Abstract
Incomplete reprogramming of pluripotent genes in cloned embryos is associated with low cloning efficiency. Epigenetic modification agents have been shown to enhance the developmental competence of cloned embryos; however, the effect of the epigenetic modification agents on pluripotent gene reprogramming remains unclear. Here, we investigated Nanog reprogramming and the expression patterns of pluripotent transcription factors during early embryo development in pigs. We found that compared with fertilized embryos, cloned embryos displayed higher methylation in the promoter and 5′-untranslated region and lower methylation in the first exon of Nanog. When 5-aza-2′-deoxycytidine (5-aza-dC) or trichostatin A (TSA) enhanced the development of porcine cloned embryos, Nanog methylation reprogramming was also improved, similar to that detected in fertilized counterparts. Furthermore, our results showed that the epigenetic modification agents improved the expression levels of Oct4 and Sox2 and effectively promoted Nanog transcription in cloned embryos. In conclusion, our results demonstrated that the epigenetic modification agent 5-aza-dC or TSA improved Nanog methylation reprogramming and the expression patterns of pluripotent transcription factors, thereby resulting in the enhanced expression of Nanog and high development of porcine cloned embryos. This work has important implications in the improvement of cloning efficiency.
Introduction
Although somatic cell nuclear transfer (SCNT) has been achieved in many species, overall cloning efficiency is still low, thus limiting the application of cloning technology in basic research, agriculture, and medicine (Galli et al., 2012; Lee and Prather 2013; Rodriguez-Osorio et al., 2012). It is generally believed that low cloning efficiency is due mainly to abnormal epigenetic reprogramming (Zhao et al., 2010). The reprogramming of pluripotent genes could influence the developmental competence of cloned embryos, because pluripotent genes play critical roles in the establishment and maintenance of pluripotency during early embryo development (Dejosez and Zwaka, 2012; Lee et al., 2013).
Nanog is one of the critical pluripotency regulators and responsible for the pluripotency of embryonic stem cells and early embryos (Pan and Thomson, 2007; Lee et al., 2013). During somatic cell reprogramming, Nanog serves as an activator of multiple target genes and can overcome reprogramming barriers (Costa et al., 2013; Theunissen et al., 2011; Zhang et al., 2011). However, in cloned embryos, Nanog expression is abnormal, probably leading to the poor development of cloned embryos (Huan et al., 2013).
Our previous studies have shown that a DNA methylation inhibitor [5-aza-2′-deoxycytidine (5-aza-dC)] or histone deacetylase inhibitor [trichostatin A (TSA)] could improve the development of cloned embryos (Huan et al., 2013; Kong et al., 2011). However, the mechanism underlying the developmental improvement of cloned embryos is still poorly understood. Because Nanog expression is regulated by epigenetic mechanisms involving DNA methylation and histone modifications and Nanog activation is essential for early embryo development (Lee et al., 2013; Miyamoto et al., 2009; Xu et al., 2013), it is thought that Nanog reprogramming must be improved efficiently in these treated embryos. However, the effect of the epigenetic modification agents on Nanog reprogramming during early embryo development remains unknown.
To understand Nanog reprogramming during SCNT, the epigenetic modification agents 5-aza-dC and TSA were employed to enhance the development of cloned embryos. We found that compared with fertilized embryos, cloned embryos displayed incomplete methylation reprogramming of Nanog, whereas Nanog methylation reprogramming was improved in 5-aza-dC– or TSA-treated embryos, similar to that detected in fertilized counterparts. Moreover, 5-aza-dC or TSA improved the expression levels of Oct4 and Sox2 and effectively promoted Nanog transcription in cloned embryos. These results demonstrated that Nanog reprogramming was associated with the development of cloned embryos and would have important implications in the improvement of cloning efficiency.
Materials and Methods
Chemicals were purchased from Sigma-Aldrich Corporation (St. Louis, MO, USA), and disposable and sterile plasticware was obtained from Nunclon (Roskilde, Denmark), unless otherwise stated.
All experiments were approved by the Animal Care Commission of Shandong Academy of Agricultural Sciences according to animal welfare laws, guidelines and policies.
Porcine fetal fibroblast cell culture
Porcine fetal fibroblast (PFF) cell culture has been described previously (Huan et al., 2013). Briefly, a 35-day-old fetus was recovered and rinsed five times with Dulbecco's phosphate buffered saline (D-PBS). After removal of the head, internal organs, and limbs, the remaining tissues were finely minced. The minced tissues were digested with a 0.25% trypsin and 0.04% EDTA solution at 37°C for 45 min, followed by dispersal in high-glucose-enriched Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 1% l-glutamine, and 1% penicillin-streptomycin. The dispersed cells were centrifuged, resuspended, and cultured in high-glucose-enriched DMEM containing 10% FBS, 1% l-glutamine, and 1% penicillin-streptomycin at 38.5°C in a 5% CO2 atmosphere and saturated humidity. Until confluence, PFFs were digested, centrifuged, and resuspended in FBS (GIBCO) containing 10% dimethyl sulfoxide and stored in liquid nitrogen until use. Prior to SCNT, PFFs were thawed, cultured, and used in three to five passages.
Oocyte collection and in vitro maturation
Oocyte maturation has been described previously (Huan et al., 2013). Briefly, porcine ovaries were collected from a local slaughterhouse and transported to the laboratory in physiological saline with antibiotics at 37°C. Follicles with a diameter between 3 and 8 mm were aspirated, and follicular contents were washed three times with HEPES-buffered Tyrode's lactate. Cumulus–oocyte complexes (COCs) with at least three uniform layers of compact cumulus cells and uniform cytoplasm were recovered, washed, and cultured in maturation medium [tissue culture medium-199 (TCM-199) supplemented with 0.57 mM cysteine, 10 ng/mL epidermal growth factor, 0.5 μg/mL follicle-stimulating hormone, 0.5 μg/mL luteinizing hormone, and 100 μL/mL porcine follicular fluid] under mineral oil at 38.5°C in 5% CO2 atmosphere and saturated humidity. After 42 h, COCs were vortexed in 1 mg/mL hyaluronidase for 3 min to remove cumulus cells. Only oocytes with a visible polar body, regular morphology, and a homogeneous cytoplasm were used.
In vitro fertilization and SCNT embryo culture, treatment, and collection
The procedures for in vitro fertilization (IVF) and SCNT have been described (Huan et al., 2013; Wei et al., 2011). Briefly, for IVF, the semen was incubated at 39°C, resuspended, and washed three times in D-PBS supplemented with 0.1% bovine serum albumin. The sperm concentration was measured using a hemocytometer, and the proportion of motile sperm was determined. The spermatozoa were diluted with modified Tris-buffered medium (mTBM) to the appropriate concentration. Matured oocytes were washed three times in mTBM, transferred into fertilization medium, and co-incubated with spermatozoa for 6 h at the rate of 1:1000. Then the embryos were washed and cultured in porcine zygote medium-3 (PZM-3) for subsequent development. For SCNT, PFFs were trypsinized, centrifuged, and resuspended in manipulation medium. The matured oocytes and PFFs were placed into manipulation medium supplemented with 7.5 μg/mL cytochalasin B. After enucleation by aspirating the first polar body and adjacent cytoplasm, donor cells were placed into the perivitelline space. Fusion and activation of the cell–cytoplast complexes were induced with two direct pulses of 1.2 kV/cm for 30 μsec in the fusion medium (0.3 M mannitol, 0.05 mM CaCl2, 0.1 mM MgCl2, and 0.5 mM HEPES), and the fusion rate was confirmed by examination with microscopy. Reconstructed embryos were cultured in PZM-3 for subsequent development.
For 5-aza-dC or TSA treatment (Huan et al., 2013; Kong et al., 2011), reconstructed embryos were treated with 25 nM (optimized) 5-aza-dC (NT-AZA) or 40 nM (optimized) TSA (NT-TSA) for 24 h, washed, and transferred into PZM-3 for further culture. For embryo collection, the one-cell, two-cell, four-cell, eight-cell, and blastocyst-stage embryos in the IVF, NT-CON (cloned), NT-AZA, and NT-TSA groups were collected at 6 h, 24 h, 48 h, 72 h, and 156 h, respectively.
Bisulfite sequencing
Bisulfite sequencing has been described in one of our previous reports (Wei et al., 2011). Briefly, pooled samples were digested with proteinase K (PK) and treated with sodium bisulfite to convert all unmethylated cytosines to uracils using an EZ DNA Methylation-Direct™ Kit (compatible with small sample inputs, Zymo Research) according to the manufacturer's protocol. For semen, the sperm was collected by centrifugation, washed in SMB solution [10 mM Tris-HCl, 10 mM EDTA, 50 mM NaCl, and 2% sodium dodecyl sulfate (SDS), pH 7.2], and incubated in SMB solution supplemented with 40 mM dithiothreitol and 0.3 mg/ml PK at 56°C for 1 h. For samples of 103 PFFs, 200 metaphase II (MII) oocytes, and 200, 100, 50, 20, and 10 pooled embryos with the zona pellucidae removed at the one-cell, two-cell, four-cell, eight-cell, and blastocyst stages, respectively, digestion was performed in M-Digestion Buffer supplemented with PK at 50°C for 20 min. After digestion of all the samples, a cytosine-to-thymine (CT) conversion reagent was added at 98°C for 10 min and 64°C for 2.5 h. Then the samples were desalted, purified, and diluted with M-Elution Buffer.
Nested PCR was carried out to amplify the target regions of Nanog using the primers described in Table S1 (Supplementary Data are available at www.liebertpub.com/cell/) and Hot Start Taq™ Polymerase (TaKaRa) with a profile of 94°C for 5 min, 45 cycles of 94°C for 30 sec, 50°C for 30 sec, and 72°C for 1 min, followed by 72°C for 10 min. Products from the first amplification reaction were used in the second PCR reaction, and the optimal annealing temperatures of inner primers (Zhao et al., 2013) were 52°C for promoter and 5′-untranslated region (5′-UTR) and 50°C for the first exon. The amplified products were verified by electrophoresis and purified using an Agarose Gel DNA Purification Kit (TaKaRa). The purified fragments were cloned into pMD18-T Vectors (TaKaRa) and subjected to sequence analysis.
Real-time quantitative PCR
Measurement of gene expression with real-time quantitative PCR (qPCR) was performed (Huan et al., 2013, Wei et al., 2011). Briefly, total RNA was extracted from 30 pooled embryos at each stage using an RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. RNA quality was confirmed by the ratios of A260/A280 (all between 1.8 and 2.0), and only RNA samples that did not show signs of degradation were used in this study. Reverse transcription was performed using a PrimeScript® RT Reagent Kit (TaKaRa). The 20-μL reaction volume contained 1 μL of 100 μM random hexamer primer, 4 μL of 5× reverse transcriptase (RT) buffer, 1 μL of RT enzyme mix, 1 μL of total RNA (<500 ng), and 13 μL of RNase-free distilled H2O. The reaction parameters were: 37°C for 15 min and 85°C for 5 sec, and the cDNA was stored at −20°C until use.
For real-time qPCR, reactions were performed in 96-well optical reaction plates (Applied Biosystems) using SYBR® Premix ExTaq™ II (TaKaRa) and a 7500 Real-Time PCR System (Applied Biosystems). Each reaction mixture (50 μL) contained 1 μL (<25 ng) of cDNA solution, 1 μL of 10 μM of each specific primer, 1 μL of 50× SYBR Green Dye, and 25 μL of 2× ExTaq. Thermal cycling conditions were 95°C for 30 sec, followed by 40 two-step cycles of 95°C for 5 sec and 60°C for 34 sec, and a dissociation stage consisting of 95°C for 15 sec, 60°C for 1 min, and 95°C for 15 sec.
The specificity of the PCR reaction was confirmed by a single peak in the dissociation curve and also by a single band in agarose gel electrophoreses. As negative controls, cDNA was omitted during the real-time reaction. For each sample, the cycle threshold (Ct) values were obtained from three replicates. The primers used for amplification of target and internal reference genes are shown in Table S1. The comparative Ct method was used for relative quantification of target gene expression levels. Each pair of primers was confirmed for equal amplification efficiency to primers of the endogenous control (18S ribosomal RNA). Ct value was calculated by the Sequence Detection System software (Applied Biosystems). The ΔCt value was defined as Ct (target gene) − Ct (18S rRNA). The ΔΔCt value was defined as ΔCt (sample) − ΔCt (calibrator). The relative expression levels of target genes were analyzed using the 2−ΔΔCt method.
Statistical analysis
Differences in data [mean±standard error of the mean (SEM)] were analyzed with the SPSS statistical software. Statistical analysis of data concerning gene expression levels was performed with one-way analysis of variance (ANOVA). For all analyses, differences were considered to be statistically significant when p<0.05.
Results
Incomplete Nanog methylation reprogramming in porcine cloned embryos
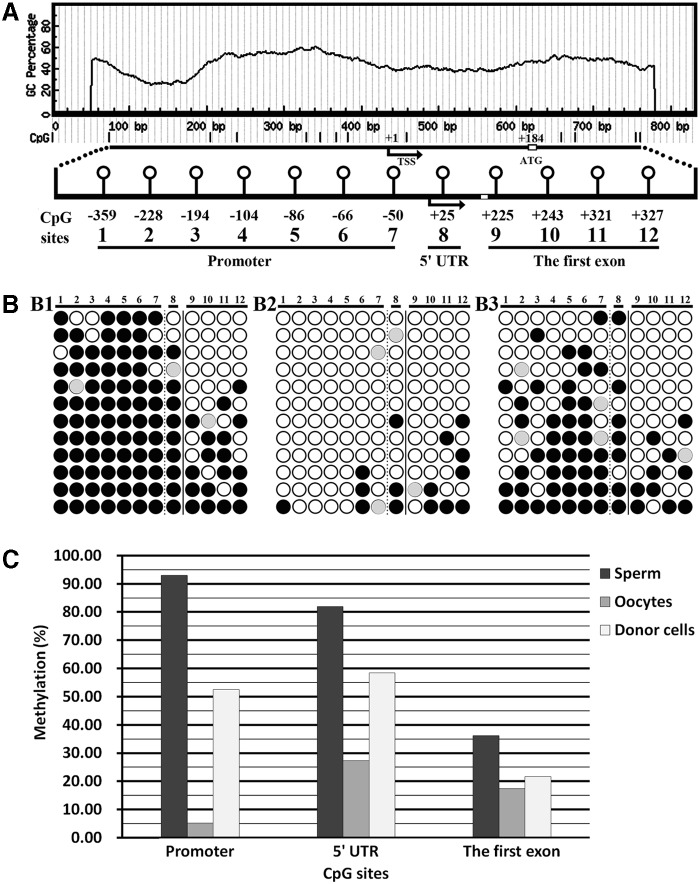
It is known that DNA methylation in a promoter or 5′-UTR silences gene expression, whereas gene body methylation positively regulates gene transcription (Szyf, 2010). Here, the distribution of CpG sites in the Nanog promoter, 5′-UTR, and the first exon was analyzed with the MethPrimer program. The result showed that no CpG island existed in the promoter; there were seven CpG sites in the promoter, one CpG site in the 5′-UTR, and four CpG sites in the first exon (Fig. 1A).
FIG. 1.
Prediction and analysis of Nanog methylation status. (A) Twelve CpG sites (seven in the promoter, one in the 5′-UTR, and four in the first exon, respectively) were analyzed in the Nanog sequence around the transcription start site (TSS) by the MethPrimer program. TSS was designated as +1. (B) Nanog methylation status in sperm (B1), oocytes (B2), and donor cells (B3). (Black or white circles) Methylated or unmethylated CpG sites; (gray circles) mutated and/or single-nucleotide polymorphism (SNP) variation at certain CpG sites. (C) Average methylation levels of the Nanog promoter, 5′-UTR, and the first exon in sperm, oocytes, and donor cells.
The methylation status of Nanog in sperm and MII oocytes was examined. The methylation levels of the promoter, 5′-UTR, and the first exon were 92.86%, 81.82%, and 36.11% in sperm and 5.16%, 27.27%, and 17.36% in MII oocytes, respectively (Fig. 1B and C). After fertilization, Nanog demethylation did not occur at the one-cell stage in comparison with the mean methylation of sperm and oocytes (Figs. 1 and 2). During the development of IVF embryos (Figs. 2 and S1), Nanog methylation levels in the promoter and 5′-UTR decreased, especially from the one-cell to two-cell stage. In the first exon, Nanog methylation levels showed an upward trend from the two-cell to blastocyst stage. Thus, IVF embryos displayed active demethylation in the promoter and 5′-UTR and a remethylation pattern in the first exon of Nanog.
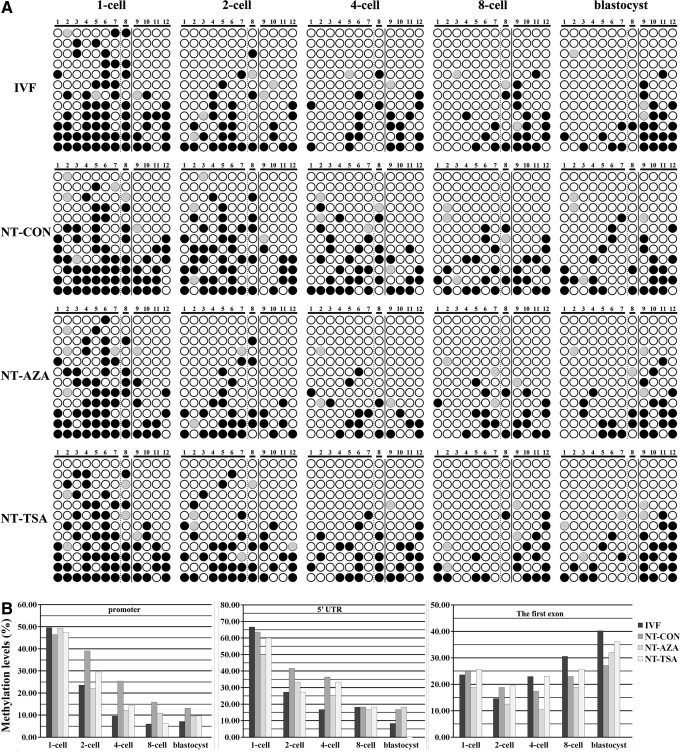
FIG. 2.
Nanog methylation status in embryos. (A) Nanog methylation status at the one-cell, two-cell, four-cell, eight-cell, and blastocyst stages of IVF, NT-CON, NT-AZA, and NT-TSA embryos. (Black and white circles) Methylated and unmethylated CpG sites, respectively; (gray circles) mutated and/or single-nucleotide polymorphism (SNP) variation at certain CpG sites. (B) Methylation status of Nanog promoter, 5′-UTR, and the first exon at the one-cell, two-cell, four-cell, 8-eight cell, and blastocyst stages of IVF, NT-CON, NT-AZA, and NT-TSA embryos.
In donor cells, Nanog methylation levels in the promoter, 5′-UTR, and first exon were 54.42%, 58.33%, and 21.53%, respectively. After SCNT, no significant differences in Nanog methylation levels were observed between donor cells and the one-cell-stage embryos (Figs. 1 and 2). In cloned embryos (Figs. 2 and S1), gradual demethylation in the promoter and 5′-UTR was observed from the one-cell to blastocyst stage and active demethylation did not occur. In the first exon, the change of Nanog methylation levels was not obvious. When compared with the individual developmental stage of IVF embryos (Fig. 2), the methylation levels of the Nanog promoter and 5′-UTR in cloned embryos were generally higher, especially at the four-cell stage. For the first exon, the methylation levels in cloned embryos from the four-cell to blastocyst stage were lower than those in fertilized counterparts. These results suggested that Nanog methylation reprogramming in porcine cloned embryos was incomplete.
Epigenetic modification agents improved Nanog methylation reprogramming in porcine cloned embryos
After cloned embryos were treated with 5-aza-dC or TSA, Nanog methylation reprogramming was investigated (Figs. 2 and S1). In the NT-AZA group, the Nanog methylation level in the one-cell-stage embryos did not differ obviously from that in donor cells. From the one-cell to blastocyst stage, particularly the one-cell to two-cell stage, the Nanog promoter and 5′-UTR underwent demethylation, and the first exon of Nanog displayed demethylation from the one-cell to four-cell stage and remethylation from the four-cell to blastocyst stage. As for the differences between the NT-AZA and NT-CON groups, Nanog demethylation was shifted earlier in the NT-AZA group and the methylation levels of Nanog were lower than those in the NT-CON group, with the exception of 5′-UTR and the first exon in blastocysts. Nanog methylation status in the NT-AZA group was closer to those in the IVF group. These results indicated that 5-aza-dC could improve Nanog methylation reprogramming in cloned embryos.
In the NT-TSA group, a trend of demethylation in the Nanog promoter and 5′-UTR and remethylation in the first exon of Nanog were observed. For Nanog methylation differences between the NT-TSA and NT-CON or NT-AZA groups, the NT-TSA group showed a much more similar methylation pattern to the IVF group. Therefore, our results showed that the epigenetic modification agents 5-aza-dC or TSA improved Nanog methylation reprogramming in porcine cloned embryos.
Epigenetic modification agents improved Nanog expression in porcine cloned embryos
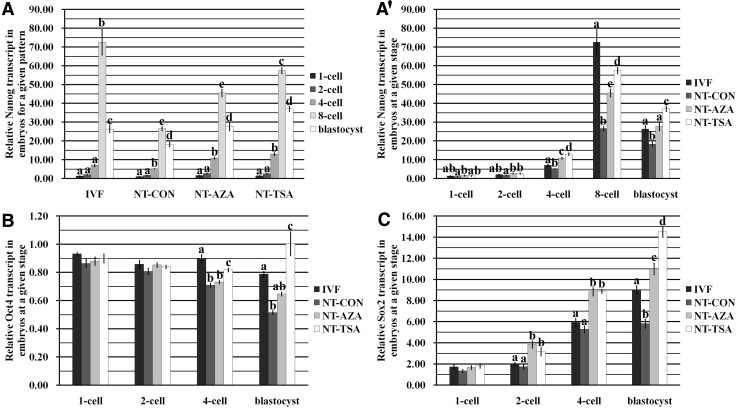
For Nanog transcription, a similar expression trend was observed in the IVF, NT-CON, NT-AZA, and NT-TSA groups, showing the initial expression at the four-cell stage, a maximum peak at the eight-cell stage, and a slight decrease at the blastocyst stage (Fig. 3A). When the individual developmental stage was compared among these groups (Fig. 3A′), Nanog transcript levels in the NT-CON group were significantly (p<0.05) lower than those in the IVF group (from the four-cell to blastocyst stage), the NT-AZA group, or the NT-TSA group (from the two-cell to blastocyst stage), and in comparison with that in the IVF group, Nanog expression in the NT-AZA or NT-TSA group showed a significant increase at the four-cell stage and a significant decrease at the eight-cell stage (p<0.05). Regarding the differences between the NT-TSA and NT-AZA groups, Nanog transcripts in the NT-TSA group showed significantly (p<0.05) higher levels from the four-cell to blastocyst stage, and a significant upregulation of Nanog expression at the blastocyst stage was also observed in the NT-TSA group in comparison with that in the IVF group (p<0.05). These results suggested that the enhanced Nanog methylation reprogramming induced by 5-aza-dC or TSA improved its expression in cloned embryos.
FIG. 3.
Transcript levels of Nanog, Oct4, and Sox2 in IVF, NT-CON, NT-AZA, and NT-TSA embryos. (A) Relative transcript levels of Nanog in IVF, NT-CON, NT-AZA, and NT-TSA embryos. (A′) Relative transcript levels of Nanog at the one-cell, two-cell, four-cell, eight-cell, and blastocyst stages of IVF, NT-CON, NT-AZA, and NT-TSA embryos. (B) Relative transcript levels of Oct4 at the one-cell, two-cell, four-cell, and blastocyst stages of IVF, NT-CON, NT-AZA, and NT-TSA embryos. (C) Relative transcript levels of Sox2 at the one-cell, two-cell, four-cell, and blastocyst stages of IVF, NT-CON, NT-AZA, and NT-TSA embryos. The transcript abundance for each gene in oocytes at the MII stage was considered as the control. The data are expressed as mean±SEM. a–dValues of a given gene in a certain group (A) or at a certain stage (A′, B, and C) in columns with different superscripts differ significantly (p<0.05).
To further explore the mechanism underlying the improved Nanog expression derived from its enhanced methylation reprogramming, transcription factors regulating Nanog expression were analyzed with the TRANSFAC and TFSEARCH programs. Sequence analysis revealed that a number of binding sites of potentially important transcription factors were present in the Nanog sequence, and the degree of methylation of Nanog CpG sites determined the binding capability of these factors, thereby regulating Nanog expression (Fig. S2).
Of course, the expression levels of these transcription factors also regulate Nanog transcription. Here, the transcription levels of two functional transcription factors (Oct4 and Sox2, identified) were investigated (Fig. 3B, C). Compared with those in the IVF group, significantly lower transcripts of Oct4 at the four-cell and blastocyst stages and Sox2 at the blastocyst stage were observed in the NT-CON group (p<0.05). After 5-aza-dC or TSA treatment, the NT-AZA group showed significantly (p<0.05) higher expression levels of Sox2 from the two-cell to blastocyst stage, and the NT-TSA group displayed significantly higher transcripts of Sox2 from the two-cell to blastocyst stage and Oct4 at the four-cell and blastocyst stages in comparison with those in the NT-CON group (p<0.05). Sox2 transcripts from the two-cell to blastocyst stage in the NT-AZA or NT-TSA group were also significantly higher than those in the IVF group, although Oct4 expression at the four-cell stage was still significantly lower (p<0.05).
When the expression levels of Oct4 and Sox2 were compared between the NT-AZA and NT-TSA groups, the NT-TSA group showed significantly (p<0.05) higher transcripts of Oct4 at the four-cell and blastocyst stages and Sox2 at the blastocyst stage, and Oct4 expression at the blastocyst stage in the NT-TSA group was also significantly higher than that in the IVF group (p<0.05). Thus, these results showed that the epigenetic modification agent 5-aza-dC or TSA improved the expression of transcription factors in cloned embryos. Overall, the improvement of Nanog methylation reprogramming and the expression and binding of transcription factors induced by 5-aza-dC or TSA enhanced Nanog expression in cloned embryos.
Discussion
Aberrant methylation reprogramming has been reported in cloned embryos (Cantone and Fisher 2013; Peat and Reik 2012; Zhao et al., 2010). Nanog is one of the critical pluripotent factors and its expression influences nuclear reprogramming efficiency, suggesting that Nanog could be a suitable marker to evaluate nuclear reprogramming in cloned embryos (Costa et al., 2013; Miyamoto et al., 2009; Stuart et al., 2014). In this study, Nanog methylation in the promoter and 5′-UTR rapidly decreased and was maintained at a low level in IVF embryos, which is consistent with a previous report (Zhao et al., 2013). After SCNT, the methylation levels of the Nanog promoter and 5′-UTR were higher than those in IVF embryos, and the methylation of the first exon was also reprogrammed inefficiently, suggesting that incomplete Nanog methylation reprogramming could be the cause of the poor development of cloned embryos. As for the reason for incomplete Nanog methylation reprogramming, it is possible that there is a mechanism that preserves the methylation pattern of donor cells against reprogramming by oocyte factors (Yamanaka et al., 2011).
Our previous studies have shown that 5-aza-dC or TSA could improve the development of porcine cloned embryos (Huan et al., 2013; Kong et al., 2011). Here, the reprogramming degree of Nanog methylation in the NT-AZA or NT-TSA group may explain the results that 5-aza-dC or TSA enhances the development of cloned embryos (Table S2 and Fig. S3). As for the improvement of Nanog methylation reprogramming after 5-aza-dC or TSA treatment, it is possible that 5-aza-dC was incorporated into the genome during DNA replication or that TSA loosened the chromatin structure, benefitting the binding of DNA demethylation-related molecules (Huan et al., 2013; Zhao et al., 2010).
Of course, other mechanisms may also exist (Huan et al., 2013; Zhao et al., 2010). In regard to the differences of Nanog methylation reprogramming between the NT-AZA and NT-TSA groups, the different manner of regulation induced by 5-aza-dC or TSA may be the cause, and histone modification possibly fits better with Nanog methylation reprogramming (Jafarpour et al., 2011; Xu et al., 2013). The results concerning the birth of cloned piglets (data not shown) could also confirm this explanation. As for the detailed mechanism of Nanog methylation reprogramming in the NT-AZA or NT-TSA group, further studies are needed.
In view of Nanog methylation status during early embryo development, our results suggest that an active demethylation mechanism exists, even though traditional bisulfite sequencing could not distinguish between 5-methylcytosine and 5-hydroxymethylcytosine (Huang et al., 2010). Due to the critical role of 5-hydroxymethylcytosine in somatic cell reprogramming (Wossidlo et al., 2011), new technologies, such as oxidative bisulfite sequencing, would be employed to investigate active demethylation of Nanog.
The improvement of Nanog methylation reprogramming after 5-aza-dC or TSA treatment should result in its effective activation in cloned embryos. The expression patterns of Nanog in cloned embryos support this view and are positively associated with the development of cloned embryos. Thus, we speculate that the improvement of Nanog expression derived from its facilitated methylation reprogramming probably enhances the developmental competence of cloned embryos. Certainly, the appropriate expression levels of other early embryo development-related genes are also essential for the development of cloned embryos (Huan et al., 2013); thus, the expression profiles of these genes are also worthy of investigation.
Regarding the improvement of Nanog expression in the 5-aza-dC or TSA treatment group, pluripotent transcription factors such as Oct4 and Sox2 should also play a key role (Kuroda et al., 2005; Rodda et al., 2005). Previous studies have shown that the expression levels and binding capability of transcription factors could regulate Nanog expression (Kuroda et al., 2005; Palacios et al., 2010; Rodda et al., 2005). Our results showed that the enhanced Nanog methylation reprogramming should benefit the binding of transcription factors (Palacios et al., 2010), and the expression levels of transcription factors Oct4 and Sox2 were positively correlated with Nanog transcription, suggesting that transcription factors regulate Nanog activation in cloned embryos. Interestingly, Nanog did not express in MII oocytes, although its methylation was low, and this may be attributed to the lack of the expression of some transcription factors (e.g., Sox2).
Thus, the expression levels and binding capability of transcription factors are crucial for Nanog transcription, and 5-aza-dC or TSA could improve this regulation. However, the methylation status of these transcription factors during nuclear reprogramming and how 5-aza-dC or TSA cooperates with these transcription factors to improve Nanog expression in cloned embryos remain unclear, needing further investigation.
In conclusion, our results showed that Nanog methylation reprogramming was incomplete in cloned embryos, and 5-aza-dC or TSA enhanced Nanog methylation reprogramming. Additionally, Nanog expression was also improved in the 5-aza-dC or TSA treatment group due to the improvement of Nanog methylation reprogramming and the expression and binding of pluripotent transcription factors, thereby resulting in the developmental improvement of cloned embryos.
Supplementary Material
Acknowledgments
This work was supported by grants from the earmarked fund for the China Agriculture Research System (grant no. CARs-37 to H.H.), the National Natural Science Fund of China (grant no. 31272586 to H.H.), the China Postdoctoral Science Foundation (grant no. 2014M551943), and the special foundation of Postdoctoral Innovation Project in Shandong Province of China (grant no. 201402044).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Cantone I., and Fisher A.G. (2013). Epigenetic programming and reprogramming during development. Nat. Struct. Mol. Biol. 20, 282–289 [DOI] [PubMed] [Google Scholar]
- Costa Y., Ding J., Theunissen T.W., Faiola F., Hore T.A., Shliaha P.V., Fidalgo M., Saunders A., Lawrence M., Dietmann S., Das S., Levasseur D.N., Li Z., Xu M., Reik W., Silva J.C, and Wang J. (2013). NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495, 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M., and Zwaka T.P. (2012). Pluripotency and nuclear reprogramming. Annu. Rev. Biochem. 81, 737–765 [DOI] [PubMed] [Google Scholar]
- Galli C., Lagutina I., Perota A., Colleoni S., Duchi R., Lucchini F., and Lazzari G. (2012). Somatic cell nuclear transfer and transgenesis in large animals: Current and future insights. Reprod. Domest. Anim. 47 (Suppl 3), 2–11 [DOI] [PubMed] [Google Scholar]
- Huan Y.J., Zhu J., Xie B.T., Wang J.Y., Liu S.C., Zhou Y., Kong Q.R., He H.B., and Liu Z.H. (2013). Treating cloned embryos, but not donor cells, with 5-aza-2′-deoxycytidine enhances the developmental competence of porcine cloned embryos. J. Reprod. Dev. 59, 442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Pastor W.A., Shen Y., Tahiliani M., Liu D.R., and Rao A. (2010). The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 5, e8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarpour F., Hosseini S.M., Hajian M., Forouzanfar M., Ostadhosseini S., Abedi P., Gholami S., Ghaedi K., Gourabi H., Shahverdi A.H., Vosough A.D., and Nasr-Esfahani M.H. (2011). Somatic cell-induced hyperacetylation, but not hypomethylation, positively and reversibly affects the efficiency of in vitro cloned blastocyst production in cattle. Cell. Reprogram. 13, 483–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q.R., Zhu J., Huang B., Huan Y.J., Wang F., Shi Y.Q., Liu Z.F., Wu M.L., and Liu Z.H. (2011). TSA improves transgenic porcine cloned embryo development and transgene expression. Yi Chuan 33, 749–756 [DOI] [PubMed] [Google Scholar]
- Kuroda T., Tada M., Kubota H., Kimura H., Hatano S.Y., Suemori H., Nakatsuji N., and Tada T. (2005). Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol. Cell. Biol. 25, 2475–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., and Prather R.S. (2013). Advancements in somatic cell nuclear transfer and future perspectives. Anim. Front. 3, 56–61 [Google Scholar]
- Lee M.T., Bonneau A.R., Takacs C.M., Bazzini A.A., DiVito K.R., Fleming E.S., and Giraldez A.J. (2013). Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K., Tsukiyama T., Yang Y., Li N., Minami N., Yamada M., and Imai H. (2009). Cell-free extracts from mammalian oocytes partially induce nuclear reprogramming in somatic cells. Biol. Reprod. 80, 935–943 [DOI] [PubMed] [Google Scholar]
- Palacios D., Summerbell D., Rigby P.W., and Boyes J. (2010). Interplay between DNA methylation and transcription factor availability: Implications for developmental activation of the mouse myogenin gene. Mol. Cell. Biol. 30, 3805–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G., and Thomson J.A. (2007). Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 17, 42–49 [DOI] [PubMed] [Google Scholar]
- Peat J.R., and Reik W. (2012). Incomplete methylation reprogramming in SCNT embryos. Nat. Genet. 44, 965–966 [DOI] [PubMed] [Google Scholar]
- Rodda D.J., Chew J.L., Lim L.H., Loh Y.H., Wang B., Ng H.H., and Robson P. (2005). Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 280, 24731–24737 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Osorio N., Urrego R., Cibelli J.B., Eilertsen K., and Memili E. (2012). Reprogramming mammalian somatic cells. Theriogenology 78, 1869–1886 [DOI] [PubMed] [Google Scholar]
- Stuart H.T., van Oosten A.L., Radzisheuskaya A., Martello G., Miller A., Dietmann S., Nichols J., and Silva J.C. (2014). NANOG amplifies STAT3 activation and they synergistically induce the naive pluripotent program. Curr. Biol. 24, 340–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. (2010). DNA methylation and demethylation probed by small molecules. Biochim. Biophys. Acta 1799, 750–759 [DOI] [PubMed] [Google Scholar]
- Theunissen T.W., van Oosten A.L., Castelo-Branco G., Hall J., Smith A., and Silva J.C. (2011). Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions. Curr. Biol. 21, 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Huan Y., Shi Y., Liu Z., Bou G., Luo Y., Zhang L., Yang C., Kong Q., Tian J., Xia P., Sun Q.Y., and Liu Z. (2011). Unfaithful maintenance of methylation imprints due to loss of maternal nuclear Dnmt1 during somatic cell nuclear transfer. PLoS One 6, e20154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M., Nakamura T., Lepikhov K., Marques C.J., Zakhartchenko V., Boiani M., Arand J., Nakano T., Reik W., and Walter J. (2011). 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2, 241. [DOI] [PubMed] [Google Scholar]
- Xu W., Li Z., Yu B., He X., Shi J., Zhou R., Liu D., and Wu Z. (2013). Effects of DNMT1 and HDAC inhibitors on gene-specific methylation reprogramming during porcine somatic cell nuclear transfer. PLoS One 8, e64705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K., Sakatani M., Kubota K., Balboula A.Z., Sawai K., and Takahashi M. (2011). Effects of downregulating DNA methyltransferase 1 transcript by RNA interference on DNA methylation status of the satellite I region and in vitro development of bovine somatic cell nuclear transfer embryos. J. Reprod. Dev. 57, 393–402 [DOI] [PubMed] [Google Scholar]
- Zhang L., Luo Y.B., Bou G., Kong Q.R., Huan Y.J., Zhu J., Wang J.Y., Li H., Wang F., Shi Y.Q., Wei Y.C., and Liu Z.H. (2011). Overexpression Nanog activates pluripotent genes in porcine fetal fibroblasts and nuclear transfer embryos. Anat. Rec. (Hoboken) 294, 1809–1817 [DOI] [PubMed] [Google Scholar]
- Zhao J., Whyte J., and Prather R.S. (2010). Effect of epigenetic regulation during swine embryogenesis and on cloning by nuclear transfer. Cell Tiss. Res. 341, 13–21 [DOI] [PubMed] [Google Scholar]
- Zhao M.T., Rivera R.M., and Prather R.S. (2013). Locus-specific DNA methylation reprogramming during early porcine embryogenesis. Biol. Reprod. 88, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.