosa, B. hypnoides and T. expeditiones contain DNA regions that contain open reading frames showing no affinity to conserved plastid genes, but instead having significant similarity to bacterial genes. These genes include rhs-family genes, and several genes involved in mobile functions, such as transposases, integrases, DNA polymerases, and phage/plasmid DNA primases. Another unexpected finding was the presence of two ORFs in T. expeditiones showing close similarity to bacterial DNA methyltransferases, a family of genes that have up till now not been found in plastid genomes. The presence of DNA regions including genes with clear bacterial affinity suggests that these regions may have been acquired through horizontal DNA transfer from bacterial donors. Siphonous green algae are characterized by giant tubular cells that contain millions of nuclei and plastids, in addition to diverse bacterial communities. It is imaginable that these horizontal DNA transfers have been facilitated by the occurrence of bacteria residing inside the host cell. Our data adds to the scarce knowledge of horizontal transfer of bacterial DNA to plastid genomes.
Methods
Collections
Plant material of Bryopsis plumosa was obtained from a culture isolated by John West from Traon Erch, Brittany, France on 5 April 2008 (culture number 4718, now maintained in the algal culture collection of the Phycology Research Group, Ghent University). The specimen was morphologically identified as B. plumosa. Molecular data, however, has indicated that many morphospecies in the genus Bryopsis (including B. plumosa) are polyphyletic. Based on analysis of rbcL sequences, the culture 4718 is a member of species clade number 4 in Hollants et al. [68]. For convenience, however, we will use the name B. plumosa throughout the paper. Plant material of Tydemania expeditiones was obtained from a silica dried specimen collected by FL from Siquijor, Philippines on 17 September 2007 (voucher specimen FL1151, deposited in the algal herbarium of Ghent University, GENT).
Genome sequencing and assembly
Total genomic DNA was extracted by using a modified CTAB extraction protocol based on Doyle & Doyle [18,69]. Sequencing of the genomic DNA was performed by Cold Spring Harbor Laboratory (Cold Spring Harbor, NY, USA) using Illumina HiSeq 2000 technology on 1/5th of a lane. The sequencing run generated 7.2 million paired-end reads (2 × 101 bp) for B. plumosa and 14.7 million paired-end reads for T. expeditiones.
Low-quality ends of the reads (Phred score < 30) and adapters were trimmed using Trim Galore! (www.bioinformatics.babraham.ac.uk/projects/trim_galore), and only reads longer than 2 × 94 bp were retained. De novo assembly was performed from the paired-end Illumina reads using three different assembly programs: (1) Velvet v. 1.2.10 [70] using a kmer = 93, (2) Geneious v. 7 (Biomatters, www.geneious.com) de novo assembler using “medium sensitivity”, and (3) CLC Genomics workbench v. 6 (www.clcbio.com) de novo assembler using word size = 63 and default parameters.
Contigs corresponding to cpDNA sequences were extracted from the total assembly using blastn similarity searches (E-value < 10-6) of a custom build blast database including all genes from published cpDNAs of Chlorophyta and Mesostigma. The three assemblies (Velvet, Geneious and CLC) yielded similar results (i.e. cpDNA contigs with identical sequences, but different in length). Scaffolds were constructed by re-assembling the cpDNA contigs using Geneious with stringent conditions (minimum overlap = 80 bp, no gaps allowed, and minimum overlap identity of 95%).
Gene annotation
Initial annotations were performed by mapping genes from published cpDNAs of Chlorophyta against the two cpDNA contigs in Geneious. This was followed by a more detailed examination of protein-coding genes, rRNA genes, tRNA genes, introns, and additional features such as sequence repeats.
For protein-coding genes, open reading frames (ORFs) were identified by getorf from the EMBOSS suite server [71]. Similarities of the protein sequences were detected using blastp 2.2.30 [72] against all non-redundant GenBank CDS translations + PDB + SwissProt + PIR + PRF (excluding environmental samples from WGS projects). Some ORFs were identified that showed no clear similarity with green algal plastid genes. In order to confirm that homologous genes were absent from green algae, blastp searcher was performed but with organisms constrained to Viridiplantae (taxid:33090).
Intron-exon boundaries were identified by translation alignment of the intron-containing genes with CDSs of orthologous genes from published chlorophytan cpDNAs. In order to identify the intron class (group I or group II), RNA secondary structures of the introns were predicted using Mfold [73], taking into account suboptimal foldings, and compared to the models of Michel et al. [74] and Michel and Westhof [75].
The boundaries of the rRNA genes and possible introns in the rRNA genes were identified based on a data set of complete rRNA genes from published green algal cpDNAs, aligned using MUSCLE [76]. tRNA genes were detected and identified using tRNAscan-SE 1.21 [77].
Repeats and inverted sequence repeats were detected using einverted, etandem and palindrome (EMBOSS suite server), and Tandem repeats finder [78].
We also re-annotated the published cpDNA of Bryopsis hypnoides (GenBank accession NC_013359) [13] because our initial comparison with B. plumosa and other species of Chlorophyta yielded some unexpected results, such as the lack of some common genes in B. hypnoides (e.g. chlI, ftsH, psaI, psaM, and rpl19).
Whole-genome alignments and analysis of genome rearrangements
The Bryopsis and Tydemania cpDNAs were aligned using the progressive Mauve algorithm in Geneious using the full alignment option and automated calculation of locally collinear block (LCB) scores [58,79]. Mauve is a general-purpose genome aligner that identifies LCBs despite small rearrangements and reversals. Minimal histories of rearrangements among the pairwise aligned genomes were predicted using UniMoG [80] under a double cut and join (DCJ) model [81,82], which allows for common rearrangement operations such as inversions, translocations, fusions, and fissions.
Phylogenomic analyses
Phylogenomic analyses were based on two data sets: 1) a 79-gene alignment of 44 taxa of Chlorophyta, including six Ulvophyceae, and 2) a 50-gene alignment of 51 taxa of Chlorophyta, including ten Ulvophyceae (Additional file 6).
Gene selection for the 79-gene alignment was based on Lemieux et al. [19], and included accD, atpA, B, E, F, H, I, ccsA, cemA, chlB, I, L, N, clpP, cysA, T, ftsH, infA, minD, petA, B, D, G, L, psaA, B, C, I, J, M, psbA, B, C, D, E, F, H, I, J, K, L, M, N, T, Z, rbcL, rpl2, 5, 12, 14, 16, 19, 20, 23, 32, 36, rpoA, B, C1, C2, rps2, 3, 4, 7, 8, 9, 11, 12, 14, 18, 19, tufA, ycf1, 3, 4, 12, 20, 47, 62. Taxon sampling included 44 taxa of Chlorophyta, based on a selection of taxa from Lemieux et al. [3,19], in addition to Ulva sp. UNA00071828 [83], and the two Bryopsis species and Tydemania. The dataset was 95% filled at the taxon × gene level. The rpoB, rpoC1 and rpoC2 sequences of B. hypnoides were excluded from the dataset because parts of these genes could not be reliably aligned with the chlorophytan orthologs.
Selection of genes for the 50-gene alignment was largely based on Fučíková et al. [18], including atpA, B, E, F, H, I, infA, petA, B, G, psaA, B, C, J, M, psbA, B, C, D, E, F, I, J, K, L, N, T, rbcL, rpl2, 5, 14, 16, 20, 23, 32, 36, rps3, 4, 7, 8, 9, 11, 12, 14, 18, 19, tufA, ycf3, 4, 12. The dataset was comprised of 51 taxa of Chlorophyta, including taxa for which complete cpDNAs or partial multigene data were available (Additional file 6). The dataset was 93% filed at the taxon × gene level. Taxa with low gene coverage included Acetabularia acetabulum (37 genes), Cephaleuros parasiticus (17 genes), Halimeda cylindracea (33 genes), Tetraselmis_sp. (11 genes), and Trentepohlia annulata (18 genes).
DNA sequences were aligned for each gene separately using the ClustalW translational alignment function [84] in Geneious with a BLOSUM cost matrix, and gap open penalty 10 and gap extend cost 0.1. Gene alignments were concatenated, and poorly aligned positions removed using the Gblocks server [85] (http://molevol.cmima.csic.es/castresana/Gblocks_server.html) by preserving the DNA codons, and using the least stringent settings: allowing smaller final blocks, gap positions within the final blocks, less strict flanking positions, and many contiguous non-conserved positions. Gblocks reduced the 79-gene alignment from 140,196 to 48,615 positions, and the 50-gene alignment from 41,166 to 27,900 positions. The resulting alignments were translated to obtain protein alignments of 16,205 amino acid positions (79-gene alignment), and 9,300 amino acid positions (50-gene alignment). For the phylogenetic analyses, only the first two codon positions were included in the nucleotide alignment (32,410 positions for the 79-gene alignment, and 18,600 positions for the 50-gene alignment).
Phylogenetic trees were inferred from the protein and nucleotide alignments using Bayesian and maximum likelihood (ML) analyses. For the protein alignments, a CPREV + Γ4 model was assumed. For the nucleotide alignments, a GTR + Γ4 model was selected, and a partitioning strategy in which codon positions were separated (2 partitions). Bayesian analyses were conducted using MrBayes v.3.2.1 [86], running each analysis for 2 million (protein alignments) or 5 million (nucleotide alignments) generations with one cold and three heated chains, and sampling every 1000 generations. For each analysis, two independent runs were performed. The first 10% of samples was discarded as burn-in based on assessment of convergence of the runs and stability of parameters using Tracer v.1.5 [87]. ML trees were inferred using RAxML v.7.3.5 [88,89] using default parameters. Branch support was assessed by bootstrapping with 500 replicates. All phylogenetic analyses were run on the CIPRES Science Gateway v3.3. [90].
Availability of supporting data
The two cpDNAs have been deposited in EMBL’s European Nucleotide Archive (ENA) (GenBank/DDBJ) under accession numbers [ENA: LN810504] (Bryopsis plumosa) and [ENA: LN810505] (Tydemania expeditiones). Phylogenetic data (alignments and phylogenetic trees) have been deposited in TreeBase under accession number 17184 (http://purl.org/phylo/treebase/phylows/study/TB2:S17184).
Acknowledgements
We thank John West for providing the Bryopsis plumosa culture, April Mitchell for help with the DNA extraction, Ana Tronholm and Trey Melton for fruitful discussions, and the two reviewers for their helpful comments that greatly contributed to improving the final version of the paper. This study was supported by The National Science Foundation under the Assembling the Tree of Life Program (DEB 1036495 and 0937978 to JMLB).
Abbreviations
- CDS
Coding DNA sequence
- cpDNA
chloroplast DNA
- DCJ
Double cut and join
- IR
Inverted repeat
- LCB
Locally collinear block
- ML
Maximum likelihood
- ORF
Open reading frame
Additional files
Read coverage graphs of the cpDNAs of Bryopsis plumosa and Tydemania expeditiones.
Re-annotated Bryopsis hypnoides cpDNA.
Amino acid alignment of psbM , rpl19 and rpl23 and corresponding trees.
petL amino acid alignment and corresponding phylogenetic tree.
Putative RNA secondary structure of atp A type II intron of Bryopsis plumosa.
Taxon sampling for the 79-gene and 50-gene datasets.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
FL and JMLB conceived the study. FL collected and cultured the samples, carried out the genome assemblies and annotations, performed the (phylo)genomic analyses, wrote the manuscript, and generated the figures. FL and JMLB read and approved the final manuscript.
Contributor Information
Frederik Leliaert, Email: frederik.leliaert@ugent.be.
Juan M Lopez-Bautista, Email: jlopez@ua.edu.
References
- 1.Lang BF, Nedelcu A. Plastid genomes of algae. In: Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria. Dordrecht: Springer Netherlands; 2012. pp. 59–87. [Google Scholar]
- 2.Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, et al. Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci. 2012;31(1):1–46. doi: 10.1080/07352689.2011.615705. [DOI] [Google Scholar]
- 3.Lemieux C, Otis C, Turmel M. Six newly sequenced chloroplast genomes from prasinophyte green algae provide insights into the relationships among prasinophyte lineages and the diversity of streamlined genome architecture in picoplanktonic species. BMC Genomics. 2014;15(1):857. doi: 10.1186/1471-2164-15-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouard J-S, Otis C, Lemieux C, Turmel M. The chloroplast genome of the green alga Schizomeris leibleinii (Chlorophyceae) provides evidence for bidirectional DNA replication from a single origin in the Chaetophorales. Genome Biol Evol. 2011;3:505–515. doi: 10.1093/gbe/evr037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bélanger AS, Brouard JS, Charlebois P, Otis C, Lemieux C, Turmel M. Distinctive architecture of the chloroplast genome in the chlorophycean green alga Stigeoclonium helveticum. Mol Genet Genomics. 2006;276(5):464–477. doi: 10.1007/s00438-006-0156-2. [DOI] [PubMed] [Google Scholar]
- 6.Wakasugi T, Nagai T, Kapoor M, Sugita M, Ito M, Ito S, et al. Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: The existence of genes possibly involved in chloroplast division. Proc Natl Acad Sci USA. 1997;94(11):5967–5972. doi: 10.1073/pnas.94.11.5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brouard J-S, Otis C, Lemieux C, Turmel M. The exceptionally large chloroplast genome of the green alga Floydiella terrestris illuminates the evolutionary history of the Chlorophyceae. Genome Biol Evol. 2010;2:240–256. doi: 10.1093/gbe/evq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Cambiaire JC, Otis C, Turmel M, Lemieux C. The chloroplast genome sequence of the green alga Leptosira terrestris: multiple losses of the inverted repeat and extensive genome rearrangements within the Trebouxiophyceae. BMC Genomics. 2007;8:213. doi: 10.1186/1471-2164-8-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turmel M, Gagnon M-C, O'Kelly CJ, Otis C, Lemieux C. The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol Biol Evol. 2009;26(3):631–648. doi: 10.1093/molbev/msn285. [DOI] [PubMed] [Google Scholar]
- 10.Civáň P, Foster PG, Embley TM, Séneca A, Cox CJ. Analyses of charophyte chloroplast genomes help characterize the ancestral chloroplast genome of land plants. Genome Biol Evol. 2014;6(4):897–911. doi: 10.1093/gbe/evu061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pombert JF, Lemieux C, Turmel M. The complete chloroplast DNA sequence of the green alga Oltmannsiellopsis viridis reveals a distinctive quadripartite architecture in the chloroplast genome of early diverging ulvophytes. BMC Biology. 2006;4:3. doi: 10.1186/1741-7007-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pombert JF, Otis C, Lemieux C, Turmel M. The chloroplast genome sequence of the green alga Pseudendoclonium akinetum (Ulvophyceae) reveals unusual structural features and new insights into the branching order of chlorophyte lineages. Mol Biol Evol. 2005;22:1903–1918. doi: 10.1093/molbev/msi182. [DOI] [PubMed] [Google Scholar]
- 13.Lü F, Xü W, Tian C, Wang G, Niu J, Pan G, et al. The Bryopsis hypnoides plastid genome: Multimeric forms and complete nucleotide sequence. PLoS One. 2011;6(2):e14663. doi: 10.1371/journal.pone.0014663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manhart JR, Kelly K, Dudock BS, Palmer JD. Unusual characteristics of Codium fragile chloroplast DNA revealed by physical and gene mapping. Mol Gen Genet. 1989;216(2–3):417–421. doi: 10.1007/BF00334385. [DOI] [PubMed] [Google Scholar]
- 15.Lehman RL, Manhart JR. A preliminary comparison of restriction fragment patterns in the genus Caulerpa (Chlorophyta) and the unique structure of the chloroplast genome of Caulerpa sertularioides. J Phycol. 1997;33(6):1055–1062. doi: 10.1111/j.0022-3646.1997.01055.x. [DOI] [Google Scholar]
- 16.Zuccarello GC, Price N, Verbruggen H, Leliaert F. Analysis of a plastid multigene data set and the phylogenetic position of the marine macroalga Caulerpa filiformis (Chlorophyta) J Phycol. 2009;45(5):1206–1212. doi: 10.1111/j.1529-8817.2009.00731.x. [DOI] [PubMed] [Google Scholar]
- 17.Cocquyt E, Verbruggen H, Leliaert F, De Clerck O. Evolution and cytological diversification of the green seaweeds (Ulvophyceae) Mol Biol Evol. 2010;27(9):2052–2061. doi: 10.1093/molbev/msq091. [DOI] [PubMed] [Google Scholar]
- 18.Fučíková K, Leliaert F, Cooper ED, Škaloud P, D'hondt S, De Clerck O, et al. New phylogenetic hypotheses for the core Chlorophyta based on chloroplast sequence data. Front Ecol Evol. 2014;2:63. [Google Scholar]
- 19.Lemieux C, Otis C, Turmel M. Chloroplast phylogenomic analysis resolves deep-level relationships within the green algal class Trebouxiophyceae. BMC Evol Biol. 2014;14:211. doi: 10.1186/s12862-014-0211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verbruggen H, Ashworth M, LoDuca ST, Vlaeminck C, Cocquyt E, Sauvage T, et al. A multi-locus time-calibrated phylogeny of the siphonous green algae. Mol Phylogenet Evol. 2009;50(3):642–653. doi: 10.1016/j.ympev.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 21.Zechman FW, Theriot EC, Zimmer EA, Chapman RL. Phylogeny of the Ulvophyceae (Chlorophyta): cladistic analysis of nuclear-encoded rRNA sequence data. J Phycol. 1990;26:700–710. doi: 10.1111/j.0022-3646.1990.00700.x. [DOI] [Google Scholar]
- 22.Turmel M, Otis C, Lemieux C. The chloroplast genomes of the green algae Pedinomonas minor, Parachlorella kessleri, and Oocystis solitatia reveal a shared ancestry between the Pedinomonadales and Chlorellales. Mol Biol Evol. 2009;26(10):2317–2331. doi: 10.1093/molbev/msp138. [DOI] [PubMed] [Google Scholar]
- 23.Turmel M, Otis C, Lemieux C. The complete chloroplast DNA sequences of the charophycean green algae Staurastrum and Zygnema reveal that the chloroplast genome underwent extensive changes during the evolution of the Zygnematales. BMC Biology. 2005;3:22. doi: 10.1186/1741-7007-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong H, Lim J-M, Park J, Sim Y, Choi H-G, Lee J, et al. Plastid and mitochondrion genomic sequences from Arctic Chlorella sp. ArM0029B. BMC Genomics. 2014;15(1):286. doi: 10.1186/1471-2164-15-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson A, Thomas G, Parkhill J, Thomson N. Evolutionary diversification of an ancient gene family (rhs) through C-terminal displacement. BMC Genomics. 2009;10(1):584. doi: 10.1186/1471-2164-10-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kung VL, Khare S, Stehlik C, Bacon EM, Hughes AJ, Hauser AR. An rhs gene of Pseudomonas aeruginosa encodes a virulence protein that activates the inflammasome. Proc Natl Acad Sci. 2012;109(4):1275–1280. doi: 10.1073/pnas.1109285109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eickbush T, Malik H. Origins and evolution of retrotransposons. In: Mobile DNA ii; 2002. [Google Scholar]
- 28.Xiong Y, Eickbush TH. Similarity of reverse transcriptase-like sequences of viruses, transposable elements, and mitochondrial introns. Mol Biol Evol. 1988;5(6):675–690. doi: 10.1093/oxfordjournals.molbev.a040521. [DOI] [PubMed] [Google Scholar]
- 29.Kazazian HH. Mobile Elements: Drivers of Genome Evolution. Science. 2004;303(5664):1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 30.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29(18):3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeltsch A. Beyond Watson and Crick: DNA Methylation and Molecular Enzymology of DNA Methyltransferases. ChemBioChem. 2002;3(4):274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 32.Turmel M, Otis C, Lemieux C. Tracing the Evolution of Streptophyte Algae and Their Mitochondrial Genome. Genome Biol Evol. 2013;5(10):1817–1835. doi: 10.1093/gbe/evt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice PA, Baker TA. Comparative architecture of transposase and integrase complexes. Nat Struct Biol. 2001;8(4):302–307. doi: 10.1038/86166. [DOI] [PubMed] [Google Scholar]
- 34.Patel PH, Suzuki M, Adman E, Shinkai A, Loeb LA. Prokaryotic DNA polymerase I: evolution, structure, and “base flipping” mechanism for nucleotide selection. J Mol Biol. 2001;308(5):823–837. doi: 10.1006/jmbi.2001.4619. [DOI] [PubMed] [Google Scholar]
- 35.Ziegelin G, Lanka E. Bacteriophage P4 DNA replication. FEMS Microbiol Rev. 1995;17(1–2):99–107. doi: 10.1111/j.1574-6976.1995.tb00191.x. [DOI] [PubMed] [Google Scholar]
- 36.Huang J, Yue J. Horizontal gene transfer in the evolution of photosynthetic eukaryotes. J Syst Evol. 2013;51(1):13–29. doi: 10.1111/j.1759-6831.2012.00237.x. [DOI] [Google Scholar]
- 37.Mackiewicz P, Bodył A, Moszczyński K. The case of horizontal gene transfer from bacteria to the peculiar dinoflagellate plastid genome. Mobile Genet Elem. 2013;3(4):e25845. doi: 10.4161/mge.25845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turmel M, Otis C, Lemieux C. The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: Insights into the architecture of ancestral chloroplast genomes. Proc Natl Acad Sci USA. 1999;96:10248–10253. doi: 10.1073/pnas.96.18.10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delwiche CF, Palmer JD. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol Biol Evol. 1996;13(6):873–882. doi: 10.1093/oxfordjournals.molbev.a025647. [DOI] [PubMed] [Google Scholar]
- 40.Moszczyński K, Mackiewicz P, Bodył A. Evidence for horizontal gene transfer from Bacteroidetes bacteria to dinoflagellate minicircles. Mol Biol Evol. 2012;29(3):887–892. doi: 10.1093/molbev/msr276. [DOI] [PubMed] [Google Scholar]
- 41.Rice D, Palmer J. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biology. 2006;4(1):31. doi: 10.1186/1741-7007-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan H, Parks N, Kozera C, Curtis BA, Parsons BJ, Bowman S, et al. Plastid genome sequence of the Cryptophyte alga Rhodomonas salina CCMP1319: Lateral transfer of putative DNA replication machinery and a test of chromist plastid phylogeny. Mol Biol Evol. 2007;24(8):1832–1842. doi: 10.1093/molbev/msm101. [DOI] [PubMed] [Google Scholar]
- 43.Hollants J, Leliaert F, Verbruggen H, Willems A, De Clerck O. Permanent residents or temporary lodgers: characterizing intracellular bacterial communities in the siphonous green alga Bryopsis. Proc R Soc B. 2013;280(1754):20122659. doi: 10.1098/rspb.2012.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aires T, Serrão EA, Kendrick G, Duarte CM, Arnaud-Haond S. Invasion is a community affair: Clandestine followers in the bacterial community associated to green algae, Caulerpa racemosa, track the invasion source. PLoS One. 2013;8(7):e68429. doi: 10.1371/journal.pone.0068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hollants J, Leroux O, Leliaert F, Decleyre H, De Clerck O, Willems A. Who Is in There? Exploration of endophytic bacteria within the siphonous green seaweed Bryopsis (Bryopsidales, Chlorophyta) PLoS One. 2011;6(10):e26458. doi: 10.1371/journal.pone.0026458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hollants J. Endophytic bacteria within the green siphonous seaweed Bryopsis: Exploration of a partnership. Ghent Ghent University; 2012.
- 47.Zhang L. Study of group II introns in Euglena chloroplast genomes: Structure, processing and evolution. Tucson: The University of Arizona; 1998. [Google Scholar]
- 48.Hallick RB, Hong L, Drager RG, Favreau MR, Monfort A, Orsat B, et al. Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res. 1993;21(15):3537–3544. doi: 10.1093/nar/21.15.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christopher DA, Cushman JC, Price CA, Hallick RB. Organization of ribosomal protein genes rpl23, rpl2, rps19, rpl22 and rps3 on the Euglena gracilis chloroplast genome. Curr Genet. 1988;14(3):275–286. doi: 10.1007/BF00376748. [DOI] [PubMed] [Google Scholar]
- 50.Tajima N, Sato S, Maruyama F, Kurokawa K, Ohta H, Tabata S, et al. Analysis of the complete plastid genome of the unicellular red alga Porphyridium purpureum. J Plant Res. 2014;127(3):389–397. doi: 10.1007/s10265-014-0627-1. [DOI] [PubMed] [Google Scholar]
- 51.Turmel M, Otis C, Lemieux C. The chloroplast genome sequence of Chara vulgaris sheds new light into the closest green algal relatives of land plants. Mol Biol Evol. 2006;23(6):1324–1338. doi: 10.1093/molbev/msk018. [DOI] [PubMed] [Google Scholar]
- 52.Kuhsel MG, Strickland R, Palmer JD. An ancient group I intron shared by eubacteria and chloroplasts. Science. 1990;250(4987):1570–1573. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- 53.Jiao HS, Hicks A, Simpson C, Stern DB. Short dispersed repeats in the Chlamydomonas chloroplast genome are collocated with sites for mRNA 3' end formation. Curr Genet. 2004;45(5):311–322. doi: 10.1007/s00294-004-0487-7. [DOI] [PubMed] [Google Scholar]
- 54.Stern DB, Gruissem W. Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987;51(6):1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- 55.Starkenburg S, Kwon K, Jha R, McKay C, Jacobs M, Chertkov O, et al. A pangenomic analysis of the Nannochloropsis organellar genomes reveals novel genetic variations in key metabolic genes. BMC Genomics. 2014;15(1):212. doi: 10.1186/1471-2164-15-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farnham PJ, Platt T. Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res. 1981;9(3):563–577. doi: 10.1093/nar/9.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Letsch MR, Lewis LA. Chloroplast gene arrangement variation within a closely related group of green algae (Trebouxiophyceae, Chlorophyta) Mol Phylogenet Evol. 2012;64(3):524–532. doi: 10.1016/j.ympev.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 58.Darling AE, Mau B, Perna NT. ProgressiveMauve: Multiple Genome Alignment with Gene Gain. Loss and Rearrangement. PLoS One. 2010;5(6):e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe S, Nakayama T. Ultrastructure and phylogenetic relationships of the unicellular green algae Ignatius tetrasporus and Pseudocharacium americanum (Chlorophyta) Phycol Res. 2007;55(1):1–16. doi: 10.1111/j.1440-1835.2006.00439.x. [DOI] [Google Scholar]
- 60.Cocquyt E, Verbruggen H, Leliaert F, Zechman F, Sabbe K, De Clerck O. Gain and loss of elongation factor genes in green algae. BMC Evol Biol. 2009;9(1):39. doi: 10.1186/1471-2148-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Škaloud P, Kalina T, Nemjová K, De Clerck O, Leliaert F. Morphology and phylogenetic position of the freshwater green microalgae Chlorochytrium (Chlorophyceae) and Scotinosphaera (Scotinosphaerales, ord. nov., Ulvophyceae) J Phycol. 2013;49(1):115–129. doi: 10.1111/jpy.12021. [DOI] [PubMed] [Google Scholar]
- 62.Gile GH, Novis PM, Cragg DS, Zuccarello GC, Keeling PJ. The distribution of Elongation Factor-1 alpha (EF-1a), Elongation Factor-Like (EFL), and a non-canonical genetic code in the Ulvophyceae: discrete genetic characters support a consistent phylogenetic framework. J Eukaryot Microbiol. 2009;56(4):367–372. doi: 10.1111/j.1550-7408.2009.00411.x. [DOI] [PubMed] [Google Scholar]
- 63.Cocquyt E, Gile G, Leliaert F, Verbruggen H, Keeling P, De Clerck O. Complex phylogenetic distribution of a non-canonical genetic code in green algae. BMC Evol Biol. 2010;10(1):327. doi: 10.1186/1471-2148-10-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leliaert F, Rueness J, Boedeker C, Maggs CA, Cocquyt E, Verbruggen H, et al. Systematics of the marine microfilamentous green algae Uronema curvatum and Urospora microscopica (Chlorophyta) Eur J Phycol. 2009;44(4):487–496. doi: 10.1080/09670260903229540. [DOI] [Google Scholar]
- 65.Marin B. Nested in the Chlorellales or independent class? Phylogeny and classification of the Pedinophyceae (Viridiplantae) revealed by molecular phylogenetic analyses of complete nuclear and plastid-encoded rRNA operons. Protist. 2012;163(5):778–805. doi: 10.1016/j.protis.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 66.Arora M, Anil AC, Leliaert F, Delany J, Mesbahi E. Tetraselmis indica (Chlorodendrophyceae, Chlorophyta), a new species isolated from salt pans in Goa. India. Eur J Phycol. 2013;48(1):61–78. doi: 10.1080/09670262.2013.768357. [DOI] [Google Scholar]
- 67.Matsumoto T, Shinozaki F, Chikuni T, Yabuki A, Takishita K, Kawachi M, et al. Green-colored plastids in the dinoflagellate genus Lepidodinium are of core chlorophyte origin. Protist. 2011;162(2):268–276. doi: 10.1016/j.protis.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 68.Hollants J, Leliaert F, Verbruggen H, De Clerck O, Willems A. Host specificity and coevolution of Flavobacteriaceae endosymbionts within the siphonous green seaweed Bryopsis. Mol Phylogenet Evol. 2013;67(3):608–614. doi: 10.1016/j.ympev.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 69.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 70.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 72.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns - a review. Gene. 1989;82(1):5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 75.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 76.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:1–19. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: Multiple Alignment of Conserved Genomic Sequence With Rearrangements. Genome Res. 2004;14(7):1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hilker R, Sickinger C, Pedersen CNS, Stoye J. UniMoG—a unifying framework for genomic distance calculation and sorting based on DCJ. Bioinformatics. 2012;28(19):2509–2511. doi: 10.1093/bioinformatics/bts440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bergeron A, Mixtacki J, Stoye J. A unifying view of genome rearrangements. In: Bücher P, Moret BE, editors. Algorithms in Bioinformatics. Berlin Heidelberg: Springer; 2006. pp. 163–173. [Google Scholar]
- 82.Yancopoulos S, Attie O, Friedberg R. Efficient sorting of genomic permutations by translocation, inversion and block interchange. Bioinformatics. 2005;21(16):3340–3346. doi: 10.1093/bioinformatics/bti535. [DOI] [PubMed] [Google Scholar]
- 83.Melton JT, Leliaert F, Tronholm A, Lopez-Bautista JM. The complete chloroplast and mitochondrial genomes of the green macroalga Ulva sp. UNA00071828 (Ulvophyceae, Chlorophyta). PLoS One. 2015:in press. [DOI] [PMC free article] [PubMed]
- 84.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 85.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 86.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 87.Rambaut A, Drummond AJ: Tracer v1.4, Available from http://beast.bio.ed.ac.uk/Tracer 2007.
- 88.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 89.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 90.Miller MA, Pfeiffer W, Schwartz T. "Creating the CIPRES Science Gateway for inference of large phylogenetic trees". New Orleans, LA: Proceedings of the Gateway Computing Environments Workshop (GCE); 2010. pp. 1–8. [Google Scholar]
- 91.Boudreau E, Turmel M, Goldschmidt-Clermont M, Rochaix JD, Sivan S, Michaels A, et al. A large open reading frame (orf1995) in the chloroplast DNA of Chlamydomonas reinhardtii encodes an essential protein. Mol Gen Genet. 1997;253(5):649–53. doi: 10.1007/s004380050368. [DOI] [PubMed] [Google Scholar]
- 92.Simpson CL, Stern DB. The treasure trove of algal chloroplast genomes. Surprises in architecture and gene content, and their functional implications. Plant Physiol. 2002;129:957–66. doi: 10.1104/pp.010908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smith DR, Burki F, Yamada T, Grimwood J, Grigoriev IV, Van Etten JL, et al. The GC-rich mitochondrial and plastid genomes of the green alga Coccomyxa give insight into the evolution of organelle DNA nucleotide landscape. PLoS One. 2011;6(8):e23624. doi: 10.1371/journal.pone.0023624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de Cambiaire JC, Otis C, Lemieux C, Turmel M. The complete chloroplast genome sequence of the chlorophycean green alga Scenedesmus obliquus reveals a compact gene organization and a biased distribution of genes on the two DNA strands. BMC Evol Biol. 2006;6:37. doi: 10.1186/1471-2148-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smith DR, Lee RW, Cushman JC, Magnuson JK, Tran D, Polle JEW. The Dunaliella salina organelle genomes: large sequences, inflated with intronic and intergenic DNA. BMC Plant Biol. 2010;10:83. doi: 10.1186/1471-2229-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Smith DR, Lee RW. The mitochondrial and plastid genomes of Volvox carteri: bloated molecules rich in repetitive DNA. BMC Genomics. 2009;10:132. doi: 10.1186/1471-2164-10-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hamaji T, Smith DR, Noguchi H, Toyoda A, Suzuki M, Kawai-Toyooka H, et al. Mitochondrial and plastid genomes of the colonial green alga Gonium pectorale give insights into the origins of organelle DNA architecture within the Volvocales. PLoS One. 2013;8(2):e57177. doi: 10.1371/journal.pone.0057177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maul JE, Lilly JW, Cui LY, de Pamphilis CW, Miller W, Harris EH, et al. The Chlamydomonas reinhardtti plastid chromosome: Islands of genes in a sea of repeats. Plant Cell. 2002;14(11):2659–79. doi: 10.1105/tpc.006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brouard J-S, Otis C, Lemieux C, Turmel M. Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): Unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer. BMC Genomics. 2008;9(1):290. doi: 10.1186/1471-2164-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]


