Abstract
We designed an in vitro signal amplification circuit that takes a short RNA input that catalytically activates the Spinach RNA aptamer to produce a fluorescent output. The circuit consists of three RNA strands: an internally blocked Spinach aptamer, a fuel strand, and an input strand (catalyst), as well as the Spinach aptamer ligand 3,5-difluoro-4-hydroxylbenzylidene imidazolinone (DFHBI). The input strand initially displaces the internal inhibitory strand to activate the fluorescent aptamer while exposing a toehold to which the fuel strand can bind to further displace and recycle the input strand. Under a favorable condition, one input strand was able to activate up to five molecules of the internally blocked Spinach aptamer in 185 min at 30 °C. The simple RNA circuit reported here serves as a model for catalytic activation of arbitrary RNA effectors by chemical triggers.
Keywords: RNA engineering, RNA circuit, Spinach aptamer, signal amplification
Genetically encoded biosensors composed exclusively of RNA may lead to a novel class of sensors that enable real-time monitoring of biomolecules in living cells. Additionally, RNA-based biosensors may be an ideal platform for sensing intracellular genetic information, such as mRNAs (mRNA) or noncoding RNAs (ncRNAs) that are specific to tissues or diseases, because the sensing mechanism can be engineered by simple base-pairing. To that end, the Spinach aptamer selected to bind 3,5-difluoro-4-hydroxylbenzylidene imidazolinone (DFHBI) and activate fluorescence by mimicking the GFP fluorophore1 has inspired a number of promising biosensors by coupling the Spinach folding with another aptamer that binds a molecule of interest.2−5
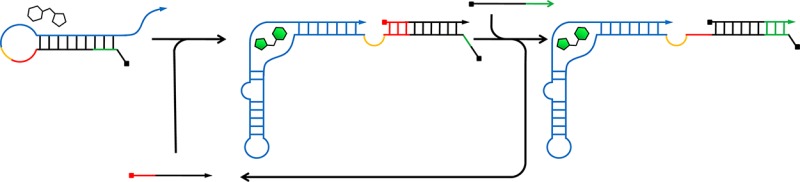
Here, we report an in vitro RNA circuit capable of signal amplification in which an input RNA strand catalytically activates multiple Spinach aptamers via nonenzymatic RNA strand displacement reactions. As depicted in Figure 1a, the circuit consists of three RNA strands: an internally blocked Spinach aptamer (Sp-I), a fuel strand (F), and a catalytic input strand (C). First, C hybridizes with the inhibitory strand within Sp-I, which allows the aptamer to bind DFHBI and become fluorescent while concomitantly exposing a short toehold sequence. Second, the fuel strand (F) which is present in excess displaces C by a toehold mediated branch migration to produce the final product Sp-IF and recycles C for further catalysis. The compact in vitro circuit exclusively composed of RNA (except for DFHBI) described here may serve as a model for catalytic signal amplification of functional RNA devices (e.g., aptamers, ribozymes, regulatory RNAs) for applications in vitro and in vivo.
Figure 1.
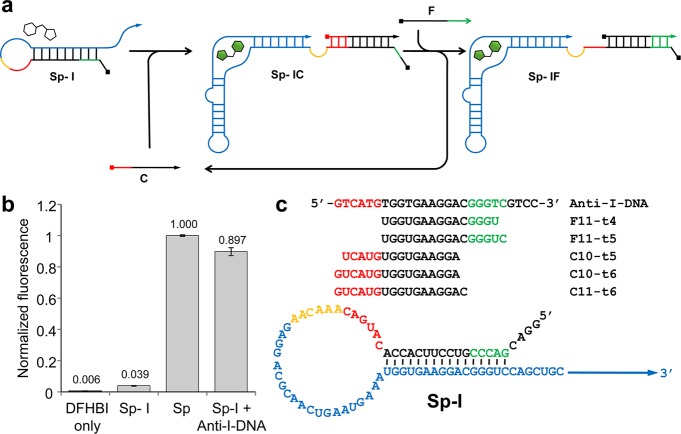
RNA signal amplification circuit design. (a) Schematic illustration of the RNA signal amplifier mechanism. The circuit consists of three RNA strands: an internally blocked Spinach aptamer (Sp-I), a fuel strand (F), and a catalytic input strand (C). C hybridizes with Sp-I using the toehold (red) to form Sp-IC allowing the Spinach aptamer domain (blue) to fold and activate fluorescence after binding DFHBI. Subsequently, F hybridizes with Sp-I via the newly exposed toehold (green) to displace C which can engage in further catalysis. (b) Fluorescence of Sp and Sp-I (500 nM) in the presence of DFHBI (3 μM). Fluorescence of Sp-I was restored in the presence of Anti-I-DNA (1500 nM). The average raw fluorescence values from triplicate experiments were obtained and normalized by the corresponding fluorescence of Sp (500 nM). (c) Sequences of the circuit components described in this work.
Results and Discussion
To implement the RNA circuit depicted in Figure 1a, the internally blocked Spinach aptamer (Sp-I) was first designed by extending the 5′ end of the Spinach aptamer with a sequence that is complementary to a part of the Spinach aptamer critical for ligand binding (Figure 1c, Supporting Information Figure S1).6,7 As expected, in vitro transcribed Sp-I exhibited diminished fluorescence in the presence of 3 μM DFHBI relative to the unmodified Spinach aptamer Sp (approximately 4% of the unmodified Spinach) (Figure 1b). Importantly, the fluorescence of Sp-I recovered to about 90% of that of Sp in the presence of 3-fold molar excess of a DNA strand complementary to the internal blocking sequence and its neighboring bases (Anti-I-DNA), demonstrating that Sp-I can be activated by sequestering the blocking sequence by hybridization (Figure 1b).
Next, we designed three catalytic input strands (C11-t6, C10-t6, and C10-t5) and two fuel strands (F11-t5, F11-t4) with different sizes (Figure 1c). Reactions containing Sp-I (500 nM) and F (1000 nM) with or without C (50 nM) were set up and the fluorescence was monitored over time at 30 °C (Figure 2a, b). The fluorescence intensity was normalized to that of the unmodified Spinach aptamer Sp (500 nM), which corresponds to the sum of the concentrations of Sp-IC and Sp-IF. The linearity of the Spinach fluorescence was confirmed for the Sp concentrations between 0 to 500 nM (Supporting Information Figure S2). Both F11-t5 and F11-t4 displayed comparable rates of uncatalyzed formation of Sp-IF likely due to transient access of the toehold. However, C11-t6 in the presence of F11-t5 exhibited the strongest activation of Sp-I (Figure 2a), and therefore, it was chosen for further analysis. Fluorescence activation was monitored in the presence of Sp-I (500 nM), F11-t5 (1000 nM), and varying concentrations of C11-t6. The reaction rate increased in a dose-dependent manner on the C11-t6 concentration. The catalytic nature of the circuit was evident, as demonstrated by the production of 126 nM activated aptamer (Sp-IC + Sp-IF) by catalysis in the presence of 25 nM C11-t6 in 185 min (Figure 2c). This corresponds to, on average, activation of 5.0 molecules of Sp-I by one molecule of C11-t6. In the absence of the fuel strand, no increase in fluorescence was observed after the rapid initial formation of the Sp-IC complex (Supporting Information Figure S3).
Figure 2.
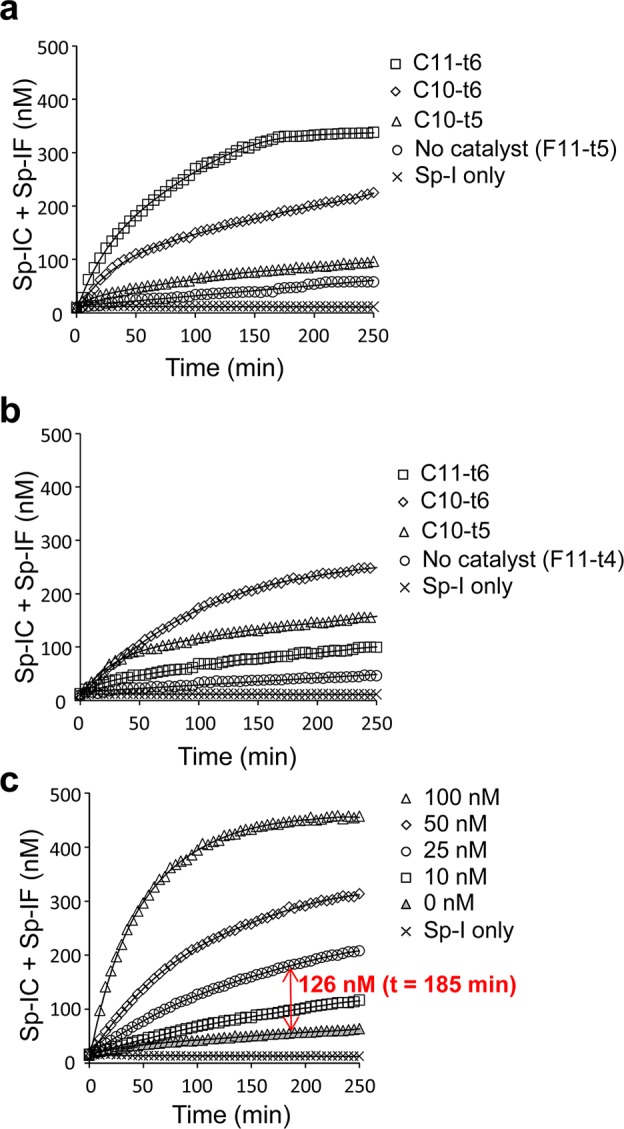
Characterization of the catalytic RNA signal amplification circuits. (a) Activation of Sp-I (500 nM) fluorescence in the presence of F11-t5 (1000 nM) with or without a catalytic strand (C11-t6, C10-t6, C10-t5; 50 nM) over time. (b) Activation of Sp-I (500 nM) fluorescence in the presence of F11-t4 (1000 nM) with or without a catalytic strand (C11-t6, C10-t6, C10-t5; 50 nM) over time. (c) Activation of Sp-I (500 nM) fluorescence in the presence of F11-t5 (1000 nM) with or without different concentrations of C11-t6 over time. (a–c) The reactions were performed as described in the Methods, and the average fluorescence values from triplicate experiments were plotted. The triplicate measurements were typically within 3% of the plotted average values, which were smaller than the data symbols shown.
Recently, a number of promising metabolite and protein sensors based on the Spinach aptamer have been designed by strategically fusing an aptamer that binds to the molecule of interest to the Spinach aptamer.2−5 Stoichiometric activation of the Spinach aptamer by a short oligonucleotide was also demonstrated.8 Catalytic activation of a fluorescent RNA aptamer such as Spinach is an attractive strategy because it can significantly improve the sensitivity of the Spinach-based sensing systems. To this end, Bhadra and Ellington recently described a related in vitro RNA circuit that combines catalytic hairpin assembly (CHA) with a fluorescent Spinach aptamer output.9 The strategy was based on the DNA CHA circuits they previously reported.10 In that work, they designed a core signal amplification circuit in which hybridization of two DNA hairpins (H1 and H2) was catalytically triggered by an input. Subsequently, the researchers designed various output modules that can be activated by the amplified H1–H2 duplex. In the recent work, Bhadra and Ellington implemented this strategy using RNA and designed an output module that activates Spinach fluorescence.9 Although their core amplification circuit exhibited impressive performance with a low uncatalyzed reaction rate as evaluated by a FRET-based output reporter, their Spinach aptamer output suffered low signal-to-noise ratio, and catalytic turnover was not explicitly demonstrated. Conceptually, the CHA architecture is attractive for its modular design, as elegantly demonstrated by the DNA implementation in vitro.10 With RNA, however, it remains to be seen if the benefits of the proposed modularity outweigh the added complexity of the circuits (more components, more reaction steps), especially for in vivo applications where control of multiple RNA species (stoichiometry, stability) can be challenging.
Also of interest, the Pierce group described “small conditional RNAs (scRNAs)” where a series of RNA-based oligonucleotides (some bases were chemically modified) were designed to conditionally produce a Dicer substrate that can induce RNA interference (RNAi) in response to a trigger RNA.11 One implementation in their recent report resembles CHA in which the product serves as a Dicer substrate.
One limitation of our current circuit is that the sequence of the catalytic strand is constrained by the Spinach sequence. Both the Pierce and the Ellington groups have demonstrated strategies to translate an arbitrary input sequence or a molecular input to trigger the catalytic reaction,9,11 which can also be adapted to the circuit described here. For example, the catalytic strand can be extended on its 5′ terminus to include a sensor sequence that also blocks the toehold domain so that the catalytic strand can be conditionally activated by a target molecule (Supporting Information Figure S4).
Our work described above and the recent related efforts by other groups9,11 strongly suggest that the robustness demonstrated by the nonenzymatic DNA signal amplification circuits10 is, in principle, achievable with RNA. However, additional efforts to improve the performance and robustness of the RNA circuits are needed to ultimately implement them as genetically encoded signal amplification circuits in living cells.
Methods
RNA Synthesis
Sp-I and Sp were prepared by in vitro transcription using T7 High Yield RNA Synthesis Kit (New England Biolabs) according to the manufacturer’s instruction. The DNA templates for in vitro transcription reactions were obtained by PCR using a plasmid in which the Spinach aptamer sequence was cloned and sequence verified. The transcripts were treated with 0.1 U/μL of TURBO DNase (Ambion) at 37 °C for 30 min to digest the template DNA, and purified using RNA Clean and Concentrator-5 kit (Zymo Research). No further purification was performed as the RNA purity was observed to be suitable by denaturing polyacrylamide gel electrophoresis (PAGE). Anti-I-DNA and the RNA catalyst and fuel strands were chemically synthesized by IDT.
RNA Circuit Reactions
The RNA circuit reactions were carried out in 10 μL volumes in black 384-well plates (Greiner Bio-One) in a buffer (25 mM Tris-HCl at pH 8.3, 37.5 mM KCl and 1.5 mM MgCl2) containing 3 μM DFHBI (Lucerna Technologies). RNA stock solutions were individually preannealed by heating to 95 °C for 3 min and rapidly cooling on ice for 30 s. Each set of reactions included wells with 500 nM Sp (+DFHBI) and wells with just DFHBI as controls. The fluorescence was monitored using Safire2 microplate reader (TECAN) set to 460 nm excitation and 510 nm emission with 20 nm bandwidth at 30 °C. The raw fluorescence values of the samples were subtracted by those of the DFHBI wells, and then normalized by the corresponding fluorescence of the 500 nM Sp wells to calculate the concentrations of the activated Sp-I species (Sp-IC and Sp-IF).
RNA and DNA Sequences Used in This Study
Sp: 5′GGAGGACGCAACUGAAUGAAAUGGUGAAGGACGGGUCCAGCUGCUUCGGCAGCUUGUUGAGUAGAGUGUGAGCUCCGUAACUAGUUGCGUCCUC 3′ Sp-I: 5′ GGACGACCCGUCCUUCACCACAUGACAAACAAGAGGACGCAACUGAAUGAAAUGGUGAAGGACGGGUCCAGCUGCUUCGGCAGCUUGUUGAGUAGAGUGUGAGCUCCGUAACUAGUUGCGUCCUC 3′ C11-t6: 5′ GUCAUGUGGUGAAGGAC 3′ C10-t6: 5′ GUCAUGUGGUGAAGGA 3′ C10-t5: 5′ UCAUGUGGUGAAGGA 3′ F11-t5: 5′ UGGUGAAGGACGGGUC 3′ F11-t4: 5′ UGGUGAAGGACGGGU 3′ Anti-I-DNA: 5′ GTCATGTGGTGAAGGACGGGTCGTCC 3′
Supporting Information Available
Supporting figures referenced in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
National Institutes of Health (GM099748)
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Paige J. S.; Wu K. Y.; Jaffrey S. R. (2011) RNA mimics of green fluorescent protein. Science 333, 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger C. A.; Wilson S. C.; Sales-Lee J.; Hammond M. C. (2013) RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J. Am. Chem. Soc. 135, 4906–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige J. S.; Nguyen-Duc T.; Song W.; Jaffrey S. R. (2012) Fluorescence imaging of cellular metabolites with RNA. Science 335, 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.; Strack R. L.; Jaffrey S. R. (2013) Imaging bacterial protein expression using genetically encoded RNA sensors. Nat. Methods 10, 873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama S.; Luo Y.; Zhou J.; Dayie T. K.; Sintim H. O. (2012) Nanomolar fluorescent detection of c-di-GMP using a modular aptamer strategy. Chem. Commun. 48, 9059–9061. [DOI] [PubMed] [Google Scholar]
- Huang H.; Suslov N. B.; Li N. S.; Shelke S. A.; Evans M. E.; Koldobskaya Y.; Rice P. A.; Piccirilli J. A. (2014) A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 10, 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner K. D.; Chen M. C.; Song W.; Strack R. L.; Thorn A.; Jaffrey S. R.; Ferre-D’Amare A. R. (2014) Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol. 21, 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra S.; Ellington A. D. (2014) A Spinach molecular beacon triggered by strand displacement. RNA 20, 1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra S.; Ellington A. D. (2014) Design and application of cotranscriptional non-enzymatic RNA circuits and signal transducers. Nucleic Acids Res. 42, e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.; Ellington A. D.; Chen X. (2011) Rational, modular adaptation of enzyme-free DNA circuits to multiple detection methods. Nucleic Acids Res. 39, e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochrein L. M.; Schwarzkopf M.; Shahgholi M.; Yin P.; Pierce N. A. (2013) Conditional dicer substrate formation via shape and sequence transduction with small conditional RNAs. J. Am. Chem. Soc. 135, 17322–17330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.