Abstract
Mycoplasma pneumoniae is a cell wall-less bacterial pathogen of the human respiratory tract that accounts for > 20% of all community-acquired pneumonia (CAP). At present the most effective means for detection and strain-typing is quantitative polymerase chain reaction (qPCR), which can exhibit excellent sensitivity and specificity but requires separate tests for detection and genotyping, lacks standardization between available tests and between labs, and has limited practicality for widespread, point-of-care use. We have developed and previously described a silver nanorod array-surface enhanced Raman Spectroscopy (NA-SERS) biosensing platform capable of detecting M. pneumoniae with statistically significant specificity and sensitivity in simulated and true clinical throat swab samples, and the ability to distinguish between reference strains of the two main genotypes of M. pneumoniae. Furthermore, we have established a qualitative lower endpoint of detection for NA-SERS of < 1 genome equivalent (cell/μl) and a quantitative multivariate detection limit of 5.3 ± 1 cells/μl. Here we demonstrate using partial least squares- discriminatory analysis (PLS-DA) of sample spectra that NA-SERS correctly identified M. pneumoniae clinical isolates from globally diverse origins and distinguished these from a panel of 12 other human commensal and pathogenic mycoplasma species with 100% cross-validated statistical accuracy. Furthermore, PLS-DA correctly classified by strain type all 30 clinical isolates with 96% cross-validated accuracy for type 1 strains, 98% cross-validated accuracy for type 2 strains, and 90% cross-validated accuracy for type 2V strains.
Introduction
The cell wall-less prokaryote Mycoplasma pneumoniae is a major cause of respiratory disease in humans, accounting for 20% to 40% of all community acquired pneumonia (CAP). M. pneumoniae is the leading cause of CAP in older children and young adults, while the incidence of infection in the very young and the elderly is on the rise [1–5]. For adults alone the annual economic burden of CAP is > $17 billion [6]. Macrolide resistance is a growing concern, particularly in children [5], and extra-pulmonary sequelae occur in up to 25% of infections. Finally, evidence continues to indicate a contributing role for M. pneumoniae infection in the onset, exacerbation, and recurrence of asthma [5].
An area of growing interest is the role of M. pneumoniae strain type in pathogenesis and disease epidemiology. Genetic diversity is relatively limited among M. pneumoniae strains, which can be categorized into two major groups (type 1 or type 2) based on variation within sequence of the P1 (MPN141) gene, although variant strains of the two are increasingly more common [7]. The P1 protein is an important virulence factor and immunogen in M. pneumoniae infection [8–10]. P1 must complex with several other proteins in order to localize to the tip of the terminal organelle, where it mediates receptor binding for attachment to the respiratory epithelium, an essential step in successful colonization of the airways [9, 11]. Variation in the P1 gene sequence is used to distinguish between type 1 and type 2 strains of M. pneumoniae, but little is known about phenotypic differences arising from this genetic variation. Perhaps notable in this regard is the periodicity of type-switching that occurs between the two major genotypes in regular patterns of four to seven years [12].
M. pneumoniae infection is transmitted through aerosolized respiratory secretions and spreads slowly but efficiently through close living quarters, with incubation periods up to three weeks [13, 14]. Symptoms tend to be non-descript, often with complex and variable clinical presentations, which makes definitive diagnosis challenging [2, 6, 15]. As a result, diagnosis is often presumptive and relies heavily on the combination of physical findings and the elimination of other possible causes [4, 5, 14]. The success rate for laboratory culture is poor, even for experienced labs, while serologic testing, historically considered the foundation for diagnosis of M. pneumoniae infection, has limited sensitivity and specificity, a high tendency for false-negatives, requires paired sera resulting in retrospective diagnosis, and must often be paired with another diagnostic method [2, 4, 5, 10, 14]. Of the currently existing alternatives, the most efficient means for detection is quantitative polymerase chain reaction (qPCR). At present, the only FDA-approved tests for the clinical detection of M. pneumoniae are the Illumigene automated detection system (Meridian Bioscience, Inc., Cincinatti, Ohio) and the FilmArray Respiratory Panel (BioFire Diagnostics Inc., Salt Lake City, Utah). The Illumigene platform uses loop-mediated isothermal amplification and is capable of detecting M. pneumoniae in both throat and nasopharyngeal swab specimens with a high degree of sensitivity and specificity. The FilmArray Respiratory Panel employs nested, multiplex qPCR with endpoint melt curve analysis on nasopharyngeal swabs to test for 21 different viral and bacterial respiratory pathogens, and is capable of detecting M. pneumoniae as low as 30 colony-forming units (CFU)/ml [16]. The current standard for M. pneumoniae genotyping is PCR-restriction fragment length polymorphism, but can also be done by nested PCR and sequencing, multilocus variable-number tandem-repeat analysis, or by qPCR and high resolution melt curve analysis [15, 17–19]. These methods for detection and genotyping exhibit high sensitivity and specificity for all known strain variants, can allow for detection in the early stages of infection, and can be performed in hospitals and reference laboratories [2, 4, 5]. However, the requirement for separate tests for detection and genotyping, as well as the cost, complexity, and expertise required, limits the practicality for widespread, point-of-care use [2, 4–6, 14]. These limitations create a critical barrier to the accurate and timely diagnosis of M. pneumoniae infection and epidemiological tracking, and a rapid, simple, diagnostic platform capable of simultaneous detection and genotyping would greatly improve the control of M. pneumoniae disease.
Vibrational spectroscopy has an inherent biochemical specificity that led to its consideration as a next-generation platform for the rapid detection, characterization, and identification of infectious agents [20–23]. Raman spectroscopy in particular has several advantages for application to biological samples, including narrow bandwidths, good spatial resolution, and the ability to analyze aqueous samples due to the absence of interference by water molecules [20, 21, 24]. Furthermore, Raman spectra provide detailed structural information on the chemical composition of a sample and can serve as a characteristic molecular fingerprint for pathogen identification [23, 24]. Despite these advantages, standard Raman spectra are inherently limited by weak signals for detection. As a result, the application of traditional Raman spectroscopy for biosensing applications was impractical and inefficient [13, 21, 24] until the discovery that sample adsorption onto nanoscopically roughened metallic surfaces results in significant enhancements in Raman signal and spectral intensity [23–25]. This enhancement, by factors up to 1014-fold, is attributed to the increased electromagnetic field for molecules in close proximity to the metallic surface [20, 21]. Surface-enhanced Raman spectroscopy (SERS) retains the advantages of standard Raman spectroscopy, in addition to markedly improved sensitivity, allowing for considerable success at whole organism molecular fingerprinting [20, 24, 26, 27].
Inconsistency and lack of reproducibility in the preparation of SERS-active substrates has hindered its widespread use for biosensing applications [20, 21, 24]. However, highly ordered silver nanorod array (NA) substrates fabricated using oblique angle deposition (OAD) yield consistent SERS enhancement factors of around 108, with less than 15% variation between substrate batches [21]. The reproducibility of NA-SERS substrates can be improved further when patterned into a multiwell format with polydimethylsiloxane (PDMS) [20]. The highly reproducible detection capabilities of NA-SERS have been well demonstrated for multiple infectious agents, including RSV, rotavirus, influenza, HIV, adenovirus, SARS coronavirus, and M. pneumoniae [13, 22, 28–30].
Hennigan et al. described an NA-SERS-based platform capable of detecting M. pneumoniae with statistically significant sensitivity and specificity in both simulated and true clinical throat swabs, with the potential to detect and type M. pneumoniae within a single test [13]. We recently determined the sensitivity of NA-SERS for M. pneumoniae detection to be < 1 genome equivalent (cell/μl) qualitatively, and to have a quantitative multivariate detection limit of 5.3 ± 1 cells/μl [31]. Initial evaluation of this biosensing platform’s capabilities indicates the potential for application as a next-generation diagnostic tool for the clinical detection of M. pneumoniae, but a more comprehensive analysis is needed prior to proceeding with clinical validation. In the present study we further explored the specificity of NA-SERS for M. pneumoniae detection with a panel of 30 M. pneumoniae isolates collected from representative global outbreaks and spanning clinically relevant genotypes. Furthermore, since NA-SERS has inherent biochemical specificity, we analyzed a panel of 12 other human commensal and pathogenic mycoplasmas to demonstrate that this biosensing platform could distinguish M. pneumoniae from its clinically relevant closest phylogenetic relatives. Finally, we evaluated the ability of the NA-SERS platform to correctly type the 30 M. pneumoniae clinical isolates relative to known reference strains of M. pneumoniae.
Methods
Preparation of M. pneumoniae controls and clinical isolates for SERS analysis
Wild type M. pneumoniae reference strains M129 (type 1) and FH (type 2) were grown, harvested, and prepared at the University of Georgia (UGA) for this study. A panel of 30 additional clinical isolates consisting of 13 type 1 strains, 11 type 2 strains, and six type 2 variant strains were grown, harvested, and prepared for SERS and quality control analysis at the Pneumonia Response and Surveillance Laboratory at the Centers for Disease Control and Prevention (CDC) in Atlanta, Georgia. P1 genotype groups were determined by the Pneumonia Response and Surveillance Laboratory at the CDC in Atlanta, GA using DNA sequence analysis, qPCR in combination with high resolution melt curve analysis, and RFLP sequencing analysis. All mycoplasma isolates and controls were cultured in SP4 medium [2, 30] in tissue culture flasks with a 1μl/ml inoculation and incubated at 37°C. Strains grown at UGA were harvested at log phase when the phenol red indicator turned an orange color upon reaching a pH of ~6.5. Strains grown at the CDC were harvested 14 days from the date of inoculation to ensure adequate growth for all isolates. At time of harvest, the spent growth medium was decanted for each flask and 0.1× volume of sterile PBS (pH 7.2) was added to wash the adherent mycoplasmas. The PBS wash was then decanted and the PBS wash repeated 3× before the cells were scraped into 1 ml sterile PBS. Cells were then syringe-passaged 10× with a 25 gauge needle and aliquots made for determination of protein content, plating on PPLO agar [32] for CFU determination (for select isolates and controls), DNA extraction for genome equivalent determination, and SERS analysis.
M. pneumoniae samples for SERS analysis were syringe-passaged 10× with a 25-gauge needle to disperse clumps, fixed with the addition of one volume of 8% formaldehyde in sterile PBS (pH 7.0), and stored at 4°C. Growth medium negative control samples were prepared in parallel under the same conditions as the M. pneumoniae reference strains as described previously [31]. At the time of SERS analysis, mycoplasma and medium-only negative control samples were diluted in sterile DI water to a concentration of 105 cells/μl, which falls within the SERS detectable range for M. pneumoniae and was found to be dilute enough to ensure that the spectra adequately represent SERS Raman spectra arising from the Ag nanorod substrate [31]. Samples were loaded onto the NA-SERS substrate immediately following this dilution.
Preparation of non-M. pneumoniae human commensal and pathogenic species for NA-SERS analysis
Twelve human commensal and pathogenic Mollicutes species closely related [33] to M. pneumoniae were grown and harvested at the University of Alabama at Birmingham (UAB). These included: Acholeplasma laidlawii (ATCC 23206), Mycoplasma amphoriforme (A39 M6123; provided by M. Balish, Miami University of Ohio), Mycoplasma fermentans (ATCC 19989), Mycoplasma genitalium (ATCC 49897), Mycoplasma hominis (ATCC Mh132), Mycoplasma orale (ATCC 23714), Mycoplasma penetrans (UAB reference strain collection, year 1995), Mycoplasma pirum (ATCC 25960), Mycoplasma salivarium (ATCC 23064), Mycoplasma spermatophilum (ATCC 49695), Ureaplasma parvum (Up1; ATCC 27813), and Ureaplasma urealyticum (Uu11; ATCC 33695). Mycoplasma buccale, Mycoplasma lipophilum, and Mycoplasma faucium were originally intended to be included in the panel, but attempts at culturing these organisms were unsuccessful. For each culture, 500 μl to 1 ml of stock culture was inoculated into approximately 30 ml of SP4, Hayflick’s, or 10B depending on the individual species’ growth requirements, and incubated until the pH indicator changed color, indicative of microbial growth and utilization of the metabolic substrate in the media, i.e., glucose, arginine, or urea. At the time of harvest the cells and spent media were poured into 50 ml polycarbonate tubes and centrifuged at 8,000 RPM for 15 min, except for Ureaplasma species, which were centrifuged for 1 hr. The supernatants were decanted and the pellets suspended in 30 ml sterile PBS. The cells were washed by centrifugation at 8,000 RPM for 15 min as above, or 10,000 RPM for 1 hr for Ureaplasma species. The supernatants were then decanted and the pellets suspended in 1 ml sterile PBS, transferred to a 1.5 ml vial, and centrifuged at 14,000 RPM for 20 min. The supernatants were again decanted and the pellets suspended in 1 ml sterile PBS and syringe-passaged using a 26-gauge needle to disperse clumps. Aliquots were made for spotting onto a blood agar plate to test for contamination, and plating for CFU and color-changing unit (CCU) determination. Two 400 μl aliquots for each were centrifuged at 14,000 RPM for 20 min, the supernatant was removed, and the pellets were frozen for shipment to UGA, where they were stored at -80°C.
For SERS and quality control analysis, cell pellets were suspended in 1 ml sterile PBS (pH 7.2) and syringe-passaged 10× with a 25 gauge needle to disperse clumps. Aliquots were then made for DNA extraction and genome equivalent determination, protein assay, and NA-SERS analysis. SERS samples were prepared by fixing 500 μl of suspended cells with 500 μl of 8% formaldehyde in sterile PBS (pH 7.0), and stored at 4°C until time of SERS analysis. At that time the samples were diluted in sterile DI water to a concentration of 103 to 104 cells/μl and then immediately loaded onto the NA-SERS substrate. A growth medium only negative control and M. pneumoniae strain M129 samples were prepared as described above for comparison.
Preparation of samples for determination of protein content and genome equivalents
All samples were analyzed for protein content via the Bicinchoninic acid assay [34]. DNA was extracted by the QIAamp DNA Blood Minikit (Qiagen, Valencia, CA) using the blood and body fluids protocol, including RNase A treatment. 200 μl of sample were used for DNA extraction, with a final elution volume of 200 μl for use to quantify DNA content and genome equivalents. Genomic DNA concentration and absorbance measurements for the Bicinchoninic acid assay were performed on a NanoDrop instrument (Model ND-1000, Thermo Scientific, Wilmington, DE) using software V3.5.2. Genome equivalents of M. pneumoniae samples were calculated from the DNA concentration obtained from this analysis and using the previously determined mass of the M. pneumoniae genome, 5.3x107 Daltons [35]. Genome equivalents for all non-M. pneumoniae samples were determined from DNA concentrations obtained from this analysis and a genome mass calculated for this study based on published genome lengths and known G+C contents from the GenBank database.
NA-SERS and chemometric analysis
NA-SERS substrates were prepared by OAD as described [21, 29, 36, 37]. Prior to their use, substrates were cleaned for 5 min in an Ar+ plasma using a plasma cleaner (Model PDC-32G, Harrick Plasma, Ithaca, NY) to remove any surface contamination [38] and then patterned into 40 3mm diameter PDMS-formed wells. 1,2-bis(4-pyridyl)ethylene (BPE; 10-4 Molar in methanol) was used as an external control to ensure consistency between substrates. Raman spectra were acquired using a Renishaw inVia Reflex multi-wavelength confocal imaging microscope (Hoffman Estates, IL). A Leicha apochromatic 5× objective (NA 0.12) illuminated a 1265 μm2 area on the substrate, which allows spatial averaging and minimizes the effect of potential random hot spots. A 785-nm near-infrared diode laser (Renishaw) operating at 10% power capacity (28 mW) provided the incoming radiation, and spectra were collected in 3 10-sec acquisitions. An internal silicon standard measurement was obtained at the beginning of each SERS analysis as an internal control for instrument performance.
All samples were applied in duplicate to the NA substrates at the concentrations specified, in a volume of 1 μl per well, and allowed to dry overnight. Spectra were collected from five random locations within each sample spot for analysis, for a total of 10 spectra per sample, and M. pneumoniae reference strain and growth medium controls were independently prepared and analyzed for each substrate. Two wells per substrate were intentionally left blank to obtain a background SERS reading on the naked nanorod substrate only. A total of three separate NA substrates were used for these experiments: two for analysis of M. pneumoniae isolates with n = 390 spectra, and one for analysis of other human and commensal Mollicutes species with n = 150 spectra, resulting in a total of n = 540 spectra. Spectra were deliberately collected from multiple locations within a single substrate, as well as from multiple substrates, to ensure any inherent variance present in the substrates did not impact the results. Raman spectra between 400–1800 cm-1 were acquired using Renishaw’s WiRE 3.4 software. Instrument settings were optimized to maximize signal and minimize saturation or sample degradation arising from laser stimulation [13, 20].
Raman spectra were first averaged using GRAMS32/A1 spectral software package (Galactic Industries, Nashua, NH) in order to assess signal-to-noise quality, and baseline-corrected using a concave rubberband algorithm which performed 10 iterations on 64 points to aid in preliminary evaluation of the spectra and peak assignment (OPUS, Bruker Optics, Inc., Billerica, MA). Chemometric analysis was carried out with MATLAB version 7.10.0 (The Mathworks, Inc., Natick, MA) using PLS-Toolbox version 7.5.1 (Eigenvector Research Inc., Wenatchee, WA). Raw spectra were pre-processed using the 1st derivative of each spectrum and a 15-point, 2nd order polynomial Savitsky-Golay algorithm. Each dataset was then vector-normalized and mean-centered. Due to the inherently complex nature of spectral data, multivariate statistical analysis of the datasets was performed using principal component analysis (PCA) and partial least squares-discriminatory analysis (PLS-DA), using the PLS Toolbox software. Unless otherwise specified, all PLS-DA models were cross-validated using a Venetian blinds algorithm with 10 data splits. All PLS-DA models in this study, excluding those for individual sample analysis, were generated using between 110–495 total spectra per model.
Results and Discussion
Detection of M. pneumoniae clinical isolates
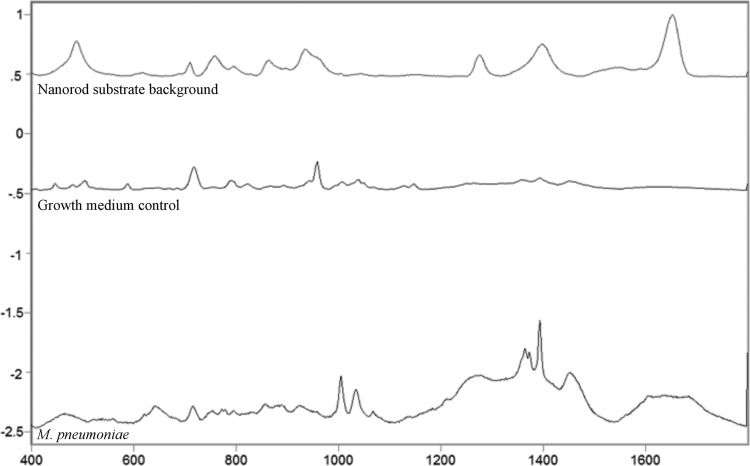
We analyzed 32 clinical isolates, including reference strains M129 (type 1) and FH (type 2), alongside a growth medium control prepared in parallel with the M. pneumoniae samples. Full details regarding isolate origin and year, P1 genotype, macrolide susceptibility, protein and DNA content, and genome equivalents for M. pneumoniae strains are given in Table 1. CFU values were determined for both reference strains and six randomly chosen additional isolates to assess cell viability at time of fixation and ranged from 1x105 to 1x107 CFU/ml. Due to the propensity for mycoplasma cells to clump, a confounding factor in using CFU values as a metric for sample content is the potential discrepancy between CFU value and actual cell number, which can differ by as much as 103-fold [39]. Therefore, protein content and genome equivalents were determined in order to better define the content of the samples at the concentration analyzed by SERS. These values fell within comparable ranges and were consistent with published values for bacterial cells [40]. Protein concentration per cell was higher for M. pneumoniae isolates harvested during stationary phase relative to those harvested during log phase (growth phase based on the color of the pH indicator in the SP4 medium), but no notable differences in genome equivalents or SERS spectra were observed between M. pneumoniae samples relative to growth phase at time of harvest (data not shown). Average SERS spectra of the nanorod substrate background, growth medium control, and M. pneumoniae samples are shown in Fig 1, with each class exhibiting a distinct band pattern, as expected.
Table 1. M. pneumoniae strain details.
| Isolate | Location | Year isolated | Genotype a | Macrolide phenotype b | Protein content (μg/μl) | DNA content (ng/μl) | Genomic equivalents (cells/μl) | fg of protein / cell |
|---|---|---|---|---|---|---|---|---|
| E1 | Egypt | 2009 | 2 | S | 0.149 | 2.0 | 4.49x106 | 26 |
| E16 | Egypt | 2010 | 1 | S | 0.057 | 1.7 | 4.34x106 | 11 |
| K3 | Kenya | 2010 | 2 | S | 0.065 | 2.3 | 6.5x106 | 10 |
| K20 | Kenya | 2010 | 1 | S | 0.044 | 1.2 | 3.39x106 | 12.9 |
| G6 | Guatemala | 2010 | 1 | S | 0.078 | 2.6 | 7.36x106 | 10.6 |
| NM2 | NM | 2010 | 1 | R | 0.074 | 1.1 | 3.11x106 | 23.8 |
| RI1 | RI | 2011 | 2 | S | 0.071 | 1.5 | 4.24x106 | 16.7 |
| OR1 | OR | 2011 | 1 | R | 0.076 | 1.3 | 3.68x106 | 20.7 |
| WV1 | WV | 2011 | 2 | S | 0.094 | 1.9 | 5.34x106 | 17.6 |
| WV9 | WV | 2012 | 1 | R | 0.071 | 1.3 | 3.68x106 | 19.3 |
| FL1 | FL | 2012 | 1 | S | 0.098 | 2.3 | 6.5x106 | 15 |
| WI11 | WI | 2014 | 2V | S | 0.085 | 1.6 | 4.53x106 | 18.8 |
| WI17 | WI | 2014 | 2V | S | 0.047 | 1.8 | 5.09x106 | 9.2 |
| CO12 | CO | 2014 | 2V | S | 0.077 | 1.6 | 4.53x106 | 16.9 |
| CO44 | CO | 2014 | 2V | R | 0.073 | 1.3 | 3.68x106 | 19.8 |
| SA18 | S. Africa | 2013 | 2V | S | 0.35 | 1.7 | 4.81x106 | 72.8 |
| SA19 | S. Africa | 2013 | 2 | S | 0.47 | 1.1 | 3.11x106 | 151.0 |
| SA22 | S. Africa | 2013 | 1 | S | 0.44 | 1.1 | 3.11x106 | 141.5 |
| 1005 | NY | 1999 | 2 | S | 0.28 | 1.5 | 4.24x106 | 66.0 |
| 1134 | IN | 1999 | 2 | S | 0.30 | 2.2 | 6.22x106 | 70.8 |
| 988 | Canada | 1992 | 1 | S | 0.12 | 2.0 | 5.65x106 | 19.3 |
| 678 | Denmark | 1962 | 1 | S | 0.12 | 3.5 | 9.9x106 | 12.1 |
| 682 | Denmark | 1988 | 2 | S | 0.16 | 2.1 | 5.94x106 | 26.9 |
| 983 | SC | 1988 | 2 | S | 0.16 | 1.6 | 4.53x106 | 35.3 |
| 386 | TX | 1994 | 2 | S | 0.10 | 2.9 | 8.2x106 | 12.1 |
| 519 | CA | 1995 | 2 | S | 0.28 | 1.5 | 4.24x106 | 63.6 |
| GA1 | GA | 2012 | 1 | S | 0.22 | 3.5 | 9.9x106 | 22.2 |
| GA3 | GA | 2012 | 2V | S | 0.46 | 3.3 | 9.33x106 | 49.3 |
| IL1 | IL | 2012 | 1 | R | 0.12 | 1.7 | 4.81x106 | 24.9 |
| IL2 | IL | 2012 | 1 | R | 0.10 | 1.8 | 5.09x106 | 19.6 |
| M129 | N/A | N/A | 1 | S | 0.169 | 4.1 | 1.13x107 | 14 |
| M129 c | N/A | N/A | 1 | S | 0.168 | 4.3 | 1.20x107 | 14 |
| FH | N/A | N/A | 2 | S | 0.046 | 2.1 | 5.94x106 | 7.7 |
| FH c | N/A | N/A | 2 | S | 0.046 | 2.2 | 6.30x106 | 7.3 |
a M. pneumoniae P1 genotype groups based upon DNA sequence analysis performed by the Pneumonia Response and Surveillance Laboratory at the CDC in Atlanta, GA.
b S, susceptible; R, resistant
c Two independent preparations of M129 and FH were examined
N/A = information not available
Fig 1. Averaged, baseline-corrected, and normalized SERS spectra for the nanorod substrate, growth medium control, and M. pneumoniae reference strain controls and clinical isolates, as indicated.
Raw spectra of the three sample classes were averaged, baseline-corrected, and normalized using GRAMS32/A1 spectral software package (Galactic Industries, Nashua, NH). For the nanorod substrate background class, n = 20; for the growth medium control class, n = 20; and for the M. pneumoniae class, n = 350.
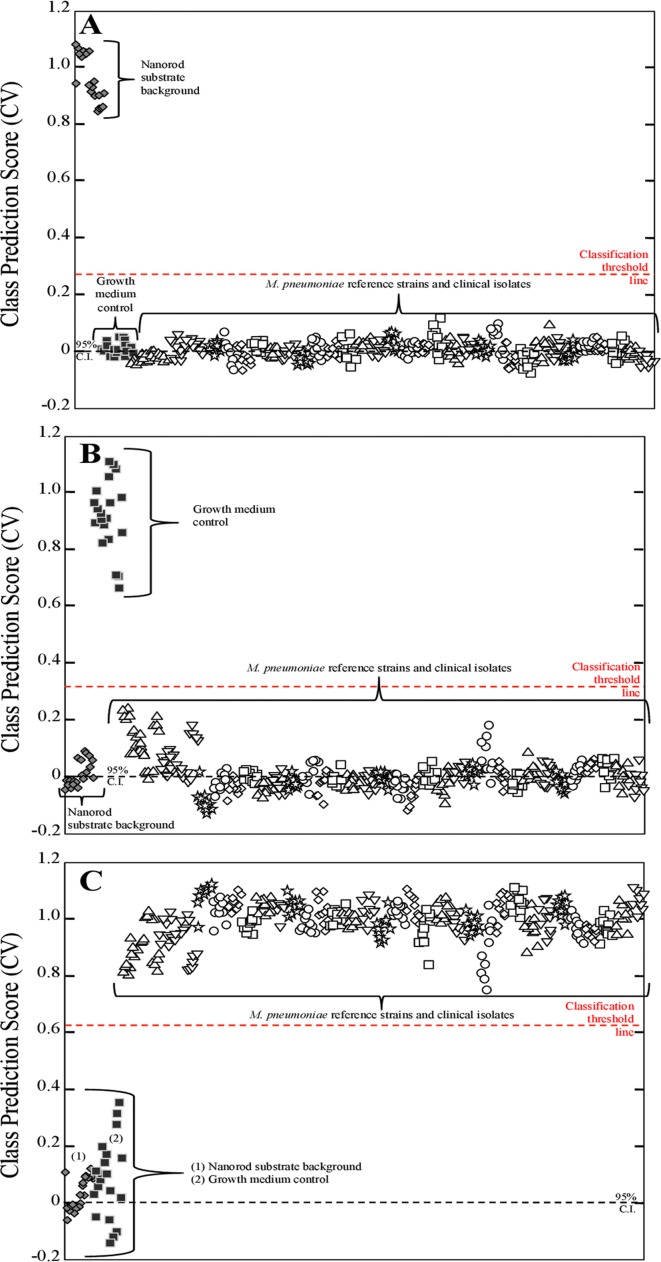
PLS-DA was applied here to determine statistically significant detection of M. pneumoniae by NA-SERS. PLS-DA is a full-spectrum, multivariate, supervised statistical method whereby prior knowledge of classes is used to yield more robust discrimination by minimizing variation within classes while emphasizing latent variables arising from spectral differences between classes [41, 42]. A PLS-DA model was generated to discriminate between three classes: the nanorod substrate background (Fig 2A); the growth medium control (Fig 2B); and all M. pneumoniae strains (Fig 2C). The inclusion of substrate background and growth medium controls allowed us to ensure that any differences in growth medium and nanorod background signal within the substrate did not affect the ability of the model to discriminate between the presence or absence of M. pneumoniae. Two nanorod substrates were used for these experiments, with each containing duplicate wells of the bare nanorod substrate, independently prepared M129, FH, and growth medium controls, and 15 additional clinical isolates of M. pneumoniae. A total of n = 390 pre-processed NA-SERS spectra collected from both substrates were included in the model, consisting of 20 nanorod substrate background spectra, 20 growth medium control spectra, 25 M129 spectra, 25 FH spectra, and 10 spectra per additional clinical isolate. The cross-validated statistics for the model show that NA-SERS correctly classified all 32 clinical isolates as M. pneumoniae regardless of global origin, year isolated, genotype, or macrolide susceptibility phenotype, and distinguished them from the substrate background and the growth medium control with 100% cross-validated sensitivity and specificity.
Fig 2. PLS-DA of 32 M. pneumoniae clinical isolates, including reference strains M129 and FH.
Each panel represents a cross-validated class prediction score for (A) class 1, substrate background; (B) class 2, growth medium control; and (C) class 3, all M. pneumoniae strains. For panels A-C, each individual shape represents a single pre-processed NA-SERS spectrum. The substrate background spectra are represented by gray diamonds, the growth medium control spectra by solid black squares, and the M. pneumoniae spectra by open shapes that differ by cluster to indicate the different individual strains and isolates. The red-dotted line indicates the classification threshold line for positive class prediction, and the black-dotted line indicates the 95% confidence interval. Cross-validated sensitivity, specificity, and class error for the panels were as follows: (A) nanorod substrate background: 1.00, 1.00, and 0, respectively; for (B) growth medium control: 1.00, 1.00, and 0, respectively; and for (C) M. pneumoniae: 1.00, 1.00, and 0, respectively. Cross-validated statistics were obtained using Venetian blinds with 10 data splits to represent the prediction performance of the PLS-DA model for M. pneumoniae detection.
Differentiation of M. pneumoniae and 12 other human commensal and pathogenic Mollicutes species
A critical question for clinical detection platforms is specificity for the pathogen of interest, particularly in the context of other organisms potentially present in a clinical sample. SERS is a structure-based technique that generates a Raman fingerprint or barcode based on the unique molecular content of the sample, and as such, the most likely organisms to generate false positives would be those most closely resembling M. pneumoniae structurally. To evaluate the specificity of the NA-SERS biosensing platform, 12 human commensal and pathogenic Mollicutes species closely related to M. pneumoniae in rpoB β-subunit nucleotide and amino acid sequence phylogenies and 16S rDNA phylogeny were chosen for analysis alongside M. pneumoniae strain M129 and a growth medium control [33]. In order to best define the content of the sample at the concentration used for SERS, analyses were done to determine total protein and DNA content, the latter allowing calculation of genome equivalents based on known genome sizes and G+C content (Table 2). These fell within comparable ranges and were consistent with published values for bacterial cells [40].
Table 2. Quality control and sample information for Mollicutes species and M. pneumoniae control cultures.
| Commensal organism | Genome Size (Bp) a | Protein content (μg/μl) | DNA content (ng/μl) | Genomic equivalents (cells/μl) | fg protein/cell | CFU/μl |
|---|---|---|---|---|---|---|
| Acholeplasma laidlawii | 1,496,992 | 0.247 | 16.2 | 1.9x106 | 130 | 4.55x105 |
| Mycoplasma amphoriforme | 1,029,022 | 0.072 | 2.4 | 4.3x105 | 167 | ND |
| Mycoplasma fermentans | 1,118,751 | 0.374 | 23.8 | 3.9x106 | 95.9 | 2.6x106 |
| Mycoplasma genitalium | 580,073 | 0.099 | 2.6 | 8.2x105 | 121 | 1.75x104 |
| Mycoplasma hominis | 665,445 | 0.454 | 10.7 | 2.9x106 | 157 | 2.2x105 |
| Mycoplasma orale | 710,549 | 0.114 | 8.1 | 2.1x106 | 54.3 | 9.9x104 |
| Mycoplasma penetrans | 1,358,633 | 0.813 | 19 | 2.6x106 | 313 | 1.7x106 |
| Mycoplasma pirum | 510,593 | 0.384 | 20 | 7.2x106 | 53.3 | 3.85x106 |
| Mycoplasma salivarium | 710,000 | 0.558 | 32.1 | 8.32x106 | 67.1 | 1x105 |
| Mycoplasma spermatophilum | 846,000 | 0.068 | 1.2 | 2.6x105 | 261 | ND |
| Ureaplasma parvum | 727,289 | 0.068 | 1.7 | 4.3x105 | 158 | 4.5x104 |
| Ureaplasma urealyticum | 874,478 | 0.067 | 2 | 4.2x105 | 159 | 5.6x103 |
| Mycoplasma pneumoniae | 816,394 | 0.061 | 1 | 2.24x106 | 27.2 | 8.4x104 |
aGenome sizes obtained from published sources [35] and known G+C contents from the GenBank database
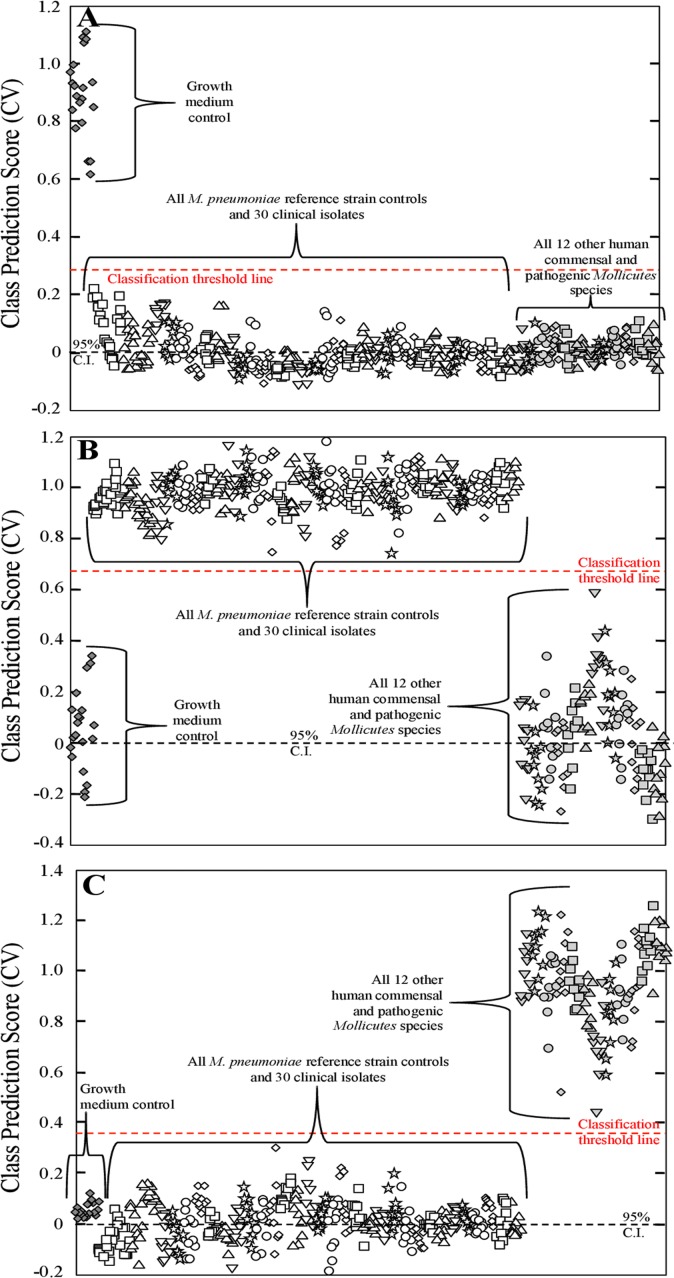
Our goal here was to develop a PLS-DA model that distinguished M. pneumoniae from a panel of closely-related other Mollicutes species that might be found in humans. A total of n = 150 pre-processed NA-SERS spectra were collected on a single nanorod substrate consisting of n = 10 substrate background spectra, n = 10 growth medium control spectra, n = 10 M. pneumoniae spectra, and 10 spectra each per other Mollicutes species. An initial PLS-DA model was generated to discriminate between two classes, the nanorod substrate background and all other biological samples, which it did with 100% cross-validated sensitivity and specificity (data not shown). The purpose of this model was to ensure that the nanorod substrate background signal was significantly different than all other samples in order to exclude the background spectra from our future models. Once we determined that the nanorod substrate background class could be excluded, a second PLS-DA model was generated using the same spectra to distinguish among three classes; the growth medium control, M. pneumoniae, and the other Mollicutes species. This model had a total of n = 140 pre-processed NA-SERS spectra, consisting of n = 10 growth medium control spectra, n = 10 M. pneumoniae spectra, and 10 spectra each per other Mollicutes species. This model distinguished the three classes with 100% cross-validated sensitivity and specificity (data not shown). Upon the successful development of a PLS-DA model to distinguish between the growth medium control, M. pneumoniae, and all other Mollicutes species, a final PLS-DA model was generated using pre-processed NA-SERS spectra from all three nanorod substrates analyzed during these experiments. This model contained a total of n = 495 spectra, consisting of 25 growth medium control spectra, 25 M129 spectra, 25 FH spectra, 10 spectra each per other M. pneumoniae clinical isolates (30 isolates total), and 10 spectra each per other Mollicutes species (12 species total). This model was also categorized into three classes: the growth medium control (Fig 3A); all M. pneumoniae clinical isolates, including reference strains (Fig 3B); and all other Mollicutes species (Fig 3C). PLS-DA distinguished all M. pneumoniae strains from all 12 other human Mollicutes species and the growth medium control with 100% cross-validated sensitivity and specificity (Fig 3A–3C).
Fig 3. PLS-DA distinguishing M. pneumoniae strains from other human commensal and pathogenic Mollicutes species.
Each panel represents a cross-validated class prediction score for (A) class 1, growth medium control; (B) class 2, all M. pneumoniae strains; and (C) class 3, all other human commensal and pathogenic Mollicutes samples. For panels A-C, each individual shape represents a single pre-processed NA-SERS spectrum. The growth medium control spectra are represented by gray diamonds, the M. pneumoniae spectra by open shapes that differ by cluster to indicate the different individual strains and isolates, and the human commensal and pathogenic Mollicutes species are represented by light gray shapes that differ by cluster to indicate the individual species. The red-dotted line indicates the classification threshold line for positive class prediction, and the black-dotted line indicates the 95% confidence interval. Cross-validated sensitivity, specificity, and class error for the panels were as follows: (A) growth medium control: 1.00, 1.00, and 0, respectively; for (B) All M. pneumoniae samples: 1.00, 1.00, and 0, respectively; and for (C) All 12 Mollicutes species: 1.00, 1.00, and 0, respectively. Cross-validated statistics were obtained using Venetian blinds with 10 data splits to represent the prediction performance of the PLS-DA model for M. pneumoniae detection.
M. pneumoniae typing capabilities of NA-SERS
A key advantage of NA-SERS for biosensing is the potential to detect and type an organism in a single test, especially of interest here since there is currently no existing platform capable of the simultaneous detection and typing of M. pneumoniae. To evaluate this capability we applied PLS-DA to the M. pneumoniae strain spectra above. Our panel of clinical isolates contained three distinct and clinically relevant genotypes of M. pneumoniae: 13 type 1 strains, 11 type 2 strains, and six type 2 variant (2V) strains. M. pneumoniae strains M129 (type 1) and FH (type 2) were used as reference strain controls, as they have been previously applied in this manner for evaluation of M. pneumoniae genotyping assays [19].
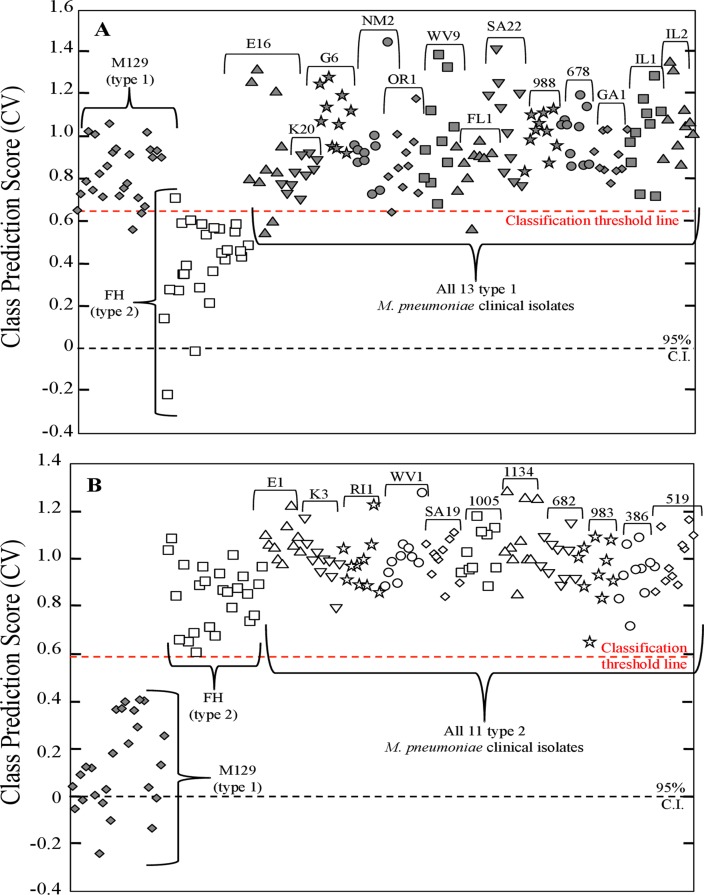
For the type 1 strains a PLS-DA model was generated using 180 pre-processed NA-SERS spectra consisting of the 25 M129 spectra and the 25 FH spectra as controls, and all spectra from the 13 other type 1 clinical isolate samples (10 spectra per isolate, 130 total spectra). The model was built to discriminate between 2 classes, either type 1 or type 2. PLS-DA was able to correctly classify all other 13 type 1 strains with the type 1 reference strain with 96.8% sensitivity and 96% specificity (Fig 4A). For the type 2 strains a second PLS-DA model was generated using 160 pre-processed NA-SERS spectra consisting of the 50 type 1 and 2 reference strain control spectra and all spectra from the 11 other type 2 clinical isolate samples (10 spectra per isolate, 110 total spectra). This model was likewise built to discriminate between 2 classes, either type 1 or type 2. PLS-DA was able to correctly classify all 11 other type 2 isolates with the type 2 reference strain control with 99.3% sensitivity and 100% specificity (Fig 4B).
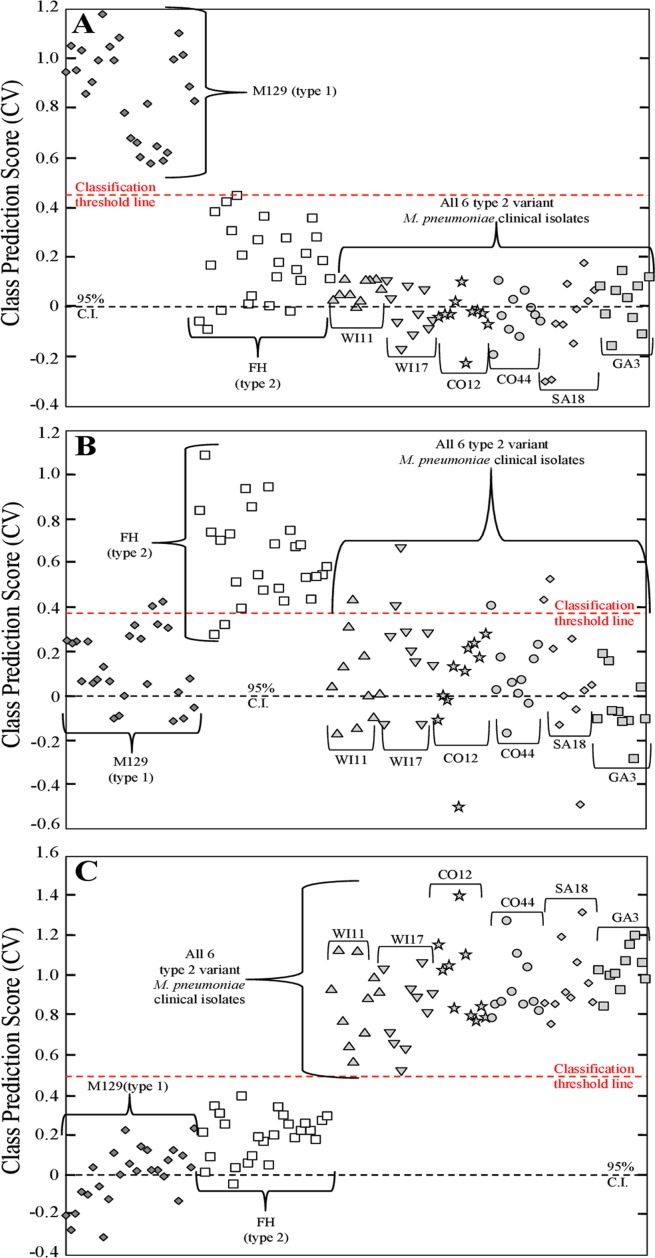
Fig 4. PLS-DA for NA-SERS typing of type 1 and 2 M. pneumoniae strains.
Cross-validated class prediction scores for (A) all 13 type 1 clinical isolates, and (B) all 11 type 2 clinical isolates. For panels A and B, each individual shape represents a single pre-processed NA-SERS spectrum. M. pneumoniae type 1 reference strain control and other clinical isolates are represented by dark gray diamonds, while the type 2 reference strain control and other clinical isolates are represented by open shapes. Shapes differ by cluster to indicate the individual clinical isolates and samples, and the strain/isolate designation is indicated above the brackets for each cluster. The red-dotted line indicates the classification threshold line for positive class prediction, and the black-dotted line indicates the 95% confidence interval. The cross-validated sensitivity, specificity and class error were obtained using Venetian blinds with 10 data splits to represent the prediction performance of models for classification of (A) type 1 strains: 0.968, 0.96, and 0.04, respectively and for (B) type 2 strains: 1.00, 0.993, and 0.004, respectively.
For type 2V clinical isolates, a third PLS-DA model was generated using 110 pre-processed NA-SERS spectra consisting of the 50 type 1 and 2 reference strain control spectra and all spectra from the type 2V clinical isolate samples (10 spectra per isolate, 110 total spectra). However, this model was built to discriminate between 3 classes, type 1 reference strain control, type 2 reference strain control, or type 2V clinical isolate spectra. A third class was necessary for classification of this genotype, as existing methods are capable of identifying variant strains as unique from type 1 and 2 isolate strains [18], and as such for clinical purposes NA-SERS typing should be able to do the same. PLS-DA correctly classified the type 1 reference strain control as distinct from the type 2 control and the type 2V clinical isolates with 100% cross-validated sensitivity and 98.8% cross-validated specificity (Fig 5A). Furthermore, PLS-DA distinguished the type 2 reference strain control from the type 1 control and 2V clinical isolates with a cross-validated sensitivity and specificity of 92% and 90.6%, respectively (Fig 5B). Lastly, PLS-DA correctly classified all six type 2V strains as distinct from the type 1 and 2 reference strain controls with 100% cross-validated sensitivity and specificity (Fig 5C). The drop in sensitivity and specificity observed for the type 2 reference strain control is likely due to the fact that these are variant strains of the type 2 parent strain, and variant strains tend to be more similar to their respective parent strains genetically than either are to the opposite strain type [18, 37].
Fig 5. PLS-DA for NA-SERS typing of type 2V M. pneumoniae clinical isolates.
Cross-validated class prediction scores for (A) class 1, the type 1 reference strain control; (B) class 2, the type 2 reference strain control; and (C) class 3, all six type 2V clinical isolates. For the panels A-C, each individual shape represents a single pre-processed NA-SERS spectrum. The M. pneumoniae type 1 reference strain control is represented by dark gray diamonds, the type 2 reference strain control is represented by open squares, and the type 2V clinical isolates are represented by light gray shapes. The light gray shapes differ by cluster to indicate the individual clinical isolates, and the strain/isolate designation is indicated above the brackets for each cluster. The red-dotted line indicates the classification threshold line for positive class prediction, and the black-dotted line indicates the 95% confidence interval. The cross-validated sensitivity, specificity and class error for panels A-C were obtained using Venetian blinds with 10 data splits to represent the prediction performance of (A) type 1 strains: 1.00, 0.988, and 0.006, respectively; for (B) type 2 strains: 0.92, 0.906, and 0.08, respectively; and for (C) type 2V strains: 1.00, 1.00, and 0, respectively.
To further evaluate the strain typing capabilities of NA-SERS, PLS-DA models were generated using the M129 and FH reference strains alongside each clinical isolate individually. Thirty PLS-DA models were built using the 25 type 1 M129 spectra and 25 type 2 FH spectra as reference strain control classes, and 10 clinical isolate spectra treated as an unknown class. For type 1 and 2 clinical isolates, two categories were used for cross-validation of the model, while for type 2V isolate strains, three categories were incorporated to cross-validate the model, as described above. For all clinical isolate types the model was cross-validated by using a Venetian blinds algorithm with seven data splits. These PLS-DA models were incorporated to simulate a potential strategy for future application of NA-SERS for M. pneumoniae genotyping wherein known strain type controls are used to predict the genotype of an unknown clinical sample. Full cross-validated statistics for all 30 PLS-DA models are given in Table 3 for all type 1 and 2 clinical isolates, and Table 4 for all type 2V clinical isolates. Overall, PLS-DA performance was consistent with the models shown in Figs 4 and 5. The only notable difference in performance was a decrease in cross-validated specificity in the individual modeling for type 1 clinical isolates K20, NM2, and FL1, but this likely arises due to the decreased sample size (n = 60) used to build the individual PLS-DA models.
Table 3. Cross-validated PLS-DA modeling statistics for the prediction performance for NA-SERS typing of individual type 1 and 2 M. pneumoniae clinical isolates.
| Isolate | P1 Type | CV Sensitivity | CV Specificity | CV Class Error |
|---|---|---|---|---|
| E16 | 1 | 0.943 | 0.92 | 0.06 |
| K20 | 1 | 1 | 0.84 | 0.08 |
| G6 | 1 | 1 | 0.92 | 0.04 |
| NM2 | 1 | 1 | 0.88 | 0.06 |
| OR1 | 1 | 0.914 | 0.92 | 0.08 |
| WV9 | 1 | 0.914 | 0.96 | 0.06 |
| FL1 | 1 | 0.971 | 0.84 | 0.09 |
| SA22 | 1 | 0.971 | 1 | 0.01 |
| 988 | 1 | 0.971 | 1 | 0.01 |
| 678 | 1 | 1 | 1 | 0 |
| GA1 | 1 | 1 | 1 | 0 |
| IL1 | 1 | 1 | 1 | 0 |
| IL2 | 1 | 0.971 | 1 | 0 |
| E1 | 2 | 1 | 0.971 | 0.01 |
| K3 | 2 | 1 | 1 | 0 |
| RI1 | 2 | 1 | 1 | 0 |
| WV1 | 2 | 1 | 1 | 0 |
| SA19 | 2 | 1 | 1 | 0 |
| 1005 | 2 | 1 | 1 | 0 |
| 1134 | 2 | 1 | 0.971 | 0.01 |
| 682 | 2 | 1 | 1 | 0 |
| 983 | 2 | 1 | 1 | 0 |
| 386 | 2 | 1 | 1 | 0 |
| 519 | 2 | 1 | 1 | 0 |
Two categories were used for cross-validation of the model, either type 1 or type 2. Clinical isolates were treated as an unknown class and cross-validated sensitivity, specificity, and class error were based on their classification prediction score with their respective reference strain control class. CV, cross-validated.
Table 4. Cross-validated PLS-DA modeling statistics for the prediction performance for NA-SERS typing of individual type 2V M. pneumoniae clinical isolates.
| Isolate | P1 Type | CV Sensitivity | CV Specificity | CV Class Error |
|---|---|---|---|---|
| 1: M129 | 1 | 1 | 0.971 | 0.01 |
| 2: FH | 2 | 0.8 | 0.943 | 0.13 |
| 3: WI11 | 2V | 1 | 0.98 | 0.01 |
| 1: M129 | 1 | 1 | 1 | 0 |
| 2: FH | 2 | 0.88 | 0.943 | 0.08 |
| 3: WI17 | 2V | 1 | 0.98 | 0.01 |
| 1: M129 | 1 | 1 | 0.971 | 0.01 |
| 2: FH | 2 | 0.84 | 1 | 0.08 |
| 3: CO12 | 2V | 1 | 1 | 0 |
| 1: M129 | 1 | 1 | 1 | 0 |
| 2: FH | 2 | 0.92 | 0.971 | 0.05 |
| 3: CO44 | 2V | 1 | 0.98 | 0.01 |
| 1: M129 | 1 | 1 | 1 | 0 |
| 2: FH | 2 | 1 | 0.943 | 0.03 |
| 3: SA18 | 2V | 1 | 1 | 0 |
| 1: M129 | 1 | 1 | 1 | 0 |
| 2: FH | 2 | 0.96 | 1 | 0.02 |
| 3: GA3 | 2V | 1 | 1 | 0 |
Three categories were incorporated to cross-validate the model: either type 1; type 2; or neither. Clinical isolates were treated as an unknown class and cross-validated sensitivity, specificity, and class error were based on their classification prediction score as neither type 1 or type 2 reference control strains (i.e. category 3).
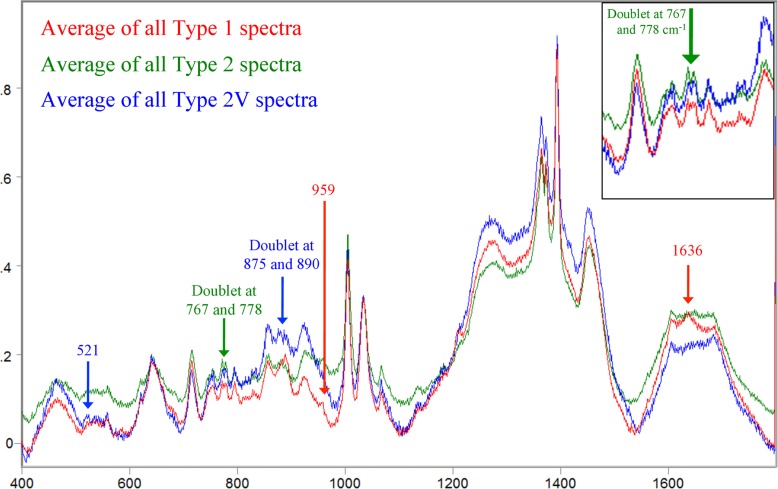
Additionally, we compared averaged, baseline-corrected, and normalized spectra of all three genotypes to look for any differences in band pattern between the three genotypes that could be contributing to the classification capabilities demonstrated in the PLS-DA modeling (Fig 6). The majority of the spectral fingerprint was identical for all three strain types, which is to be expected since they are all the same species and classify as such in the PLS-DA models shown in Figs 2 and 3. However, several visible differences in band pattern were present in the spectra for each genotype of M. pneumoniae, which could account for the ability of NA-SERS to distinguish between the three genotypes with statistically significant sensitivity and specificity. The averaged type 1 spectrum had two unique peaks, one at 1636 cm-1 that does not appear in the averaged type 2 or 2V spectra, and one at 959 cm-1 which appeared as more distinct and shifted slightly right in the type 1 spectrum when compared to the type 2 spectrum, and did not appear in the type 2V spectrum. The averaged type 2 strain spectrum was very similar to the type 1 strain spectrum aside from the differences mentioned above and the presence of a doublet at 767 and 778 cm-1, which appeared as more distinct than that present in the type 2V spectrum and as a broad singlet in the type 1 spectrum. The averaged type 2V spectrum appeared to be the most distinct of the three, with a doublet at 875 and 890 cm-1 that appeared as a single peak at 890 in type 1 and 2 spectra, and a small peak at 521 that was also absent in type 1 and 2 spectra. While these spectral differences were extremely subtle, chemometric analysis is highly capable of discerning differences such as these with substantial discriminatory classification power [43].
Fig 6. Comparison of averaged, baseline-corrected, and normalized SERS spectra for type 1, type 2, and type 2V genotypes.
Raw spectra of all type 1 (n = 155), type 2 (n = 135), and type 2V (n = 60) clinical isolates and controls were averaged, baseline-corrected, and normalized using GRAMS32/A1 spectral software package (Galactic Industries, Nashua, NH). Red, average spectrum of all type 1 M. pneumoniae strains; green, average spectrum of all type 2 M. pneumoniae strains; blue, average spectrum of all type 2V M. pneumoniae strains. Peaks unique to a specific genotype of M. pneumoniae are indicated by arrows and identified above the spectral fingerprint. Type 1 peaks, red arrows; type 2 peaks, green arrows; and type 2V peaks, blue arrows. Inset at top right of image depicts zoomed-in view of the type 2 doublet at 767 and 778 cm-1.
Although little is known about the phenotypic effects of strain type beyond observable differences in biofilm formation [44], the genotypic differences between them are well characterized. Briefly, homologous recombination within the p1 gene of repetitive element sequences located both in and outside the p1 gene contributes to sequence variation between strain types [7]. Nucleotide and amino acid sequencing of 60 M. pneumoniae isolates indicates that trinucleotide short sequence repeats (SSR’s) coding for serine can be found in all strain types anywhere from 5–14 times, but appear to be most prevalent in type 1 strains [7]. Serine repeats may form a hinge structure and lead to downstream conformational differences in the P1 protein between the different strain types, which could potentially affect its interaction with the host as a surface antigen [45, 46]. In addition, 14 of the 60 isolates in the Zhao et al. study had point mutations in several variant strains corresponding to amino acid changes in P1 to glutamine, proline, asparagine, and isoleucine residues [7].
In our study, the peaks unique to the type 1 spectrum are commonly associated with vibrational mode bonds present in lysine (959 cm-1) and amide I or alpha helix (1636 cm-1) molecular structures [47–49]. The peaks unique to the type 2 spectral fingerprint located at 767 and 778 cm-1 are commonly associated with vibrational modes found in histidine, tryptophan, or carbohydrate bonds [48–50]. Finally, the peaks unique to the type 2V clinical isolates found at 521, 875 and 890 cm-1 are frequently associated with bonds present in histidine, tryptophan, ribose, indole, asparagine, methionine, glutamine, and S-S and C-C stretching vibrational modes [48–50]. Interestingly, the unique peaks present in the average spectra for the strain types analyzed in this study are predominately associated with protein backbone, amino acid residue, and DNA bond vibrations. Furthermore, spectral features in the averaged spectrum of the 2V variant strains are consistent with the point mutations identified in the Zhao et al. study [7], and our overall spectral interpretation of the averaged spectra for each strain type is consistent with what is known about the differences between strain types of M. pneumoniae infection.
Unsupervised chemometric analysis of M. pneumoniae strain types and Mollicutes species
We applied PCA to supplement the PLS-DA modeling of sample spectra and evaluate the total variance present in our M. pneumoniae typing and other human commensal and pathogenic Mollicutes datasets. PCA is an unsupervised form of chemometric analysis which reduces the dimensionality of the dataset and facilitates establishing patterns and grouping of similar spectra without a priori knowledge of sample class [43]. PCA explains successively smaller proportions of the variance, with the first few principal components explaining the greatest percentage of total variance present in the dataset [51].
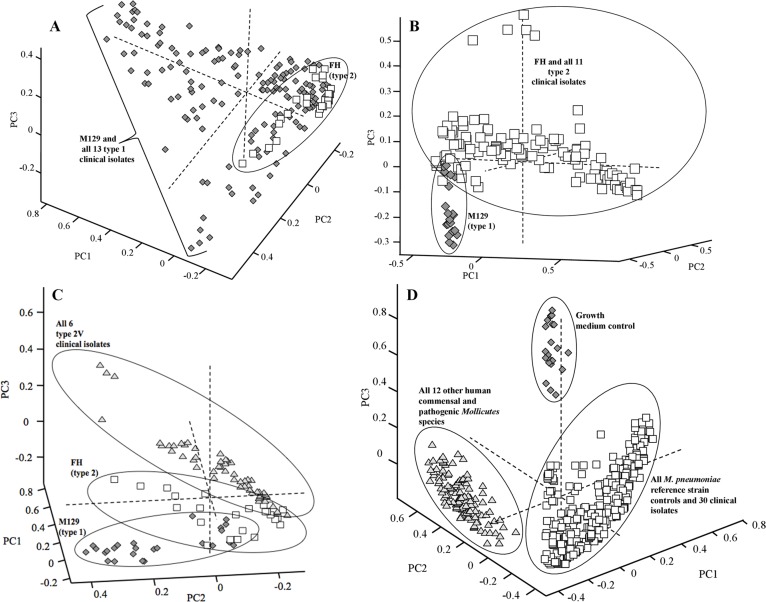
Pre-processed SERS spectra from M. pneumoniae reference strain type 1 and 2 controls and all other type 1 clinical isolates were used to generate a PCA plot comparing principle components 1, 2, and 3, which captured 54.3% of the total variance present in the 180 spectra used to build the model (Fig 7A). Type 2 control strain FH clustered in the bottom right corner, and the clustering pattern for all type 1 strains was predominately below and to the left, though some overlap between the two strain types was present. The PCA model of the type 1 clinical isolates supports the PLS-DA modeling of the spectra shown in Fig 4A.
Fig 7. Principle component analysis of M. pneumoniae strain typing and other human commensal and pathogenic Mollicutes species.
For all panels, each individual shape represents a single sample spectrum. PC scores plots of 1 vs. 2 vs. 3 of: (A) M. pneumoniae reference strains and all 13 other type 1 clinical isolates; (B) M. pneumoniae reference strains and all 11 other type 2 clinical isolates; (C) M. pneumoniae type 1 reference strain, type 2 reference strain, and all six type 2V clinical isolates; and (D) growth medium control, all M. pneumoniae strains, and all 12 other human commensal and pathogenic Mollicutes species. For panels A-C, dark gray diamonds represent type 1 sample spectra whereas open squares represent the type 2 sample spectra. In panel C, type 2V clinical isolate spectra are represented by light gray triangles. For panel D, growth medium control spectra are represented by dark gray diamonds, M. pneumoniae spectra by open squares, and all 12 other Mollicutes species by light gray triangles. For panels A-D, clustering of samples is indicated by black circles or brackets.
A second PCA model was built using pre-processed SERS spectra (n = 160) consisting of type 1 and 2 reference strain controls and all other type 2 clinical isolates of M. pneumoniae. Principal components 1–3 captured 58.0% of the total variance and when plotted orthogonally showed a distinct separation between the type 1 reference strain control and all type 2 reference strain and other isolates, with very little overlap of clusters (Fig 7B). PCA modeling for the type 2 clinical isolate dataset was consistent with the PLS-DA modeling of the data shown in Fig 4B.
A third PCA model was built using the pre-processed SERS spectra from the type 2V clinical isolate dataset (n = 110). Principal components 1–3 captured 54.1% of the total variance and when plotted orthogonally showed distinctly separated clusters for the type 1 control, the type 2 control, and the type 2V clinical isolates, with some overlap present between the type 2 and type 2V clusters (Fig 7C). This clustering pattern further supports the PLS-DA classification performance shown in Fig 5.
Finally, a PCA model was built using the full M. pneumoniae and Mollicutes species dataset consisting of pre-processed spectra from all 3 nanorod array substrates (n = 495). Principal components 1–3 captured 50.1% of the total variance and when plotted orthogonally showed three distinctly separated clusters for growth medium control spectra, all M. pneumoniae spectra, and all other Mollicutes species spectra, with no overlap between clusters (Fig 7D). This supports the PLS-DA model of the data shown in Fig 3.
Species-level discrimination by NA-SERS
Our final question of interest was the ability of the platform to discriminate among the 13 different species analyzed in this study. However, PLS-DA classification performance diminishes as the number of classes in question increases, due to the underlying algorithms applied by the modeling, and as such, attempts to classify all 13 species within a single PLS-DA model failed to yield statistically significant accuracy. In order to overcome this limitation and address the question in a clinically relevant classification system, individual pair-wise PLS-DA modeling was employed. For this analysis, we chose to focus on the nine other human commensal and pathogenic mycoplasma species most closely related phylogentically to M. pneumoniae. Individual PLS-DA models were built for each of the nine mycoplasma species to distinguish among three categories: the growth medium control (1), M. pneumoniae (2), or the mycoplasma species in question. Each model contained a total of n = 30 spectra, with 10 spectra representing each category. Furthermore, for each category eight of the 10 spectra were of known sample class, whereas two out of the 10 spectra were treated as unknowns and classified based on their resemblance to category 1, 2, or 3 spectra. Cross-validation of the prediction capability for each model was done using a Venetian blinds algorithm with five data splits. Models were designed this way to simulate the prediction of a potential unknown clinical sample as mycoplasma-negative, M. pneumoniae-positive, or positive for one of the other human commensal or pathogenic mycoplasma species. The cross-validated sensitivities and specificities for all nine models are given in Table 5. Overall, the cross-validated sensitivity and specificity was 90% or greater for all nine models, which is very promising for further application of NA-SERS for species-level discrimination and prediction.
Table 5. Cross-validated PLS-DA modeling statistics for the prediction performance for species discrimination between M. pneumoniae and nine other human commensal and pathogenic mycoplasma species individually.
| Isolate | CV Sensitivity | CV Specificity | CV Class Error |
|---|---|---|---|
| 1: GMC | 1 | 1 | 0 |
| 2: M129 | 1 | 1 | 0 |
| 3: M. amphoriforme | 1 | 0.95 | 0.025 |
| 1: GMC | 1 | 1 | 0 |
| 2: M129 | 1 | 1 | 0 |
| 3: M. fermentans | 1 | 1 | 0 |
| 1: GMC | 1 | 1 | 0 |
| 2: M129 | 1 | 1 | 0 |
| 3: M. genitalium | 1 | 1 | 0 |
| 1: GMC | 0.9 | 1 | 0.05 |
| 2: M129 | 1 | 1 | 0 |
| 3: M. hominis | 1 | 1 | 0 |
| 1: GMC | 1 | 1 | 0 |
| 2: M129 | 1 | 1 | 0 |
| 3: M. orale | 1 | 1 | 0 |
| 1: GMC | 1 | 1 | 0 |
| 2: M129 | 1 | 0.95 | 0.025 |
| 3: M. penetrans | 1 | 1 | 0 |
| 1: GMC | 1 | 1 | 0 |
| 2: M129 | 1 | 1 | 0 |
| 3: M. pirum | 1 | 1 | 0 |
| 1: GMC | 1 | 1 | 0 |
| 2: M129 | 1 | 1 | 0 |
| 3: M. salivarium | 1 | 0.9 | 0.05 |
| 1: GMC | 1 | 1 | 0 |
| 2: M129 | 1 | 1 | 0 |
| 3: M. spermatophilum | 1 | 0.95 | 0.025 |
For species discrimination, three categories were incorporated to cross-validate the model: either category 1: growth medium control (GMC); category 2: M. pneumoniae (M129); or category 3: other mycoplasma species in question. Eight spectra from each category were of known class and two spectra from each category were treated as unknowns to generate a cross-validated prediction model. Cross-validated sensitivity, specificity, and class error were based on the classification prediction score for each category.
Conclusions
M. pneumoniae is a significant human respiratory tract pathogen in both incidence and public health impact, but diagnostic strategies are complicated by the atypical and complex presentation of disease, non-descript symptoms, the requirement for separate tests for detection and genotyping, and the challenges posed by direct culture. Serologic testing was historically the gold standard for diagnosis but suffers from severe limitations that make it both unreliable and impractical for rapid detection. Advances in qPCR technologies have overcome many issues with sensitivity and reliability, but the cost of reagents and requirement for technical expertise are still high, and independent tests must be done for detection and genotyping, limiting diagnosis by qPCR to hospital or advanced laboratory facilities and making it impractical for point-of-care use.
We previously established that this NA-SERS biosensing platform is capable of statistically significant detection of M. pneumoniae in true and simulated throat swabs and has a qualitative endpoint of detection for M. pneumoniae of < 1 cell/μl, a sensitivity exceeding that of qPCR [13, 31]. Here, NA-SERS showed statistically significant specificity for M. pneumoniae detection regardless of clinical isolate origin, year of isolation, macrolide susceptibility phenotype, or strain type, and was also able to distinguish all M. pneumoniae clinical isolates and control strains from 12 other human commensal and pathogenic Mollicutes species. Furthermore, NA-SERS discriminated between the two major strain types of M. pneumoniae with a high degree of statistically significant accuracy and correctly identified variant strains as different from the two major genotypes. Most importantly, NA-SERS was capable of detecting and strain-typing M. pneumoniae within a single test and thus has the potential to facilitate tracking epidemiological trends, such as type-switching and outbreak periodicity [12]. Studies with clinical samples are ongoing, and the effect of the presence of a clinically relevant sample background, for example from a patient’s throat swab, on the ability of this platform to identify strain types or distinguish M. pneumoniae from other human commensal and pathogenic Mollicutes species remains to be determined. Furthermore, assessment of alternative methods of chemometric analysis to account for the increased number of classes and modeling complexity for species-level discrimination by NA-SERS is necessary. In addition, future clinical application of this technology will require collection of a larger spectral library of isolates and background controls. However, from a point-of-care clinical standpoint, the ability to detect M. pneumoniae rapidly is critical to informing appropriate treatment regimens consistent with the responsible use of antimicrobials. This feature is underscored by the availability of handheld Raman instruments having the potential for point-of-care use [52–54]. In combination with the minimal sample preparation requirements and expedient detection, NA-SERS shows great promise as a platform for future application to point-of-care M. pneumoniae diagnostics.
Data Availability
All spectral data are available as CSV file from the Dryad database (datadryad.org; doi:10.5061/dryad.s5h20).
Funding Statement
Funding provided by Public Health Service grants AI096364 to DCK from the National Institute of Allergy and Infectious Disease (www.niaid.nih.gov) and GM102546 to RAD from the National Institute of General Medical Sciences (www.nigms.nih.gov). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chalker V, Stocki T, Litt D, Bermingham A, Watson J, Fleming D, et al. (2012) Increased detection of Mycoplasma pneumoniae infection in children in England and Wales, October 2011 to January 2012. Euro Surveill 17. [PubMed] [Google Scholar]
- 2. Daxboeck F, Krause R, Wenisch C (2003) Laboratory diagnosis of Mycoplasma pneumoniae infection. Clin Microbiol Infect 9: 263–273. [DOI] [PubMed] [Google Scholar]
- 3. Kung CM, Wang HL (2007) Seroprevalence of Mycoplasma pneumoniae in healthy adolescents in Taiwan. Jpn J Infect Dis 60: 352–354. [PubMed] [Google Scholar]
- 4. Loens K, Goossens H, Ieven M (2010) Acute respiratory infection due to Mycoplasma pneumoniae: current status of diagnostic methods. Eur J Clin Microbiol Infect Dis 29: 1055–1069. 10.1007/s10096-010-0975-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Waites KB, Balish MF, Atkinson TP (2008) New insights into the pathogenesis and detection of Mycoplasma pneumoniae infections. Future Microbiol 3: 635–648. 10.2217/17460913.3.6.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thibodeau KP, Viera AJ (2004) Atypical pathogens and challenges in community-acquired pneumonia. Am Fam Physician 69: 1699–1706. [PubMed] [Google Scholar]
- 7. Zhao F, Cao B, Li J, Song S, Tao X, Yin Y, et al. (2011) Sequence analysis of the P1 adhesin gene of Mycoplasma pneumoniae in clinical isolates collected in Beijing in 2008 to 2009. J Clin Microbiol 49: 3000–3003. 10.1128/JCM.00105-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerstenecker B, Jacobs E (1990) Topological mapping of the P1-adhesin of Mycoplasma pneumoniae with adherence-inhibiting monoclonal antibodies. J Gen Microbiol 136: 471–476. [DOI] [PubMed] [Google Scholar]
- 9. Jacobs E, Pilatschek A, Gerstenecker B, Oberle K, Bredt W (1990) Immunodominant epitopes of the adhesin of Mycoplasma pneumoniae . J Clin Microbiol 28: 1194–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thurman KA, Walter ND, Schwartz SB, Mitchell SL, Dillon MT, Baughman AL, et al. (2009) Comparison of laboratory diagnostic procedures for detection of Mycoplasma pneumoniae in community outbreaks. Clin Infect Dis 48: 1244–1249. 10.1086/597775 [DOI] [PubMed] [Google Scholar]
- 11. Krause DC (1998) Mycoplasma pneumoniae cytadherence: organization and assembly of the attachment organelle. Trends Microbiol 6: 15–18. [DOI] [PubMed] [Google Scholar]
- 12. Jacobs E (2012) Mycoplasma pneumoniae: now in the focus of clinicians and epidemiologists. Euro Surveill 17. [PubMed] [Google Scholar]
- 13. Hennigan SL, Driskell JD, Dluhy RA, Zhao Y, Tripp RA, Waites KB, et al. (2010) Detection of Mycoplasma pneumoniae in simulated and true clinical throat swab specimens by nanorod array surface-enhanced Raman spectroscopy. PLoS One 5: e13633 10.1371/journal.pone.0013633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Winchell JM, Thurman KA, Mitchell SL, Thacker WL, Fields BS (2008) Evaluation of three real-time PCR assays for detection of Mycoplasma pneumoniae in an outbreak investigation. J Clin Microbiol 46: 3116–3118. 10.1128/JCM.00440-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dumke R, Luck PC, Noppen C, Schaefer C, von Baum H, Marre R, et al. (2006) Culture-independent molecular subtyping of Mycoplasma pneumoniae in clinical samples. J Clin Microbiol 44: 2567–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanack K, Amiott E, Nolte F, Saliminia H, Rogers B, Poritz M, et al. (2011) Analytical and Clnical Evaluation of the FilmArray Respiratory Panel. Idaho Technology, Inc. [Google Scholar]
- 17. Dumke R, Schurwanz N, Jacobs E (2008) Characterisation of subtype- and variant-specific antigen regions of the P1 adhesin of Mycoplasma pneumoniae . Int J Med Microbiol 298: 483–491. [DOI] [PubMed] [Google Scholar]
- 18. Schwartz SB, Mitchell SL, Thurman KA, Wolff BJ, Winchell JM (2009) Identification of P1 variants of Mycoplasma pneumoniae by use of high-resolution melt analysis. J Clin Microbiol 47: 4117–4120. 10.1128/JCM.01696-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schwartz SB, Thurman KA, Mitchell SL, Wolff BJ, Winchell JM (2009) Genotyping of Mycoplasma pneumoniae isolates using real-time PCR and high-resolution melt analysis. Clin Microbiol Infect 15: 756–762. 10.1111/j.1469-0691.2009.02814.x [DOI] [PubMed] [Google Scholar]
- 20. Abell JL, Driskell JD, Dluhy RA, Tripp RA, Zhao YP (2009) Fabrication and characterization of a multiwell array SERS chip with biological applications. Biosens Bioelectron 24: 3663–3670. 10.1016/j.bios.2009.05.039 [DOI] [PubMed] [Google Scholar]
- 21. Driskell JD, Shanmukh S, Liu Y, Chaney SB, Tang XJ, Zhao YP, et al. (2008) The use of silver nanorod arrays prepared by oblique angle deposition as surface-enhanced Raman scattering substrates. J Phys Chem C 112: 895–901. [Google Scholar]
- 22. Liu YJ, Zhang ZY, Zhao Q, Dluhy RA, Zhao YP (2009) Surface Enhanced Raman Scattering from an Ag Nanorod Array Substrate: The Site Dependent Enhancement and Layer Absorbance Effect. J Phys Chem C 113: 9664–9669. [Google Scholar]
- 23. Willemse-Erix DF, Scholtes-Timmerman MJ, Jachtenberg JW, van Leeuwen WB, Horst-Kreft D, Bakker Schut TC, et al. (2009) Optical fingerprinting in bacterial epidemiology: Raman spectroscopy as a real-time typing method. J Clin Microbiol 47: 652–659. 10.1128/JCM.01900-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harz M, Rösch P, Popp J (2009) Vibrational spectroscopy—A powerful tool for the rapid identification of microbial cells at the single-cell level. Cytometry Part A 75A: 104–113. [DOI] [PubMed] [Google Scholar]
- 25. Otto A (2002) What is observed in single molecule SERS, and why? J Raman Spectrosc 33: 593–598. [Google Scholar]
- 26. Golightly RS, Doering WE, Natan MJ (2009) Surface-Enhanced Raman Spectroscopy and Homeland Security: A Perfect Match? ACS Nano 3: 2859–2869. 10.1021/nn9013593 [DOI] [PubMed] [Google Scholar]
- 27. Shanmukh S, Jones L, Driskell JD, Zhao Y, Dluhy RA, Tripp RA (2006) Rapid and Sensitive Detection of Respiratory Virus Molecular Signatures Using a Silver Nanorod Array SERS Substrate. Nano Letters 6: 2630–2636. [DOI] [PubMed] [Google Scholar]
- 28. Chu H, Huang YJ, Zhao Y (2008) Silver Nanorod Arrays as a Surface-Enhanced Raman Scattering Substrate for Foodborne Pathogenic Bacteria Detection. Appl Spectrosc 62: 922–931. 10.1366/000370208785284330 [DOI] [PubMed] [Google Scholar]
- 29. Driskell JD, Zhu Y, Kirkwood CD, Zhao Y, Dluhy RA, Tripp RA (2010) Rapid and sensitive detection of rotavirus molecular signatures using surface-enhanced Raman spectroscopy. PLoS One 5: e10222 10.1371/journal.pone.0010222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Granato PA, Poe L, Weiner LB (1983) New York City medium for enhanced recovery of Mycoplasma pneumoniae from clinical specimens. J Clin Microbiol 17: 1077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Henderson KC, Sheppard ES, Rivera-Betancourt OE, Choi JY, Dluhy RA, Thurman KA, et al. (2014) The multivariate detection limit for Mycoplasma pneumoniae as determined by Nanorod Array-Surface Enhanced Raman Spectroscopy and comparison with limit of detection by qPCR. Analyst 139: 6426–6434. 10.1039/c4an01141d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lipman RP, Clyde WA Jr, Denny FW (1969) Characteristics of virulent, attenuated, and avirulent Mycoplasma pneumoniae strains. J Bacteriol 100: 1037–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim K-S, Ko K-S, Chang M-W, Hahn TW, Hong SK, Kook Y-H (2003) Use of rpoB sequences for phylogentic study of Mycoplasma species. FEMS Microbiol Lett 226: 299–305. [DOI] [PubMed] [Google Scholar]
- 34. Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, et al. (1987) Measurement of protein using bicinchoninic acid. Anal Biochem 150: 76–85. [DOI] [PubMed] [Google Scholar]
- 35. Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li BC, Herrmann R (1996) Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae . Nucleic Acids Res 24: 4420–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leverette CL, Jacobs SA, Shanmukh S, Chaney SB, Dluhy RA (2006) Aligned silver nanorod arrays as substrates for surface-enhanced infrared absorption spectroscopy. Appl Spectrosc 60: 906–913. [DOI] [PubMed] [Google Scholar]
- 37. Zhao YP, Chaney SB, Shanmukh S, Dluhy RA (2006) Polarized surface enhanced Raman and absobance spectra of aligned silver nanorod arrays. J Phys Chem B 110: 3153–3157. [DOI] [PubMed] [Google Scholar]
- 38. Negri P, Marotta NE, Bottomly LA, Dluhy RA (2011) Removal of surface contamination and self-assembled monolayers (SAMs) from silver (Ag) nanorod substrates by plasma cleaning with argon. Appl Spectrosc 65: 66–74. 10.1366/10-06037 [DOI] [PubMed] [Google Scholar]
- 39. Smith PF (1971) The Biology of Mycoplasmas. New York, New York: Academic Press, Inc. [Google Scholar]
- 40. Zubkov MV, Fuchs BM, Eilers H, Burkill PH, Amann R (1999) Determination of total protein content of bacterial cells by SYPRO staining and flow cytometry. Appl Environ Microbiol 65: 3251–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barker M, Rayens W (2003) Partial least squares for discrimination. J Chemom 17: 166–173. [Google Scholar]
- 42. Musumarra G, Barresi V, Condorelli DF, Fortuna CG, Scire S (2004) Potentialities of multivariate approaches in genome-based cancer research: identification of candidate genes for new diagnostics by PLS discriminate analysis. J Chemom 18: 125–132. [Google Scholar]
- 43.Adams MJ, Barnett NW (2004) Chemometrics in Analytical Spectroscopy.—X001.
- 44. Simmons WL, Daubenspeck JM, Osborne JD, Balish MF, Waites KB, Dybvig K (2013) Type 1 and type 2 strains of Mycoplasma pneumoniae form different biofilms. Microbiol 159: 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mrázek J (2006) Analysis of distribution indicates diverse functions of simple sequence repeats in Mycoplasma genomes. Mol Biol Evol 23: 1370–1385. [DOI] [PubMed] [Google Scholar]
- 46. Flores SC, Lu LJ, Yang J, Carriero N, Gerstein MB (2007) Hinge Atlas: relating protein sequence to sites of structural flexibility. BMC Bioinformatics 8: 167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Culha M, Adigüzel A, Yazici MM, Kahraman M, Sahin F, Güllüce M (2008) Characterization of thermophilic bacteria using Surface-Enhanced Raman scattering. Appl Spectrosc 62: 1226–1232. 10.1366/000370208786401545 [DOI] [PubMed] [Google Scholar]
- 48. Podstawka E, Ozaki Y, Proniewicz LM (2004) Adsorption of S-S containing proteins on a colloidal silver surface studied by Surface-enhanced Raman spectroscopy. Appl Spectrosc 58: 1147–1156. [DOI] [PubMed] [Google Scholar]
- 49. Stewart S, Fredericks PM (1998) Surface-enhanced Raman spectroscopy of amino acids adsorbed on an electrochemically prepared silver surface. Spectrochim Acta A 55: 1641–1660. [Google Scholar]
- 50. Mrozek MF, Weaver MJ (2002) Detection and identification of aqueous saccharides by using surface-enhanced Raman spectroscopy. Anal Chem 74: 4069–4075. [DOI] [PubMed] [Google Scholar]
- 51. Beebe KR, Pell RJ, Seasholtz MB (1998) Chemometrics: a Practical Guide. New York: John Wiley & Sons. [Google Scholar]
- 52. Cullum BM, Mobley J, Chi Z, Stokes DL, Miller GH, Vo-Dinh T (2000) Development of a compact, handheld Raman instrument with no moving parts for use in field analysis. Rev Sci Instrum 71: 1602–1608. [Google Scholar]
- 53. Moore DS, Scharff RJ (2009) Portable Raman explosives detection. Anal Bioanal Chem 393: 1571–1578. 10.1007/s00216-008-2499-5 [DOI] [PubMed] [Google Scholar]
- 54. Jehlička J, Culka A, Vandenabeele P, Edwards HGM (2011) Critical evaluation of a handheld Raman spectrometer with near infrared (785 nm) excitation for field identification of minerals. Spectrochim Acta A 80: 36–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All spectral data are available as CSV file from the Dryad database (datadryad.org; doi:10.5061/dryad.s5h20).