Abstract
Background
Neisseria gonorrhoeae (GC) is a Gram-negative pathogen that most commonly infects mucosal surfaces, causing sexually transmitted urethritis in men and endocervicitis in women. Serious complications associated with these infections are frequent and include pelvic inflammatory disease, ectopic pregnancy, and infertility. The incidence of gonorrhea cases remains high globally while antibiotic treatment options, the sole counter measures against gonorrhea, are declining due to the remarkable ability of GC to acquire resistance. Evaluating of potential drug targets is essential to provide opportunities for developing antimicrobials with new mechanisms of action. We propose the GC Obg protein, belonging to the Obg/CgtA GTPase subfamily, as a potential target for the development of therapeutic interventions against gonorrhea, and in this study perform its initial functional and biochemical characterization.
Results
We report that NGO1990 encodes Obg protein, which is an essential factor for GC viability, associates predominantly with the large 50S ribosomal subunit, and is stably expressed under conditions relevant to infection of the human host. The anti-Obg antisera cross-reacts with a panel of contemporary GC clinical isolates, demonstrating the ubiquitous nature of Obg. The cellular levels of Obg reach a maximum in the early logarithmic phase and remain constant throughout bacterial growth. The in vitro binding and hydrolysis of the fluorescent guanine nucleotide analogs mant-GTP and mant-GDP by recombinant wild type and T192AT193A mutated variants of Obg are also assessed.
Conclusions
Characterization of the GC Obg at the molecular and functional levels presented herein may facilitate the future targeting of this protein with small molecule inhibitors and the evaluation of identified lead compounds for bactericidal activity against GC and other drug-resistant bacteria.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-015-0453-1) contains supplementary material, which is available to authorized users.
Keywords: Neisseria gonorrhoeae, Drug resistance, Obg proteins, GTPase, Drug target, Mant guanine nucleotides
Background
Neisseria gonorrhoeae (GC) is a Gram-negative bacterium and a human-specific pathogen that causes gonorrhea. This sexually transmitted disease remains a global health burden. The World Health Organization estimated 106.1 million new cases in adults in 2008, which was a 21 % increase compared to 2005 [1]. The disease usually manifests as cervicitis, urethritis, proctitis, conjunctivis, or pharyngitis. A significant proportion of women (≥50 %) and some men (≤10 %) undergo asymptomatic infections and therefore many cases remain undiagnosed [2]. Untreated or inadequately treated gonorrhea often has serious long-term health consequences including endometritis, pelvic inflammatory disease, ectopic pregnancy, epididymitis, and infertility [2–4]. The serious sequelae of gonorrhea are exacerbated by a significant increase of the risk of HIV acquisition [5]. Pharmaceutical interventions against GC infections are limited to antibiotic regimens, as a preventive anti-gonorrhea vaccine does not exist. Antibiotic therapies, however, have been continually challenged by the remarkable ability of the bacteria to acquire and retain resistance [6]. Treatment failures associated with the current emergence of GC with decreased susceptibility to the last effective treatment option, third-generation cephalosporins, are concerning and emphasize the pressing need for the development of alternative antimicrobial strategies to combat drug-resistant gonorrhea [1, 6–14].
Here, we focus on biochemical and functional characterization of the GC homolog of conserved bacterial Obg GTPases, NGO1990 (henceforth ObgGC), as a target for the discovery of anti-gonorrhea compounds. Obg proteins (also recognized as YhbZ or CgtA) belong to the OBG-HflX superfamily within the TRAFAC (translational factors) class of P-loop (phosphate-binding loop) GTPases (reviewed in [15, 16]). The OBG family is comprised of four subfamilies: Obg, Nog1, DRG, and YchF. The family exists in all three domains of life with bacteria possessing Obg and YchF, archea having two Obg proteins and YchF, and eukaryotes commonly encoding four Obg proteins and YchF [15–18]. Structurally, the bacterial Obg proteins contain highly conserved N-terminal- and central-domains, and C-terminal domain that can vary in length and sequence, or as in Chlamydia, may even be absent [16, 19]. The N-terminal domain is glycine-rich and has a unique fold, the Obg fold [20]. The signature GTP-binding domain shares overall topology with the small Ras-like GTPases, but the biochemical features of the Obg proteins are distinct from those of eukaryotic Ras-like proteins [21–26].
The name Obg originates from spo0B-associated GTP-binding protein of Bacillus subtilis, in which the obg gene was identified as a part of the spo0B operon [27]. Since its identification in 1989, Obg homologs have been demonstrated to be essential for viability not only in B. subtilis, but also Streptomyces coelicolor, Staphylococcus pneumoniae, S. aureus, Haemophilus influenzae, Caulobacter crescentus, Escherichia coli, Vibrio harveyi and V. cholerae [27–33]. These findings strongly suggest that Obg proteins are crucial for the survival of both gram-positive and gram-negative bacteria. The depletion of cellular Obg levels results in species-specific pleiotropic effects on bacterial physiology, including alterations of ribosome maturation and DNA synthesis; cell division and morphology; and induction of general, as well as (p)ppGpp-mediated stringent stress responses [15, 16, 34]. Growing lines of evidence support the link between Obg and ribosome function. Obg predominantly associates with the 50S ribosomal particles, and long-term Obg depletion results in reduced levels of 70S monosomes, ribosomal proteins S1, S14, S21 and L10, as well as a perturbed polyribosome profile [17, 22, 32, 35–37]. Recent studies suggest that in E. coli cultured under standard laboratory growth conditions, Obg acts as a checkpoint in the final steps of 50S subunit assembly and, via an interplay with (p)ppGpp, might modulate the production of large ribosomal particle in response to environmental cues [34]. Introduction of the temperature-sensitive variant of obg, G80E, in C. crescentus caused a decline in both the growth rate and the amount of 50S subunits, even under permissive conditions [38]. Additional ribosome defects were not observed in non-permissive temperatures; however, the bacteria rapidly halted cell cycle progression and lost viability. Thus, the essential nature of Obg likely does not result directly from its function in the late stages of 50S subunit assembly [15, 38]. Obg might provide a key molecular nexus between different metabolic pathways to regulate cellular processes in response to the energy status of the cell [16]. All Obg proteins characterized to date bind GDP and GTP, and display relatively slow GTP hydrolysis, which can be moderately stimulated in the presence of purified 50S ribosomal particles [22, 24–26, 34, 39]. The mechanistic insights into how Obg participates in different metabolic paths and stress responses remain to be elucidated. Nevertheless, the Obg proteins appear to be promising molecular targets for the development of broad-spectrum antibiotics against drug-resistant bacterial infections because of their essential nature, conservation, and strong link with pivotal physiological processes.
Results and discussion
Local gene context and ObgGC domain architecture
In virtually all bacteria the obg gene has been reported to be physically linked to rplU and rpmA, encoding 50S ribosomal proteins L21 and L27, respectively [15]. The inspection of the genetic organization of the obg region in available completed genome sequences of Neisseriaceae, however, revealed that this arrangement is not followed. In GC strain FA1090, the putative protein NGO1990, annotated as Obg, is the last ORF in a cluster comprised of NGO1989 and NGO1988. NGO1989 is a hypothetical protein present also in GC DGI18 and PID24-1, whereas NGO1988, encoding a homolog of the rRNA small subunit methyltransferase I, is located upstream of obg in other Neisseriaceae.
Analysis of the predicted amino acid sequence of NGO1990 from FA1090 revealed a typical structure of Obg GTPases (Fig. 1) and significant similarities to other Obg proteins (Table 1). The N-terminal domain of ObgGC (amino acids 3–158) contains 26 glycine residues, similar to the B. subtilis Obg [20]. The central, GTP-binding domain (residues 160–348), includes five conserved G motifs (G1-G5) and two switch elements (switch I and switch II) that determine the active or inactive state of the G protein [15, 16]. As expected, the C-terminal domain shows the lowest conservation when compared to Obg homologs from different bacterial species (Additional file 1: Figure S1). Nevertheless, this region of ObgGC contains clusters of acidic residues, a feature characteristic of Obg proteins [17, 33]. This charged C-terminus has been shown to be important for Obg association with 50S ribosomal particles in C. crescentus as well as GTP and GDP binding in V. harveyi [17, 26].
Fig. 1.

ObgGC domain architecture. The individual structural domains of ObgGC are shown in green. The N-terminal domain (amino acids 3–158) is glycine-rich. The central, GTP-binding domain (residues 160–348) includes two switch elements (switch I and switch II) and five conserved G motifs (G1-G5; indicated in blue boxes). The C-terminal domain contains clusters of acidic residues. The conserved T192 and T193 residues within the G2 motifs and introduced substitutions are designated in red
Table 1.
Comparison of the amino acid sequences of the N. gonorrhoeae Obg protein with Obg homologs
| Organism | Accession number | Protein length | Region aligned | % identity | % similarity |
|---|---|---|---|---|---|
| Neisseria gonorrhoeae NCCP11945 | B4RQP4 | 384 | 1-384 | 100 | 100 |
| Neisseria meningitidis MC58 | Q9JXE5 | 384 | 1-384 | 98 | 98 |
| Neisseria lactamica 020-06 | E4ZAV0 | 384 | 1-384 | 97 | 97 |
| Neisseria weaveri LMG 5135 | G2DJV3 | 384 | 1-384 | 85 | 91 |
| Escherichia coli K12 | P42641 | 390 | 1-344 | 56 | 71 |
| Caulobacter crescentus NA1000/CB15N | B8GYI7 | 354 | 1-320 | 52 | 67 |
| Bacillus subtilis 168 | P20964 | 428 | 2-328 | 49 | 68 |
| Chlamydia trachomatis D/UW-3/Cx | O84423 | 335 | 2-335 | 43 | 62 |
| Homo sapiens | Q9H4K7* | 406 | 72-364 | 40 | 58 |
| Homo sapiens | A4D1E9** | 308 | 77-293 | 35 | 52 |
| Saccharomyces cerevisiae 204508 | P38860 | 518 | 295-488 | 40 | 61 |
*ObgH1 (GTP binding protein 5, GTBP5; mitochondrial ribosome-associated GTPase 2, MTG2)
**ObgH2 (GTP binding protein 10, GTPBP10)
Purification of Obg variants and evaluation of anti-ObgGC antisera
To begin the characterization of ObgGC, N- and C-terminally His-tagged versions of the wild type NGO1990, N-His-ObgGC and C-His-ObgGC (respectively), were overexpressed in E. coli BL21(DE3), and purified. The purified recombinant N-His-ObgGC was subsequently used to obtain polyclonal rabbit anti-ObgGC antisera. The antisera specifically recognized both the native and recombinant versions of ObgGC (Fig. 2a). The purified proteins migrated in SDS-PAGE more slowly than the native protein and accordingly with the deduced molecular mass of ObgGC (41.998 kDa) with the addition of the histidine epitope. Further, the antibodies cross-reacted with Obg homologs in the N. meningitidis serogroup B strains MC58 and NZ98/254 but failed to recognize Obg from N. weaveri and E. coli despite their 85 and 56 % identity to ObgGC, respectively (Fig. 2b and Table 1). It is possible that the anti-ObgGC antisera bind to the highly variable C-terminal domain of ObgGC.
Fig. 2.
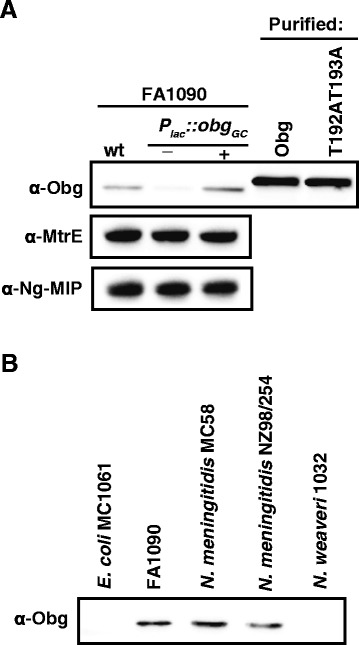
Validation of polyclonal rabbit anti-ObgGC antisera. (a) The polyclonal rabbit anti-ObgGC antibodies were used to probe the whole-cell lysates derived from wild type FA1090 and isogenic Plac::obg GC as well as purified variants of ObgGC. The bacteria were harvested following 2 h of growth in GCBL with (+) and without (−) 100 μM IPTG, and the samples were matched by equivalent OD600 units. Purified recombinant proteins (40 ng) include wild type ObgGC with N-terminal 6 × His tag, (N-His-ObgGC) and N-His-ObgGC with T192AT193A substitutions. (b) Samples of whole-cell lysates derived from various Neisseria species, as indicated, were harvested from GCB and matched by equivalent OD600 units. All samples were separated in 4-20 % Mini-PROTEAN TGX precast gels, the proteins were transferred onto the nitrocellulose membrane and probed with polyclonal rabbit anti-ObgGC antisera raised against N-His-ObgGC
ObgGC binds GTP and GDP
GTPases cycle between being turned “on” in the GTP-bound state and turned “off” in the GDP-bound state (Fig. 3a). In each state, G proteins undergo conformational changes and downstream effectors sense the GTP-bound protein complexes. Switch-off involves the exchange of GTP for GDP or hydrolysis of the γ-phosphate of GTP. The fluorescent N-methyl-3’-O-anthranoyl (mant) guanine nucleotide analogs, mant-GTP and mant-GDP, have been widely utilized for examining the nucleotide binding and GTP hydrolysis of various G-proteins including Obg homologs. The highly environmentally sensitive fluorescence of the mant group enables detection of nucleotide-protein interaction [22, 24, 26, 38, 40–43]. The binding of GTP to Obg requires the presence of physiological Mg2+ concentrations in C. crescentus, E. coli, and V. harveyi, whereas Obg-GDP complexes form over a wide range of Mg2+ concentrations [22, 24, 26]. The total intracellular Mg2+ content is about 100 mM in E. coli and B. subtilis and includes bound and free Mg2+, with the latter ranging from 1–5 mM [44–46].
Fig. 3.
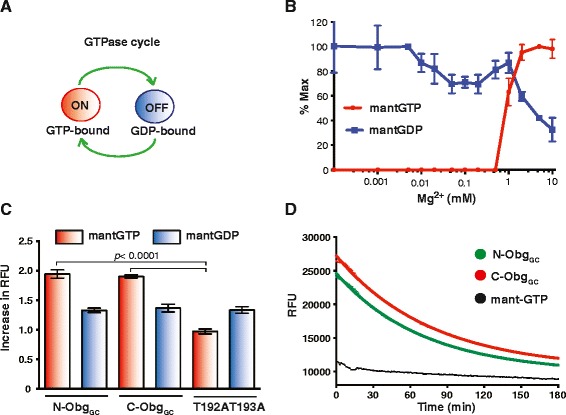
Biochemical properties of ObgGC. a GTPase cycle. Obg GTPases oscillate between active (ON, GTP-bound) and inactive (OFF, GDP-bound) states. b ObgGC binds mant-GTP and mant-GDP with guanine nucleotide-specific Mg2+ dependence. Binding of mant-GTP (red circles) and mant-GDP (blue squares) to N-His-ObgGC was assessed in the presence of varying concentrations of Mg2+. The averages with SEM from three independent experiments are shown. c Increase in Relative Fluorescence Units (RFU) of mant-GTP (red bars) and mant-GDP (blue bars) upon addition of different ObgGC variants: recombinant wild type ObgGC with N- and C-terminal 6 × His tag (N-His-ObgGC and C-His-ObgGC, respectively), and N-His-ObgGC with T192AT193A substitutions. The data shows averages with corresponding SEM of at least eight experiments performed on separate occasions. d Hydrolysis of mant-GTP by N-His-ObgGC (green) and C-His-ObgGC (red) was monitored by recording the decrease in fluorescence that is coupled to the conversion of mant-GTP-Obg to mant-GDP-Obg complexes. Data from at least four experiments were fitted to a single exponential decay equation. The fluorescence intensity of the mant-GTP in the absence of protein served as a control and is shown in black
To determine whether ObgGC requires Mg2+ to optimally bind mant-nucleotides, N-His-ObgGC was incubated with increasing concentrations of Mg2+ and either mant-GTP or mant-GDP. The ObgGC binding profiles obtained for both nucleotides differed noticeably similarly to that observed for other Obg family members [22, 24, 26]. The optimal formation of mant-GTP-ObgGC complexes occurred between 5 and 10 mM Mg2+, as indicated by maximal fluorescence (Fig. 3b, red circles). The binding of mant-GDP to ObgGC did not require Mg2+ and was inhibited at above 1 mM concentrations (Fig. 3b, blue squares).
Subsequently, binding of mant-GTP and mant-GDP was assessed for both N-and C-His-ObgGC, as addition of a six-histidine epitope to the C-terminus of the V. harveyi Obg completely abolished interaction with GTP and resulted in a weak binding of GDP [26]. Likewise, the C. crescentus Obg containing influenza virus hemagglutynin tag demonstrated a reduction in protein function [17]. In contrast, the C-His-ObgGC showed very similar properties to N-His-ObgGC (Fig. 3c). Binding of either variants of ObgGC to mant-GTP and mant-GDP led to 1.9- and 1.3-fold enhancement in mant-nucleotide fluorescence, respectively. These results suggest that subtle perturbations to the C-terminus of ObgGC are not detrimental to protein function. The C. abortus Obg naturally lacks the C-terminal domain, yet the protein is a functional GTPase, with similar activity to other Obg proteins, and binds to the 50S large ribosomal particle [19].
mantGTP hydrolysis by ObgGC
We next examined the GTPase activity of purified N-His-ObgGC and C-His-ObgGC by monitoring the decrease in fluorescence that is associated with the single-turnover conversion of bound mant-GTP to bound mant-GDP (Fig. 3d). The peak of fluorescence was recorded for 3 h at 1 min intervals. The reduction in fluorescence was fitted to a single exponential decay with a first-order rate constant, kh, of 2.3 × 10−4 s−1and 2.1 × 10−4 s−1, or half life (T1/2) of 49.1 min and 53.2 min, for N-His-ObgGC and C-His-ObgGC, respectively. Therefore, the GTP hydrolysis rates of both ObgGC variants are very similar and are approximately twenty times slower than that of the V. harveyi Obg and two-fold slower than C. crescentus and E. coli Obg proteins, respectively [22, 24, 26]. These differences may reflect distinct Obg control or function in distantly related bacterial species.
Alteration of switch I element of ObgGC abolishes GTP but not GDP binding abilities
The two adjacent threonine residues, T192 and T193, which coordinate Mg2+, are ubiquitously present within the G2 domain of Obg proteins (Fig. 1). However, their function has been assessed only in C. crescentus [47]. To address their importance for guanine nucleotide binding in ObgGC, the double T192AT193A mutant protein with N-terminal-His epitope was constructed and purified. Compared with the wild type ObgGC, the mutated protein exhibited completely impaired mant-GTP binding, whereas a 1.3-fold increase in fluorescence was observed in the presence of mant-GDP, indicating unaffected formation of Obg-GDP complexes (Fig. 3c). A similar effect was observed in the C. crescentus Obg, and T193 was identified as the pivotal residue. The obg T193A allele was not able to support C. crescentus growth, which demonstrated that the Obg GTPase activity was a prerequisite for cell viability [47].
Depletion of ObgGC has deleterious effect on GC survival
To examine whether ObgGC plays a critical function in GC physiology, we used an allelic exchange approach and placed NGO1990 under the control of the isopropyl-β-D-thiogalactoside (IPTG)-inducible promoter, Plac, in its native chromosomal locus in GC FA1090. The resulting conditional knockout strain, FA1090 Plac::obgGC, failed to grow when inoculated directly from the freezer stocks onto the gonococcal base agar solid medium (GCB) lacking IPTG, whereas robust bacterial growth was observed in the presence of the inducer (Fig. 4a).
Fig. 4.

ObgGC is essential for GC viability. a The FA1090 conditional obg knockout strain, Plac::obg GC, failed to grow when plated from freezer stocks onto GCB without (−) 100 μM IPTG, whereas abundant growth was observed on media supplemented with the inducer (+). b, c FA1090 cells carrying chromosomal Plac::obg GC were collected from GCB agar plates supplemented with 100 uM IPTG, washed, divided, and grown in GCBL in the presence or absence of IPTG for 3 h. At this experimental time point (indicated by the blue arrow), the bacteria were harvested, washed again and growth was continued for 6 h in liquid media with (+) or without (−) IPTG, as indicated. Culture density was measured as Optical Density at OD600 (b). Cell viability was monitored every hour after the second inoculation by spotting serial dilutions onto GCB with IPTG (c). Experiments were performed in biological triplicates and means and SEM are presented. d Representative immunoblot showing ObgGC levels over time in FA1090 Plac::obg GC grown in the presence (+) and absence (−) of IPTG. The samples were collected every hour after back dilution (as indicated), matched by the same OD600 units, and whole cell lysates were probed with anti-ObgGC antisera. e Cultures of FA1090 Plac::obg GC grown in the liquid media in presence (+) and absence (−) of IPTG were serially diluted and spotted on GCB with (+) and without (−) the inducer. The 2- and 4-h time points from back dilution are shown
Subsequently, to examine the effect of ObgGC depletion on GC viability over time, non-piliated and translucent colonies of FA1090 Plac::obgGC, harvested from GCB supplemented with 100 μM IPTG, were washed, suspended to the same OD600 of 0.1, divided, and cultured in gonococcal base liquid (GCBL) medium in the presence or absence of IPTG. After 2 h under repressive conditions, ObgGC was still detectable by immunoblotting (Fig. 2a) and the bacterial proliferation rate was indistinguishable from the permissive condition (Fig. 4b). Similarly, depletion of C. crescentus Obg using the Pxyl promoter and repressive growth conditions (glucose instead of xylose) resulted in much lower but detectable levels of Obg even 12 h after a carbon shift [17].
Prolonged culturing of GC is not feasible because the bacteria undergo autolysis shortly after reaching stationary phase [48–52]. Therefore to ensure significant reduction in the amount of ObgGC, the conditional knockout strain FA1090 Plac::obgGC was first treated as described above, cultures were collected 3 h after initial inoculation (indicated by an arrow in Fig. 4b), washed, and back diluted into fresh GCBL with or without the inducer. Culture density and bacterial viability, measured as Optical Density at 600 nm (OD600, Fig. 4b) and Colony-Forming Units (CFUs, Fig. 4c), respectively, were monitored every hour. Growth kinetics of FA1090 Plac::obgGC cultured in the presence of IPTG (Fig. 4a) closely followed the pattern observed in parental wild type strain (Fig. 5a). In contrast, under non-permissive conditions, the culture density and bacterial viability were decreased significantly (Fig. 4b and c, respectively), concomitant with the reduction in ObgGC level (Fig. 4d). At 6 h of the experiment, the ObgGC-depleted culture contained on average 3.46 × 105 live bacterial cells, whereas 1.87 × 108 were present in permissive conditions (Fig. 4c). Further, FA1090 Plac::obgGC grown in liquid media in the presence of IPTG was unable to survive upon plating on GCB lacking the inducer (Fig. 4e), confirming our prior observations (Fig. 4a).
Fig. 5.
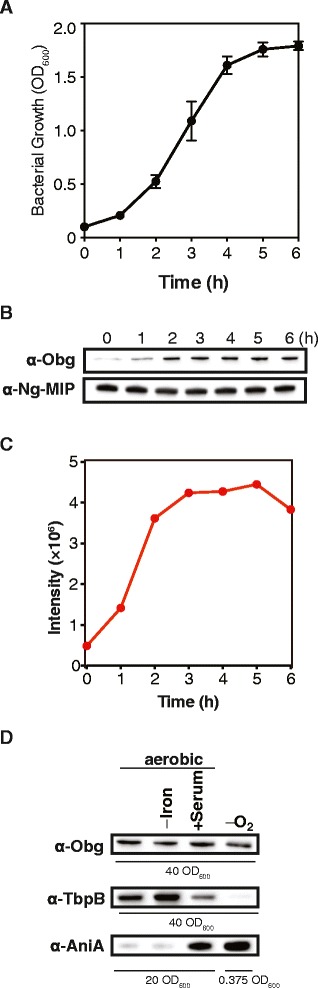
Expression of ObgGC. The growth of FA1090 (a) and ObgGC amounts (b) were examined during regular aerobic conditions in GCBL by measurements of bacterial turbidity (OD600) and immunoblotting analyses of whole cell lysates every hour. The graph shows means with corresponding SEM from biological triplicate experiments. Samples were matched by equivalent OD600 units and representative immunoblots are shown. Immuoblotting with anti-Ng-MIP antisera was used as a loading control. c The immunoblot probed with anti-ObgGC antisera was scanned and subjected to densitometric analysis. To quantify the intensity of the ObgGC protein bands, the volume tool, local background subtraction, and linear regression methods were used. d The expression of ObgGCwas assessed in whole cell lysates derived from GC cultured on GCB aerobically, in iron-limited conditions, in the presence of 7.5 % normal human sera, and anaerobically in the presence of nitrite as a terminal electron acceptor. Immunoblotting analyses with anti-TbpB and anti-AniA antisera were used as controls for iron-depleted [54] and anaerobic [55] growth conditions, respectively. Samples of whole-cell lysates were matched by the same OD600 units (40 or 20 as indicated) with the exception of detection of AniA during anaerobic growth conditions, where 0.375 OD600 units were used
Together, these studies demonstrate that Obg plays a pivotal function in GC physiology, as the depletion of ObgGC caused loss of GC viability.
Expression of ObgGC
The expression of Obg protein varies in different examined bacterial species. For instance, in S. coelicolor Obg is expressed in a growth-dependent manner with a sharp decline right after the beginning of aerial mycelium development and at the end of vegetative growth [28], whereas constant levels of Obg are maintained throughout the C. crescentus life cycle [30].
To examine the expression of ObgGC, wild type FA1090 was maintained under routine aerobic cultivation in GCBL. Bacterial proliferation was monitored by measurements of cell density at OD600 within 6 h of the experiment (Fig. 5a). Every hour GC samples were collected and the whole cell lysates were probed with anti-ObgGC antisera. The same samples were also examined using antibodies against an unrelated protein, Ng-MIP (Fig. 5b) and by SDS-PAGE coupled with colloidal coomassie staining (Additional file 2: Figure S2B) as loading controls. Immunoblotting and densitometry analyses (Fig. 5b and c, respectively) showed that ObgGC reached maximum expression in the early logarithmic phase of GC growth at OD600 ~ 0.5 (2 h from the start of the experiment), and remained constant until stationary phase.
We also asked whether the conditions that more closely resemble clinical infection, such as anoxia, iron deprivation and, in the event of disseminated infection, exposure to human serum [2, 53], influence expression of ObgGC. As expected, immunoblotting analysis showed increased levels of TbpB and AniA, which are well-recognized protein markers for iron-limited [54] and anaerobic [55] growth conditions, respectively (Fig. 5d). In contrast, the cellular concentrations of ObgGC remained unaltered during growth of wild type FA1090 on GCB aerobically, in iron-limited conditions, in the presence of 7.5 % normal human sera, and anaerobically in the presence of nitrite as a terminal electron acceptor.
Subcellular localization of ObgGC
The subcellular fractionation experiments showed that Obg was localized in the cytosol [35] and partially associated with the crude cell envelopes in E. coli [56], whereas in S. coelicolor, immunoelectron microscopy indicated that Obg was associated with the cytoplasmic membrane [28]. In addition, a growing number of reports show that different members of the Obg family cofractionate primarily with the 50S ribosomal subunit [17, 22, 32, 57]; however, in Mycobacterium tuberculosis, Obg is present in the 30S, 50S, and 70S ribosomal fractions [37].
To examine the cellular localization of ObgGC, wild type FA1090 was cultured under standard laboratory conditions in GCBL and harvested at the mid-exponential phase of growth. The bacterial cells were lysed and the cell envelope proteins were separated from the cytosolic proteome by a sodium carbonate extraction procedure [58]. The crude cell envelopes were treated with sarkosyl to solubilize the inner membrane proteins, and the outer membrane was recovered by ultracentrifugation [59]. The same total amounts of purified subproteomes (15 μg) were separated by SDS-PAGE and probed with anti-ObgGC antisera (Fig. 6a). The outer membrane protein, MtrE, which contains an extended periplasmic tunnel [60], was used as the cell envelope protein marker. As expected, MtrE was absent in the cytosol and enriched in the sarkosyl-insoluble fraction, whereas the vast majority of ObgGC was present in the cytosolic protein fractions (Fig. 6a). A faint band of the same molecular weight was also detected in the cytoplasmic membrane. Many cytoplasmic proteins are repeatedly identified in different cell envelope proteomics studies and are often considered “contaminants” [61]. However, recent thorough sequential biochemical fractionations of E. coli, combined with mass spectrometry, demonstrated that many of these proteins, including Obg, form an actual peripheral inner membrane proteome linked via functional and/or structural oligomeric complexes [62].
Fig. 6.

Obg is a primarily cytoplasmic protein that predominantly fractionates together with the 50S ribosomal particles. a Samples of isolated subproteome fractions (as indicated; 15 μg) obtained from wild type FA1090 cultured under regular aerobic laboratory growth conditions were separated in 4-20 % gradient gels and probed with either polyclonal rabbit anti-ObgGC antisera or monoclonal mouse anti-MtrE antibodies. b FA1090 cell lysate was subjected to separation on a 10-47 % step sucrose gradient by ultracentrifugation at 174,000 × g for 4 h and monitored by UV absorbance at 254 nm. The positions of the 30S and 50S ribosomal subunits, the 70S monosomes, and polyribosomes are indicated. The level of ObgGC in the collected fractions was detected by immunoblotting with polyclonal rabbit anti-ObgGC antibodies. A representative polyribosome profile and immunoblot are shown
Next, we addressed whether ObgGC cofractionates with the ribosomes by ultracentrifugation of GC cell lysates through sucrose gradients and analysis of the polyribosomes profiles. Most of the cellular proteins accumulated at the top of the gradient followed by the peaks for small and large ribosomal subunits, the 70S monosomes, and the polyribosomes (Fig. 6b). Immunoblotting analysis with anti-ObgGC antisera showed that under these conditions, the greatest amounts of ObgGC were in 50S fractions and at the top of the gradient.
Based on these results, we conclude that ObgGC is largely localized to the cytosol and primarily associates with the 50S ribosomal particle and not with the 70S monosomes or with translating ribosomes. Association of a part of the ObgGC cellular pool with the cytoplasmic membrane may have functional implications, as Obg has been shown to be involved in key cellular processes such as ribosome maturation, DNA synthesis, cell division and morphology. Obg could be recruited to the membrane-bound complexes on demand, depending on the metabolic status of the bacterial cell.
ObgGC is expressed by contemporary clinical isolates of GC
Finally, the conservation of the predicted amino acid sequence of ObgGC was assessed using the completed genome sequences of strains FA1090 (Gen Bank accession number AE004969) and NCCP11945 (Gen Bank accession number CP001050), as well as the draft genome sequences of 14 different GC strains (downloaded from the Broad Institute website http://www.broadinstitute.org/annotation/genome/neisseria_gonorrhoeae/MultiHome.html). These analyses demonstrated that ObgGC is 100 % identical among 14 strains and has a single amino acid change in the GC isolates designated as DGI2 and PID18 (Additional file 3: Figure S3).
Subsequently, to examine expression of ObgGC, a diversified panel of GC isolates was utilized. This panel included common laboratory strains MS11, F62, and 1291, as well as 32 strains isolated from different gonorrhea patients from distinct geographical areas and at different time points. The anti-ObgGC antibodies detected a band of the same size in all examined GC isolates, albeit the level of expression varied between some strains (Fig. 7).
Fig. 7.
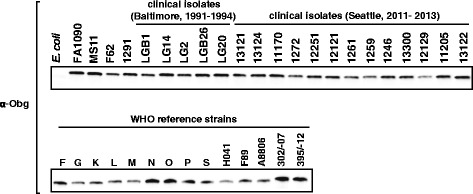
The polyclonal rabbit anti-ObgGC antibodies cross-react with cell lysates of 36 diverse GC strains. Samples of whole-cell lysates derived from various GC isolates (as indicated) harvested from GCB were matched by equivalent OD600 units and resolved in 4-20 % Tris-glycine precast gels. The proteins were transferred onto nitrocellulose membrane and probed with polyclonal rabbit anti-ObgGC antisera
Conclusions
Targeting essential proteins and critical cellular processes that are widely conserved remains an attractive avenue in antibacterial drug discovery programs. Compounds interfering with ribosome function and biogenesis and thus inhibiting different aspects of protein synthesis are among the most clinically useful antibiotics in spite of evolutionary conservation of bacterial and mitochondrial ribosomes [63].
Here, we show for the first time that ObgGC is a GTPase essential for GC viability, mainly associated with the 50S large ribosomal subunit, abundant during different growth phases as well as under environmental conditions relevant to infection, and conserved in GC isolates. Together, these findings underscore the potential of ObgGC as a target for the development of therapeutics against gonorrhea.
Methods
Bacterial strains, plasmids, and growth conditions
Strain of GC FA1090 [64] was primarily used in this study. Additionally, we employed: MS11 [65], 1291 [66], F62 [67], FA19 [68], isolates LGB1, LG14, LG20, and LG26 collected from two public health clinics in Baltimore between the years 1991–1994 [58], 13 strains derived from different patients seen at the Public Health–Seattle & King County STD clinic in 2011–2013 (Wierzbicki, et al., manuscript in preparation), as well as 14 WHO reference strains [69, 70]. Clinical isolates were kindly provided by Olusegun O. Soge, and King K. Holmes (Departments of Medicine and Global Health, University of Washington, Seattle, WA) and Magnus Unemo (Örebro University Hospital, Örebro, Sweden).
GC was maintained on GCB Medium Base agar plates with Kellogg’s supplements in 5 % CO2 atm at 37 °C or in GCBL Medium Base Broth containing Kellogg’s supplements and sodium bicarbonate (at a final concentration of 0.042 %) at 37 °C [48, 71, 72]. To achieve iron limited conditions, GCB without ferric nitrate in Kellogg’s Supplements and with deferoxamine mesylate salt (Desferal, Sigma) at 5 μM final concentration was utilized [73]. In addition, when stated in the text, GCB supplemented with 7.5 % normal human serum [74] was used to grow GC.
The E. coli strain NEB5α was used for genetic manipulations. Bacteria were streaked from −80 °C on Luria-Bertani (LB) agar supplemented with kanamycin (50 μg/mL) when needed. E. coli strains were cultured in LB medium at 37 °C. All media utilized in this study were purchased from Difco.
Construction of recombinant wild type and mutated versions of ObgGC
All oligonucleotide primers designed and used in this study were synthesized by IDT DNA Technologies. Relevant restriction sites within primers are underlined. Recombinant N-His-ObgGC and C-His-ObgGC were generated by amplifying the obg gene from genomic DNA using two pairs of primers, respectively: obg-f 5’GAATTCCATATGAAATTCATCGACGAAGCAAAA3’ and obg-r 5’GAATTCAAGCTTTTACTCCGGCTTAAACACG3’; as well as obg-c-f 5’GACTCCATGGAATTCATCGACGAAGCAAAAATCG3’ and obg-c-r 5’GACTAAGCTTCTCCGGCTTAAACACGCC3’. The corresponding PCR products were digested with NdeI-HindIII or NcoI-HindIII (respectively) and cloned into similarly digested pET28a(+) to create plasmids pET-N-His-ObgGC and pET-C-His-ObgGC. Mutagenesis of T192 and T193 residues into alanine were accomplished using the QuickChange II site directed mutagenesis kit (Agilent). Mutations were introduced using oligonucleotides: T192AT193A-f 5’GTTCGGATGCAGGGCGGCGAAGGGGTAGTTGG3’ and T192AT193A-r 5’CCAACTACCCCTTCGCCGCCCTGCATCCGAAC3’, and pET-N-His-ObgGC as the DNA template. The presence of the desired mutations was confirmed by sequencing at the Center for Genome Research and Biocomputing at Oregon State University.
Protein purification
The E. coli BL21(DE3) was used as a host for expression of all ObgGC variants. Bacteria harboring N-His-ObgGC, C-His-ObgGC, or T192AT193A were cultured in LB at 37 °C until the cultures reached OD600 of ~ 0.5, and the expression of individual ObgGC variants was induced by the addition of IPTG to a final concentration of 1 mM. Cells were pelleted 3 h after induction and resuspended in lysis buffer (20 mM Tris–HCl pH 8.0, 10 mM imidazole, 450 mM NaCl). The cell lysis was carried out by passing the suspension five times through a French pressure cell press at 12,000 psi. Bacterial debris was removed by centrifugation and the clarified crude cell extracts were loaded onto HisPur Cobalt Resin (Thermo) equilibrated with lysis buffer. Columns were subsequently washed with solution containing 20 mM Tris–HCl pH 8.0, 20 mM imidazole, and 450 mM NaCl. Proteins were eluted with the same buffer containing 250 mM imidazole. The eluate was dialyzed two times for an hour and overnight against 20 mM Tris–HCl pH 8.0. The purified proteins were concentrated using Microsep Advance Centrifugal Devices with 10 K molecular cutoff (PALL). Protein concentrations were determined using the Bradford method with a Protein Assay Kit (BioRad). Glycerol was added to purified proteins to a final concentration of 10 % and proteins were stored at −80 ° C until further use.
Polyclonal rabbit anti-ObgGC antisera
The polyclonal anti-ObgGC antibodies were prepared by Pacific Immunology (Ramona, CA, USA) using the purified N-His-ObgGC, two New Zealand White rabbits, and a 13-week antibody production protocol approved by IACUC Animal Protocol #1, in a certified animal facility (USDA 93-R-283) and the NIH Animal Welfare Assurance Program (#A4182-01).
Biochemical assays
The binding and hydrolysis of guanidine nucleotides were executed as described [26]. All fluorescence measurements were performed at 37 °C using a Synergy HT plate reader (BioTek). To determine the concentration of Mg2+ required for binding of mant-GTP or mant-GDP (Life Technologies), N-His-ObgGC (2 μM) was incubated with either mant nucleotide (0.3 μM) and increasing concentrations of Mg2+ (from 0 to 10 mM) in binding buffer B containing 50 mM Tris–HCl pH 8.0, 50 mM KCl, 2 mM dithiothreitol, 10 μM ATP, and 10 % (wt/vol) glycerol. The experiments were repeated on three separate occasions and the data is presented as the percentage of maximal measured relative fluorescence units.
The ability to form GTP-and GDP-Obg complexes was studied using 0.3 μM mant-GTP or mant-GDP analogs in binding buffer B with or without 5 mM Mg2+, respectively, and 2 μM purified ObgGC protein (N-His-ObgGC, T192AT193A, or C-His-ObgGC). These studies were carried out with at least eight biological replicates, and means with corresponding standard error of the mean (SEM) are reported.
To assess the GTP hydrolysis rate of ObgGC, N-His-ObgGC or C-His-ObgGC (16 μM) was prebound to 0.3 μM mant-GTP in binding buffer B supplemented with 5 mM MgCl2. The decrease in fluorescence associated with conversion of mant-GTP-Obg complexes into mant-GDP-Obg was recorded in 1 min intervals for 3 h. Data were fitted to a single exponential decay equation using GraphPad Prism 6.0f (Graph Pad Software). The single turnover rate constant and the half-life of hydrolysis were obtained from at least four independent experiments.
Construction of conditional obgGC mutant strain
The conditional obgGC mutant strain, FA1090 Plac::obgGC, was constructed using a similar approach as we previously described [58]. Briefly, a gene encoding the Obg homolog in GC, ObgGC (NGO1990), including an upstream region containing its indigenous ribosome-binding site (16 bp) was amplified with primers 5’CAAACAAGAGCATTTAATG3' and 5'AAGTTGGGCCGGCCTTACTCCGGCTTAAAC3'. To place the obgGC under the control of the Plac promoter, the resulting 1189 bp PCR product was digested with FseI and sub-cloned into ScaI-FseI digested pGGC4. This vector contains an IPTG-inducible promoter, which enables the controlled expression of a cloned gene [75]. The upstream region of obgGC was amplified with primers 5'ACTAGTGAATTCGCCTTGCTGTCGCTTTG3' and 5'ATCGATGGTACCTTGGTTTTAAATAGGGTTTCAGGC3'. The obtained 531 bp product was digested with EcoRI and KpnI, and cloned into pUC18K [76], yielding the pUC18K-obgGC-up. Next, the DNA fragment containing lacI repressor gene, Plac promoter and obgGC gene carried on the pGCC4 was amplified with primers 5'ACTCAATAGGATCCTCACTGCCCGCTTTCCAG3' and 5'CATAAGCAGTCGACTTCAGACGGCGGAGACGGCGGTAATCAGG3'. The PCR product was purified, digested with BamHI-SalI and cloned into pUC18K [76], generating pUC18K-Plac::obgGC. This final construct encompassing nonpolar kanamycin resistance cassette apha-3 [76] flanked by homologous regions for recombination and allelic exchange was used to introduce the mutation onto the FA1090 chromosome. The plasmid was linearized by digestion with NdeI and piliated bacteria were transformed with 0.1 μg of plasmid DNA in liquid media as described [77] with the exception that the growth media were supplemented with 100 μM IPTG. The resulting GC transformants were selected on GCB agar containing 40 μg/mL kanamycin and 100 μM IPTG. The FA1090 Plac::obgGC clones were verified by PCR with primers 5’GCCTTGCTGTCGCTTTG3' and 5’ GGAGACGGCGGTAATCAGG3' and immunoblotting analysis with anti-ObgGC antibodies.
ObgGC depletion studies
To assess the consequence of ObgGC depletion on GC viability, the GC FA1090 Plac::obgGC strain was plated from freezer stocks onto GCB supplemented with 100 μM IPTG. The following day, single nonpiliated colonies were passaged onto fresh GCB plates containing 100 μM IPTG and incubated approximately 18 h in 5 % CO2 atm at 37 °C. The bacteria were swabbed from plates, suspended in GCBL to OD600 of 0.1, and washed two times with GCBL that was pre-warmed to 37 °C to ensure removal of IPTG. Equal amounts of bacterial suspensions were divided into two flasks and cultured aerobically with and without 100 μM IPTG for 3 h at 37 °C. Subsequently, the cultures were diluted to OD600 of 0.1 into fresh GCBL with and without 100 μM IPTG. For measurements of OD600, bacterial viability (CFUs), and immunoblotting analysis, samples were withdrawn every hour and processed as described [58]. Experiments were performed in biological triplicates and mean values with corresponding SEM are presented.
Subcellular fractionation
Non-piliated and translucent GC colonies of wild type FA1090 were swabbed from solid media, suspended in 500 mL of GCBL to OD600 of 0.1 and cultured at 37 °C with aeration (220 rpm) to OD600 of 0.6 - 0.8. Cells were harvested by centrifugation (20 min, 6000 × g) and the crude cell envelope fraction was separated from the cytosolic proteins using a sodium carbonate extraction procedure and subsequent ultracentrifugation steps [58]. The inner membrane proteins were solubilized using 2 % Sarkosyl in 20 mM Tris–HCl pH 7.5 according to the method described by Leuzzi et al. [59]. The Sarkosyl-insoluble outer membrane fractions were recovered by ultracentrifugation for 1 h at 100,000 × g and 4 °C. The pellet was suspended in PBS containing 1 % SDS. The total protein amount in each isolated subproteome fraction was assessed using a Protein Assay Kit (Bio Rad).
Preparation of GC cell lysates for ribosome profiles
Wild type FA1090 bacteria were incubated in GCBL medium until the mid logarithmic phase of growth (OD600 ~ 0.5 to 0.7). Chloramphenicol was added to a final concentration of 100 μg/mL one minute before harvesting. Cells were harvested at 4,000 × g for 10 min at 4 °C and immediately frozen at −80 °C. After thawing on ice, bacteria were resuspended in 1 mL of lysis buffer comprised of 10 mM Tris–HCl pH 7.5, 10 mM MgCl2, 30 mM NH4Cl, 100 μg/mL chloramphenicol [78]. Subsequently, an equal volume of glass beads (100 μm; Electron Microscopy Sciences) was added to the solution and bacterial cells were lysed by vortexing every minute for 10 min with a 1 min cooling interval on ice. Lysates were clarified by centrifugation for 15 min at 21,000 × g and 4 °C.
Polyribosome fractionation
The isolated GC cell lysates in amounts corresponding to 15 OD260 units were overlaid on top of a 10 to 47 % step sucrose gradient as described previously [79]. Ribosomal subunits were separated by centrifugation in a Beckman SW41 rotor at 174,000 × g for 4 h and 4 ° C. Separated ribosomal subunits were fractionated using an Econo Pump and Econo UV Monitor (Bio-Rad). UV traces were recordered using a Model 1325 Econo Recorder (Bio-Rad) and 500 μL fractions were collected. Protein samples were precipitated with 15 % trichloroacetic acid. The resulting precipitates were solubilized in SDS loading buffer, and after separation in 10-20 % Criterion Tris-Tricine TGX (BioRad) acrylamide gel, all collected fractions were subjected to immunoblotting with anti-ObgGC antisera as described below.
SDS-PAGE and immunoblotting
Whole cell lysates were obtained from GC grown in GCBL with aeration and on GCB plates maintained under growth conditions as stated in the text. When bacterial colonies reached approximately the same size, all strains were harvested, suspended in pre-warmed GCBL, and the cell density was examined by OD600 measurement. Fractions containing either cytoplasmic, inner- or outer- membrane proteins (15 μg of proteins loaded per lane), ribosomal particles, or whole cell lysates matched by equivalent OD600 units, were prepared in SDS sample buffer in the presence of 50 mM dithiotreitol and separated in either 10-20 % Criterion Tris-Tricine TGX (BioRad) or 4-20 % Mini-PROTEAN TGX precast gels (Bio-Rad). The proteins were transferred onto 0.2 μm nitrocellulose membrane (Bio-Rad) using a Trans-blot Turbo (Bio-Rad). A solution of 5 % milk in phosphate buffered saline pH 7.0 (PBS, Li-Core) supplemented with 0.1 % Tween 20 (PBST) was used for blocking. Following 1 h of incubation, polyclonal rabbit antisera against ObgGC (1:5,000), polyclonal anti-AniA antibodies (1:10,000; [58]),monoclonal mouse anti-MtrE antisera (1:10,000; a gift of Ann Jerse, Uniformed Services University, Bethesda), monoclonal mouse anti-Ng-MIP antibodies (1:10,000; a gift of Mariagrazia Pizza, Novartis Vaccines, Italy), or polyclonal rabbit anti-TbpB antisera (1:1,000; a gift of Cynthia Cornelissen, Virginia Commonwealth University, Richmond) diluted in PBST as indicated in parenthesis were added to the membranes. The horseradish peroxidase conjugate of goat anti-rabbit IgG antisera (BioRad) or goat anti-mouse IgG antibody (ThermoFisher Scientific), correspondingly, were utilized as secondary antibodies at 1:10,000 dilution. The reactions were developed using Clarity Western ECL-Substrate (BioRad) and a Chemi-DocTM MP System (BioRad) was used for western blot imaging.
Densitometry
The immunoblot probed with anti-ObgGC antisera was scanned using the ChemiDocTM system (BioRad) and subjected to densitometric analysis using Image LabTM 5.0 software (BioRad). To quantify the intensity of the ObgGC protein bands, the volume tool (rectangle), local background subtraction, and linear regression were used.
Statistical analyses
Statistical analyses were conducted using GraphPad Prism 6.0f (Graph Pad Software) and an unpaired Student’s t-test was used to analyze the data.
Availability of supporting data
The data supporting the results of this article are included within its Additional files 1, 2 and 3: Figures S1-S3.
Acknowledgements
This work was supported by bridge funding from the College of Pharmacy (OSU) and in part by the RO1AI117235 to AES. We are grateful to Olusegun O. Soge and King K. Holmes, as well as Magnus Unemo for kindly providing GC clinical isolates. We acknowledge Peter Beernink and Dan Graanoff for sharing the N. meningitidis isolates, and Daniel Rockey for providing N. weaveri. We also thank Ann Jerse, Cynthia Cornelissen, and Mariagrazia Pizza for generous gifts of anti-MtrE, anti-TbpB, and anti-Ng-MIP antibodies, respectively.
Abbreviations
- A
Absorbance
- GC
Neisseria gonorrhoeae
- GCB
Gonococcal base agar
- GCBL
Gonococcal base liquid medium
- OD
Optical density
- IPTG
Isopropyl thio-β-D-galactopyranoside
- PBS
Phosphate buffered saline
- PBST
Phosphate buffered saline supplemented with 0.1 % Tween
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SEM
Standard error of the mean
Additional files
Sequence alignment of Obg from N. gonorrhoeae and N. meningitidis with characterized members of Obg subfamily from various bacterial species. Identical residues are indicated by a star (*); G motifs (from G1 to G5) are underlined. Accession numbers of compared Obg homologs are as follows: M. tuberculosis (Mt) (CCE37910), S. coelicolor (Sc) (BAA13498), D. radiodurans (Dr) (NP_293810), T. thermophilus (Tt) (YP_145047), B. subtilis (Bs) (AAA22505), C. crescentus (Cc) (NP_419134), N. gonorrhoeae (Ng) (YP_209010), N. meningitidis (Nm) (NP_275074), E. coli (Ec) (NP_417650), V. cholerae (Vc) (NP_230091).
Loading controls for immunoblotting experiments. Samples of whole-cell lysates were prepared for SDS-PAGE as described in the text, separated in precast gradient gels and the protein profiles were visualized using colloidal coomassie. Loaded OD600 units in individual experiments matched the corresponding samples used in immunoblotting analyses and are indicated below each gel. (A) Loading controls for immunoblotting experiment presented in Figure 2 A. (B) Loading controls for immunoblotting analysis shown in Figure 2 B. (C) Loading controls for experiment shown in Figure 5 B. (D) Loading controls for experiment in Figure 5 D for whole-cell lysates probed with anti-Obg and anti-TbpB antisera. (D) Loading controls for immunoblotting experiment with anti-AniA antisera presented in Figure 5 D.
Comparison of predicted amino acid sequences of ObgGC between different GC isolates.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AES designed the studies. RAZ engineered the recombinant wild type- and mutated- versions of ObgGC, performed bioinformatics analyses, polysome profiles, immunoblotting, and densitometry. IHW constructed the conditional FA1090 Plac::obg GC knockout strain and conducted ObgGC depletion studies, as well as subcellular fractionations. RAZ and BIB purified recombinant proteins. BIB and IHW performed SDS-PAGE coupled with colloidal coomassie staining and immunoblottings. AES, RAZ, and BIB conducted the biochemical analyses of recombinant versions of ObgGC. All authors interpreted the data. AES wrote the manuscript. All authors read and agreed to the content of the manuscript.
Contributor Information
Ryszard A. Zielke, Email: Ryszard.Zielke@oregonstate.edu
Igor H. Wierzbicki, Email: igorw@onid.oregonstate.edu
Benjamin I. Baarda, Email: baardab@onid.oregonstate.edu
Aleksandra E. Sikora, Email: Aleksandra.Sikora@oregonstate.edu
References
- 1.World Health Organization: Global action plan to control the spread and impact of antimicrobial resistance in Neisseria gonorrhoeae. In. Geneva: World Health Organization; 2012: 32 p.
- 2.Edwards JL, Apicella MA. The molecular mechanisms used by Neisseria gonorrhoeae to initiate infection differ between men and women. Clin Microbiol Rev. 2004;17(4):965–981. doi: 10.1128/CMR.17.4.965-981.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woods CR. Gonococcal infections in neonates and young children. Semin Pediatr Infect Dis. 2005;16(4):258–270. doi: 10.1053/j.spid.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Campbell DG. A case of angina pectoris with terminal uraemia. Can Med Assoc J. 1928;18(4):421. [PMC free article] [PubMed] [Google Scholar]
- 5.Fleming DT, Wasserheit JN. From epidemiological synergy to public health policy and practice: the contribution of other sexually transmitted diseases to sexual transmission of HIV infection. Sex Transm Infect. 1999;75(1):3–17. doi: 10.1136/sti.75.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unemo M, Shafer WM. Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev. 2014;27(3):587–613. doi: 10.1128/CMR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohnishi M, Golparian D, Shimuta K, Saika T, Hoshina S, Iwasaku K, et al. Is Neisseria gonorrhoeae initiating a future era of untreatable gonorrhea?: detailed characterization of the first strain with high-level resistance to ceftriaxone. Antimicrob Agents Chemother. 2011;55(7):3538–3545. doi: 10.1128/AAC.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokoi S, Deguchi T, Ozawa T, Yasuda M, Ito S, Kubota Y, et al. Threat to cefixime treatment for gonorrhea. Emerg Infect Dis. 2007;13(8):1275–1277. doi: 10.3201/eid1308.060948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unemo M, Shipitsyna E, Domeika M. Recommended antimicrobial treatment of uncomplicated gonorrhoea in 2009 in 11 East European countries: implementation of a Neisseria gonorrhoeae antimicrobial susceptibility programme in this region is crucial. Sex Transm Infect. 2010;86(6):442–444. doi: 10.1136/sti.2010.042317. [DOI] [PubMed] [Google Scholar]
- 10.Ison CA, Hussey J, Sankar KN, Evans J, Alexander S. Gonorrhoea treatment failures to cefixime and azithromycin in England. Euro Surveill. 2010;2011:16(14). [PubMed] [Google Scholar]
- 11.Allen VG, Mitterni L, Seah C, Rebbapragada A, Martin IE, Lee C, et al. Neisseria gonorrhoeae treatment failure and susceptibility to cefixime in Toronto. Canada JAMA. 2013;309(2):163–170. doi: 10.1001/jama.2012.176575. [DOI] [PubMed] [Google Scholar]
- 12.Tapsall J, Read P, Carmody C, Bourne C, Ray S, Limnios A, et al. Two cases of failed ceftriaxone treatment in pharyngeal gonorrhoea verified by molecular microbiological methods. J Med Microbiol. 2009;58(Pt 5):683–687. doi: 10.1099/jmm.0.007641-0. [DOI] [PubMed] [Google Scholar]
- 13.Unemo M, Nicholas RA. Emergence of multidrug-resistant, extensively drug-resistant and untreatable gonorrhea. Future Microbiol. 2012;7(12):1401–1422. doi: 10.2217/fmb.12.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ison CA, Deal C, Unemo M: Current and future treatment options for gonorrhoea. Sex Transm Infect 2013, 89 Suppl 4:iv52-56. [DOI] [PubMed]
- 15.Verstraeten N, Fauvart M, Versees W, Michiels J. The universally conserved prokaryotic GTPases. Microbiol Mol Biol Rev. 2011;75(3):507–542. doi: 10.1128/MMBR.00009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kint C, Verstraeten N, Hofkens J, Fauvart M, Michiels J. Bacterial Obg proteins: GTPases at the nexus of protein and DNA synthesis. Crit Rev Microbiol. 2014;40(3):207–224. doi: 10.3109/1040841X.2013.776510. [DOI] [PubMed] [Google Scholar]
- 17.Lin B, Thayer DA, Maddock JR. The Caulobacter crescentus CgtAC protein cosediments with the free 50S ribosomal subunit. J Bacteriol. 2004;186(2):481–489. doi: 10.1128/JB.186.2.481-489.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blombach F, Brouns SJ, van der Oost J. Assembling the archaeal ribosome: roles for translation-factor-related GTPases. Biochem Soc Trans. 2011;39(1):45–50. doi: 10.1042/BST0390045. [DOI] [PubMed] [Google Scholar]
- 19.Polkinghorne A, Vaughan L. Chlamydia abortus YhbZ, a truncated Obg family GTPase, associates with the Escherichia coli large ribosomal subunit. Microb Pathog. 2011;50(3–4):200–206. doi: 10.1016/j.micpath.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Buglino J, Shen V, Hakimian P, Lima CD. Structural and biochemical analysis of the Obg GTP binding protein. Structure. 2002;10(11):1581–1592. doi: 10.1016/S0969-2126(02)00882-1. [DOI] [PubMed] [Google Scholar]
- 21.Kukimoto-Niino M, Murayama K, Inoue M, Terada T, Tame JR, Kuramitsu S, et al. Crystal structure of the GTP-binding protein Obg from Thermus thermophilus HB8. J Mol Biol. 2004;337(3):761–770. doi: 10.1016/j.jmb.2004.01.047. [DOI] [PubMed] [Google Scholar]
- 22.Wout P, Pu K, Sullivan SM, Reese V, Zhou S, Lin B, et al. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J Bacteriol. 2004;186(16):5249–5257. doi: 10.1128/JB.186.16.5249-5257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan J, Jakob U, Bardwell JC. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J Bacteriol. 2002;184(10):2692–2698. doi: 10.1128/JB.184.10.2692-2698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin B, Covalle KL, Maddock JR. The Caulobacter crescentus CgtA protein displays unusual guanine nucleotide binding and exchange properties. J Bacteriol. 1999;181(18):5825–5832. doi: 10.1128/jb.181.18.5825-5832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welsh KM, Trach KA, Folger C, Hoch JA. Biochemical characterization of the essential GTP-binding protein Obg of Bacillus subtilis. J Bacteriol. 1994;176(23):7161–7168. doi: 10.1128/jb.176.23.7161-7168.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sikora AE, Datta K, Maddock JR. Biochemical properties of the Vibrio harveyi CgtAV GTPase. Biochem Biophys Res Commun. 2006;339(4):1165–1170. doi: 10.1016/j.bbrc.2005.11.129. [DOI] [PubMed] [Google Scholar]
- 27.Trach K, Hoch JA. The Bacillus subtilis spo0B stage 0 sporulation operon encodes an essential GTP-binding protein. J Bacteriol. 1989;171(3):1362–1371. doi: 10.1128/jb.171.3.1362-1371.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okamoto S, Ochi K. An essential GTP-binding protein functions as a regulator for differentiation in Streptomyces coelicolor. Mol Microbiol. 1998;30(1):107–119. doi: 10.1046/j.1365-2958.1998.01042.x. [DOI] [PubMed] [Google Scholar]
- 29.Zalacain M, Biswas S, Ingraham KA, Ambrad J, Bryant A, Chalker AF, et al. A global approach to identify novel broad-spectrum antibacterial targets among proteins of unknown function. J Mol Microbiol Biotechnol. 2003;6(2):109–126. doi: 10.1159/000076741. [DOI] [PubMed] [Google Scholar]
- 30.Maddock J, Bhatt A, Koch M, Skidmore J. Identification of an essential Caulobacter crescentus gene encoding a member of the Obg family of GTP-binding proteins. J Bacteriol. 1997;179(20):6426–6431. doi: 10.1128/jb.179.20.6426-6431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arigoni F, Talabot F, Peitsch M, Edgerton MD, Meldrum E, Allet E, et al. A genome-based approach for the identification of essential bacterial genes. Nat Biotechnol. 1998;16(9):851–856. doi: 10.1038/nbt0998-851. [DOI] [PubMed] [Google Scholar]
- 32.Sikora AE, Zielke R, Datta K, Maddock JR. The Vibrio harveyi GTPase CgtAV is essential and is associated with the 50S ribosomal subunit. J Bacteriol. 2006;188(3):1205–1210. doi: 10.1128/JB.188.3.1205-1210.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shah S, Das B, Bhadra RK. Functional analysis of the essential GTP-binding-protein-coding gene cgtA of Vibrio cholerae. J Bacteriol. 2008;190(13):4764–4771. doi: 10.1128/JB.02021-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng B, Mandava CS, Guo Q, Wang J, Cao W, Li N, et al. Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol. 2014;12(5) doi: 10.1371/journal.pbio.1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato A, Kobayashi G, Hayashi H, Yoshida H, Wada A, Maeda M, et al. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes to cells : devoted to molecular & cellular mechanisms. 2005;10(5):393–408. doi: 10.1111/j.1365-2443.2005.00851.x. [DOI] [PubMed] [Google Scholar]
- 36.Michel B. Obg/CtgA, a signaling protein that controls replication, translation, and morphological development? Dev Cell. 2005;8(3):300–301. doi: 10.1016/j.devcel.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Sasindran SJ, Saikolappan S, Scofield VL, Dhandayuthapani S. Biochemical and physiological characterization of the GTP-binding protein Obg of Mycobacterium tuberculosis. BMC Microbiol. 2011;11:43. doi: 10.1186/1471-2180-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datta K, Skidmore JM, Pu K, Maddock JR. The Caulobacter crescentus GTPase CgtAC is required for progression through the cell cycle and for maintaining 50S ribosomal subunit levels. Mol Microbiol. 2004;54(5):1379–1392. doi: 10.1111/j.1365-2958.2004.04354.x. [DOI] [PubMed] [Google Scholar]
- 39.Wittinghofer A, Vetter IR. Structure-function relationships of the G domain, a canonical switch motif. Annu Rev Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- 40.John J, Sohmen R, Feuerstein J, Linke R, Wittinghofer A, Goody RS. Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry. 1990;29(25):6058–6065. doi: 10.1021/bi00477a025. [DOI] [PubMed] [Google Scholar]
- 41.Remmers AE, Posner R, Neubig RR. Fluorescent guanine nucleotide analogs and G protein activation. J Biol Chem. 1994;269(19):13771–13778. [PubMed] [Google Scholar]
- 42.Nomanbhoy TK, Leonard DA, Manor D, Cerione RA. Investigation of the GTP-binding/GTPase cycle of Cdc42Hs using extrinsic reporter group fluorescence. Biochemistry. 1996;35(14):4602–4608. doi: 10.1021/bi951743d. [DOI] [PubMed] [Google Scholar]
- 43.Lin B, Maddock JR. The N-terminal domain of the Caulobacter crescentus CgtA protein does not function as a guanine nucleotide exchange factor. FEBS Lett. 2001;489(1):108–111. doi: 10.1016/S0014-5793(00)02402-9. [DOI] [PubMed] [Google Scholar]
- 44.Alatossava T, Jutte H, Kuhn A, Kellenberger E. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J Bacteriol. 1985;162(1):413–419. doi: 10.1128/jb.162.1.413-419.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moncany ML, Kellenberger E. High magnesium content of Escherichia coli B. Experientia. 1981;37(8):846–847. doi: 10.1007/BF01985672. [DOI] [PubMed] [Google Scholar]
- 46.Akanuma G, Kobayashi A, Suzuki S, Kawamura F, Shiwa Y, Watanabe S, et al. Defect in the formation of 70S ribosomes caused by lack of ribosomal protein L34 can be suppressed by magnesium. J Bacteriol. 2014;196(22):3820–3830. doi: 10.1128/JB.01896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin B, Skidmore JM, Bhatt A, Pfeffer SM, Pawloski L, Maddock JR. Alanine scan mutagenesis of the switch I domain of the Caulobacter crescentus CgtA protein reveals critical amino acids required for in vivo function. Mol Microbiol. 2001;39(4):924–934. doi: 10.1046/j.1365-2958.2001.02285.x. [DOI] [PubMed] [Google Scholar]
- 48.Morse SA, Bartenstein L. Factors affecting autolysis of Neisseria gonorrhoeae. Proc Soc Exp Biol Med. 1974;145(4):1418–1421. doi: 10.3181/00379727-145-38025. [DOI] [PubMed] [Google Scholar]
- 49.Elmros T, Sandstrom G, Burman L. Autolysis of Neisseria gonorrhoeae. Relation between mechanical stability and viability Br J Vener Dis. 1976;52(4):246–249. doi: 10.1136/sti.52.4.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hebeler BH, Young FE. Mechanism of autolysis of Neisseria gonorrhoeae. J Bacteriol. 1976;126(3):1186–1193. doi: 10.1128/jb.126.3.1186-1193.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hebeler BH, Young FE. Autolysis of Neisseria gonorrhoeae. J Bacteriol. 1975;122(2):385–392. doi: 10.1128/jb.122.2.385-392.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elmros T, Burman LG, Bloom GD. Autolysis of Neisseria gonorrhoeae. J Bacteriol. 1976;126(2):969–976. doi: 10.1128/jb.126.2.969-976.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Falsetta ML, Steichen CT, McEwan AG, Cho C, Ketterer M, Shao J, et al. The composition and metabolic phenotype of Neisseria gonorrhoeae biofilms. Front Microbiol. 2011;2:75. doi: 10.3389/fmicb.2011.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cornelissen CN. Identification and characterization of gonococcal iron transport systems as potential vaccine antigens. Future Microbiol. 2008;3(3):287–298. doi: 10.2217/17460913.3.3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoehn GT, Clark VL. Isolation and nucleotide sequence of the gene (aniA) encoding the major anaerobically induced outer membrane protein of Neisseria gonorrhoeae. Infect Immun. 1992;60(11):4695–4703. doi: 10.1128/iai.60.11.4695-4703.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kobayashi G, Moriya S, Wada C. Deficiency of essential GTP-binding protein ObgE in Escherichia coli inhibits chromosome partition. Mol Microbiol. 2001;41(5):1037–1051. doi: 10.1046/j.1365-2958.2001.02574.x. [DOI] [PubMed] [Google Scholar]
- 57.Jiang M, Sullivan SM, Wout PK, Maddock JR. G-protein control of the ribosome-associated stress response protein SpoT. J Bacteriol. 2007;189(17):6140–6147. doi: 10.1128/JB.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zielke RA, Wierzbicki IH, Weber JV, Gafken PR, Sikora AE. Quantitative proteomics of the Neisseria gonorrhoeae cell envelope and membrane vesicles for the discovery of potential therapeutic targets. Mol Cell Proteomics. 2014;13(5):1299–1317. doi: 10.1074/mcp.M113.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leuzzi R, Serino L, Scarselli M, Savino S, Fontana MR, Monaci E, et al. Ng-MIP, a surface-exposed lipoprotein of Neisseria gonorrhoeae, has a peptidyl-prolyl cis/trans isomerase (PPIase) activity and is involved in persistence in macrophages. Mol Microbiol. 2005;58(3):669–681. doi: 10.1111/j.1365-2958.2005.04859.x. [DOI] [PubMed] [Google Scholar]
- 60.Lei HT, Chou TH, Su CC, Bolla JR, Kumar N, Radhakrishnan A, et al. Crystal structure of the open state of the Neisseria gonorrhoeae MtrE outer membrane channel. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0097475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poetsch A, Wolters D. Bacterial membrane proteomics. Proteomics. 2008;8(19):4100–4122. doi: 10.1002/pmic.200800273. [DOI] [PubMed] [Google Scholar]
- 62.Papanastasiou M, Orfanoudaki G, Koukaki M, Kountourakis N, Sardis MF, Aivaliotis M, et al. The Escherichia coli peripheral inner membrane proteome. Mol Cell Proteomics. 2013;12(3):599–610. doi: 10.1074/mcp.M112.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tenson T, Mankin A. Antibiotics and the ribosome. Mol Microbiol. 2006;59(6):1664–1677. doi: 10.1111/j.1365-2958.2006.05063.x. [DOI] [PubMed] [Google Scholar]
- 64.Connell TD, Black WJ, Kawula TH, Barritt DS, Dempsey JA, Kverneland K, Jr, et al. Recombination among protein II genes of Neisseria gonorrhoeae generates new coding sequences and increases structural variability in the protein II family. Mol Microbiol. 1988;2(2):227–236. doi: 10.1111/j.1365-2958.1988.tb00024.x. [DOI] [PubMed] [Google Scholar]
- 65.Meyer TF, Mlawer N, So M. Pilus expression in Neisseria gonorrhoeae involves chromosomal rearrangement. Cell. 1982;30(1):45–52. doi: 10.1016/0092-8674(82)90010-1. [DOI] [PubMed] [Google Scholar]
- 66.Apicella MA, Breen JF, Gagliardi NC. Degradation of the polysaccharide component of gonococcal lipopolysaccharide by gonococcal and meningococcal sonic extracts. Infect Immun. 1978;20(1):228–234. doi: 10.1128/iai.20.1.228-234.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sparling PF. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J Bacteriol. 1966;92(5):1364–1371. doi: 10.1128/jb.92.5.1364-1371.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maness MJ, Sparling PF. Multiple antibiotic resistance due to a single mutation in Neisseria gonorrhoeae. J Infect Dis. 1973;128(3):321–330. doi: 10.1093/infdis/128.3.321. [DOI] [PubMed] [Google Scholar]
- 69.Unemo M, Fasth O, Fredlund H, Limnios A, Tapsall J. Phenotypic and genetic characterization of the 2008 WHO Neisseria gonorrhoeae reference strain panel intended for global quality assurance and quality control of gonococcal antimicrobial resistance surveillance for public health purposes. J Antimicrob Chemother. 2009;63(6):1142–1151. doi: 10.1093/jac/dkp098. [DOI] [PubMed] [Google Scholar]
- 70.Ohnishi M, Saika T, Hoshina S, Iwasaku K, Nakayama S, Watanabe H, et al. Ceftriaxone-resistant Neisseria gonorrhoeae. Japan Emerg Infect Dis. 2011;17(1):148–149. doi: 10.3201/eid1701.100397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kellogg DS, Jr, Peacock WL, Jr, Deacon WE, Brown L, Pirkle DI. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J Bacteriol. 1963;85:1274–1279. doi: 10.1128/jb.85.6.1274-1279.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spence JM, Wright L, Clark VL: Laboratory maintenance of Neisseria gonorrhoeae. Curr Protoc Microbiol 2008, Chapter 4:Unit 4A 1. [DOI] [PubMed]
- 73.Chen CJ, Sparling PF, Lewis LA, Dyer DW, Elkins C. Identification and purification of a hemoglobin-binding outer membrane protein from Neisseria gonorrhoeae. Infect Immun. 1996;64(12):5008–5014. doi: 10.1128/iai.64.12.5008-5014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cardinale JA, Clark VL. Expression of AniA, the major anaerobically induced outer membrane protein of Neisseria gonorrhoeae, provides protection against killing by normal human sera. Infect Immun. 2000;68(7):4368–4369. doi: 10.1128/IAI.68.7.4368-4369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Skaar EP, Lazio MP, Seifert HS. Roles of the recJ and recN genes in homologous recombination and DNA repair pathways of Neisseria gonorrhoeae. J Bacteriol. 2002;184(4):919–927. doi: 10.1128/jb.184.4.919-927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Menard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175(18):5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dillard JP: Genetic manipulation of Neisseria gonorrhoeae. Curr Protoc Microbiol 2011, Chapter 4:Unit4A 2. [DOI] [PMC free article] [PubMed]
- 78.Jiang M, Datta K, Walker A, Strahler J, Bagamasbad P, Andrews PC, et al. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J Bacteriol. 2006;188(19):6757–6770. doi: 10.1128/JB.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sheidy DT, Zielke RA. Analysis and expansion of the role of the Escherichia coli protein ProQ. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0079656. [DOI] [PMC free article] [PubMed] [Google Scholar]


